Abstract
Protein arginine methyltransferases (PRMTs) catalyze symmetric and asymmetric methylation on arginine residues of multiple protein targets including histones and have essential roles in organismal development and disease. PRMT5 mediates symmetric di-methylation (sDMA) of arginine 2 (H3R2me2) and arginine 8 on histone 3 (H3R8me2), arginine 3 on histones 2A and 4 (H2A/H4R3me2) as well as several non-histone substrates like Sm proteins. Here, we found that selective inhibition of Prmt5 in mesenchymal stromal cells (MSCs) led to a reduction in colony forming units (CFUs) and increased osteoblast differentiation. Prmt5 inhibition blocked global symmetric dimethylation of H3R8 and H4R3 but not H3R2. Genome-wide expression analysis by total RNA sequencing of mesenchymal stromal cells undergoing osteogenic differentiation revealed significant reduction in the intrinsic expression of several interferon-stimulated genes (ISGs) upon Prmt5 inhibition. Effects of Prmt5 inhibition on basal ISG expression and osteogenic differentiation was effectively blocked by exogenous activation of type I IFN signaling. Together, these results indicate important functions for Prmt5 in the regulation of basal interferon gene expression in MSCs and in control of differentiation potential of MSCs during osteogenic differentiation.
1. Introduction
Protein arginine methylation is a conserved and ubiquitous modification that plays important roles in multiple biological processes. In mammals, monomethyl and asymmetric or symmetric dimethyl arginine modifications are catalyzed by protein arginine methyltransferase family (PRMTs) [1]. Symmetric dimethyl arginine (sDMA) on histones and other non-histone protein substrates is brought about by class II PRMTs, PRMT5, PRMT9 and PRMT7. Prmt5 is required for most of the cellular sDMA modification on histones H2A/H4R3, H3R8 and H3R2 as well as several non-histone substrates including Sm proteins [2], P53, SPT5 etc. [3, 4]. Specific interaction of PRMT5 with cooperator of PRMT5 (Co-Pr5) is required for PRMT5 nuclear functions especially in symmetric arginine dimethylation on histone H4 (H4R3me2s) [5]. Prmt5 deletion is embryonic lethal in mouse and is required for proliferation and/or differentiation of several stem cell lineages including ES cells [6], skeletal muscle stem cells [7–9], adipose stem cells [10], hematopoietic stem and progenitors [11], chondrocyte progenitors [12] among others. PRMT5 is also required for important cellular processes such as maintenance of chromatin states [13], cell cycle and RNA splicing [14]. Prmt5 expression is elevated in many transformed cells and its silencing or blockade slowed cancer cell growth [15, 16]. Recently, a specific and potent inhibitor of PRMT5 has been shown to effectively inhibit tumor growth in vitro and in vivo [17]. Here we studied the effect of pharmacological blockade of PRMT5 in two different mesenchymal stromal cell lines in steady state and during osteogenic differentiation. PRMT5 inhibition led to increased osteoblast differentiation and down modulation of several genes implicated in basal interferon signaling pathway including several Gbp family genes. This downregulation is accompanied by promoter specific decreases in PRMT5 mediated arginine histone methylation (H3R8me2s and H4R3me2s) at interferon stimulated gene loci. Together, our results indicate an important role for PRMT5 in basal interferon pathway gene expression, MSC proliferation and critical regulator of osteogenic differentiation.
2. Materials and Methods
2.1. Cell lines and treatments
W-20–17 (W-20) murine bone marrow stromal cells were described in [18] and were kindly provided by V.Rosen. ST-2 cells were purchased from RIKEN. Proliferating ST-2 and W-20 cell lines were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). Mouse embryonic fibroblasts were established from 13.5 dpc embryos and maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 15% fetal bovine serum (Gibco). Osteoblast differentiation was induced by culturing the stromal cell lines in osteogenic differentiation medium (OM) containing 10 mM beta-glycerol phosphate(sigma) and 100 μg/ml ascorbic acid (Sigma Aldrich) for 8–10 days with medium changes every 48 h. For PRMT5 inhibition, mesenchymal stromal cells (with or without differentiation medium) were treated with either DMSO (Sigma Aldrich) or PRMT5 inhibitor, GSK3235025 (Cayman chemical) at a final concentration of 0.625 μM or 1.25 μM for 10 days. Cells were replenished with fresh medium containing either DMSO or PRMT5 inhibitor every 48h. For exogenous activation of type I IFN signaling, cells were either treated alone or in combination with vehicle, or PRMT5 inhibitor and type 1 IFN agonist RO8191, at a concentration of 50 μM.
2.2. Real time PCR quantification
Total RNA was prepared from cultured cells using RNeasy kit (Qiagen). DNA contamination was eliminated by DNaseI digestion. Complementary DNA was prepared using multiscript reverse transcription system (Applied biosystems). The reverse transcribed cDNA was subjected to qPCR using a Sybr green based detection system (Qiagen). Relative levels of transcripts were normalized to NAT1 levels and quantified based on 2−ΔΔCT method. The primers were used in the study were described in supplementary table 2. A minimum of 3–5 independent biological replicates were analyzed.
2.3. Alkaline Phosphatase staining
W-20 or ST-2 cells were seeded on to 6 well or 12 well plates and cultured in osteogenic differentiation medium (OM) with medium changes every 48 hrs. In the experiments that included inhibitor treatment (see figure legend for details), OM also included DMSO or GSK3235025 alone or in combination with RO8191. On day 10 post incubation, cells were washed twice with PBS and fixed in 4% (v/v) paraformaldehyde (sigma) for 5 min. Cells were washed and incubated with alkaline phosphatase substrate solution (SigmaFast BCIP-NBT; Sigma Aldrich) for 10–20 min in dark.
2.4. Alizarin red staining
For Alizarin red staining, cells were cultured and differentiated as described above and on day 20 post culturing, cells were washed twice with PBS and fixed in 4% (v/v) paraformaldehyde (sigma) for 15 min. Cells were washed 5 times with deionized water and incubated with 2% alizarin red solution for 20 min and washed 5 times with water to remove excess staining solution.
2.5. Immunofluorescence analysis
Cell lines (W-20, ST-2) were grown on coverslips and treated with either vehicle control (DMSO) or PRMT5 inhibitor (GSK325025) for 10 days. Cells were fixed with 4% paraformaldehyde, followed by permeabilization with 1% Triton X-100. Cells were incubated for 2 h with the following antibodies, Histone H3R8 symmetric dimethyl (H3R8me2s; Epigentek- Cat # A-3706), Histone H4R3 symmetric dimethyl (H4R3me2s; Sigma Aldrich- Cat # SAB4300870), Histone H3R2 symmetric dimethyl (H3R2me2s; Epigentek- Cat # A-3705). Appropriate fluorescent-tagged secondary antibodies were used to detect antibody specific signals.
2.6. Western blotting analysis
Total cell lysates were prepared using RIPA lysis buffer supplemented with protease inhibitor cocktail. Equal amounts of protein samples were separated on a 4–12% polyacrylamide gel (Bio-Rad), blotted onto polyvinylidene difluoride membrane, and probed with different antibodies as indicated [anti-Prmt5 (Santa cruz- Cat # 376937), anti-Gapdh (Santa cruz- Cat # 25778), anti-H3R8me2s (Epigentek- Cat # A-3706), anti-H4R3me2s (Sigma aldrich- Cat # SAB4300870), anti-H3R2me2s (Epigentek- Cat # A-3705), anti-H3K4me3 (Cell signaling technology- Cat #: 9751), anti-Histone H3 (Cell signaling technology- Cat #: mAb 4499). Appropriate horseradish peroxidase conjugated secondary antibodies were used and signals were developed using chemiluminescent substrates (Amersham ECL prime detection reagents). A minimum of 2 or more independent biological experiments were analyzed. Band densities were calculated using ImageJ software.
2.7. Chromatin Immunoprecipitation (ChIP) analysis
Chromatin Immunoprecipitation (ChIP) analyses were performed using EZ-ChIP kit according to manufacturer’s protocol (Millipore). Briefly, cells were cross-linked with paraformaldehyde, lysed and chromatin were prepared by sonication. Immunoprecipitation of chromatins were performed using antibodies specific to H3R8me2s (Epigentek- Cat # A-3706), H4R3me2s (Sigma aldrich- Cat # SAB4300870). DNA were purified from the immunoprecipiated chromatin and subsequently analyzed by qPCR using promoter specific primers (Supplementary table 2). Data were analyzed following percent input method as described below. ΔCt [normalized ChIP] = (Ct [ChIP] - (Ct [Input] - Log2 (Input dilution factor))), where Input Dilution Factor = (fraction of the input chromatin saved)−1. In our analyses, 1/100th of the chromatin used for immunoprecipitation was used as input. Finally, the percentage (Input %) value for each sample is calculated as follows: Input % = 100/2 ΔCt [normalized ChIP]. The “Input %” value represents the enrichment of certain histone modification on specific region. Three independent biological replicates were analyzed.
2.8. RNA sequencing analysis
RNA sequencing was performed on total RNA isolated from W-20 stromal cells grown in osteogenic differentiation medium for 10 days treated with either DMSO or PRMT5 inhibitor (1.25 uM) (GSK3235025) from 2 independent experiments. Library preparation and sequencing were performed by the Biopolymers Facility at Harvard medical school. Data was processed using a standard RNA -seq pipeline that used STAR aligner to align the reads to mm10. Cufflinks suite and cuffdiff2 was used to calculate differential expression [19–21]. List of genes that were 1.5 fold up or downregulated (log2FC) in PRMT5 inhibitor treated samples with a q value (FDR corrected P value of the test statistic) of < 0.05 were chosen for further analysis. Same gene lists were used to identify gene set enrichment analysis as described in enrichr [22]. Results of Reactome pathway term [23] output for the up- or down-regulated genes sorted based on P values <0.01 are indicated as bar graphs.
2.9. Statistical analysis
All comparisons were performed with a two-tailed unpaired Student’s t test. The data are expressed as mean ± SEM. For data that are normalized, data analyses were performed following the method described by Valcu and Valcu [24]. Briefly, values for control and experimental samples were divided by the mean of the control sample, which conserves the distribution and the relative variance of the samples, allowing the subsequent use of a t test: mean (k * X) = k * mean(X); std error(k * X) = k * std error(X), where X is the sample and k = 1/mean of the control and is a normalization factor. p values are indicated by * for p<0.05; ** for p<0.01; *** for p<0.001.
3. Results and Discussion
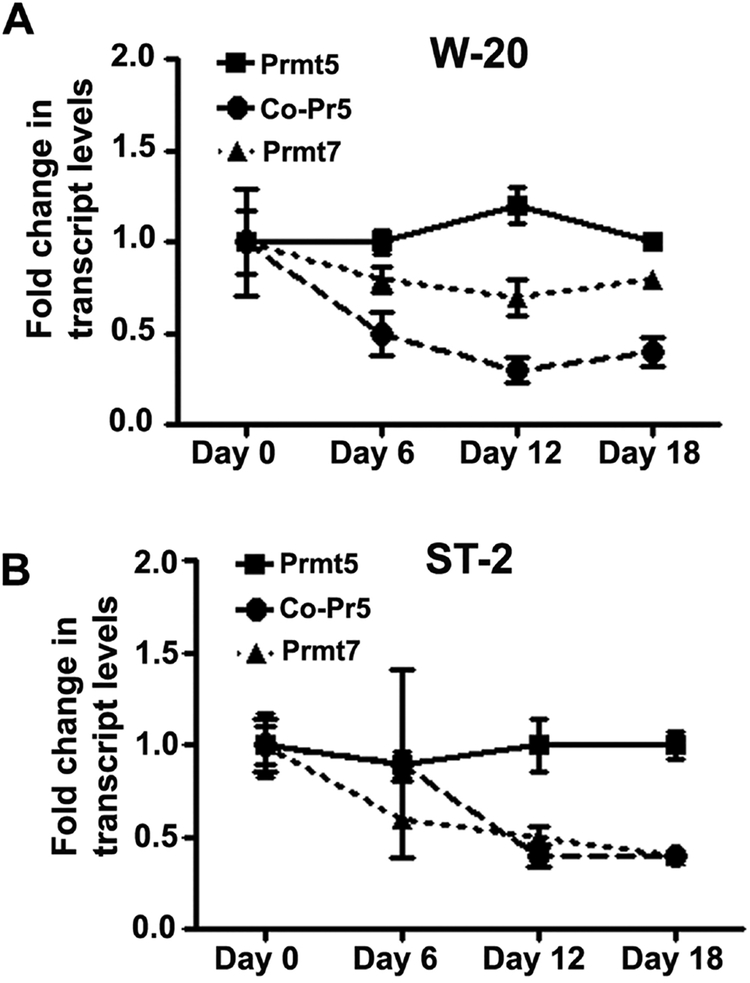
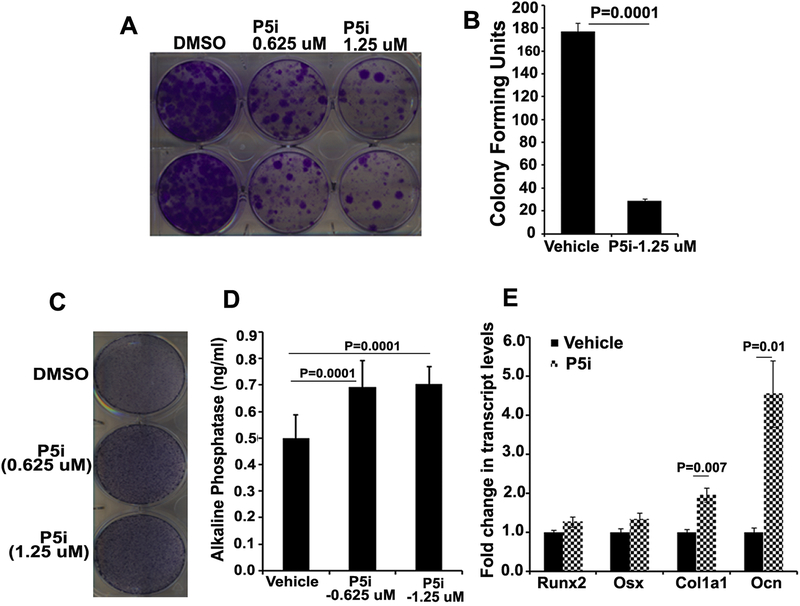
To understand the function and modulation of PRMT5 in mesenchymal stromal cells (MSCs) in steady state and during osteogenic differentiation, expression kinetics of Prmt5, Co-Pr5 and Prmt7 was investigated in two mesenchymal stromal cell lines, W-20 and ST-2. Prmt7 encodes for protein arginine methyl transferase that has specific activity against histones and capable of generating both monomethyl as well as symmetric dimethyl arginines similar to Prmt5 [25, 26]. All three genes, Prmt5, Prmt7 and Co-Pr5 are expressed in MSCs during steady state as well as during osteogenic differentiation (Fig 1). To delineate the biological functions of PRMT5 expression on proliferation and differentiation in MSCs, ST-2 and W-20 cells were treated with GSK3235025, a selective and potent inhibitor of PRMT5. Blockade of PRMT5 in W-20 and ST-2 mesenchymal stromal cells resulted in decreased colony forming units (CFUs) compared to vehicle only treatment (Fig 2A and B). We also analyzed the effects of PRMT5 inhibition on the osteogenic differentiation of the MSC by culturing W-20 and ST-2 cells in the presence of osteogenic induction medium containing 10 mM beta-glycerol phosphate and 100 μg/ml ascorbic acid. Alkaline phosphatase staining of MSCs cultured in OM in the presence of vehicle (DMSO) or GSK3235025 (P5i) on Day 10 post-treatment showed significantly increased enzyme activity in PRMT5 inhibitor treated cells (Fig 2C and D, Fig S1A). Quantitative RT-PCR assessment of osteogenic differentiation markers revealed significant increases in the transcript levels of alpha-1 type I collagen (Col1a1) and Osteocalcin (Ocn) and an increasing trend in the transcript levels of Runt Related Transcription Factor 2 (Runx2) and Osterix (Osx) upon PRMT5 inhibition compared to vehicle control at Day 10 (Fig 2E, Fig S1B). Thus, blockade of PRMT5 reduced MSC proliferation but aided their differentiation into osteogenic lineage.
Figure 1. Gene expression analyses of protein arginine methyl transferases (Prmt5, Prmt7) and cooperator of Prmt5(Co-Pr5) in W-20 and ST-2 mesenchymal stromal cells in steady state and during osteoblast differentiation.
Kinetics of Prmt5, Prmt7 and Co-Pr5 gene expression analyzed by qRT-PCR in undifferentiated state (Day 0) as well as during osteogenic differentiation (Day 6, Day 12 and Day 18) in (A) W-20 and (B) ST-2 cells (Both Co-Pr5 and Prmt7: D0 vs D12 as well as D0 vs. D18, p =<0.01). Results are from three independent biological experiments.
Figure 2. Effect of pharmacological inhibition of PRMT5 in mesenchymal stromal cells (MSCs) during steady state and following osteoblast differentiation.
Enzymatic activity of PRMT5 in ST-2 and W-20 cells was inhibited using GSK3235025, a selective and potent inhibitor (P5i). PRMT5 inhibition led to decrease in colony forming units (CFU) of ST-2 cells as assessed by crystal violet staining (a representative image is shown, N=4) (A) and CFU quantification (N=4) (B). Inhibition of Prmt5 during differentiation of W-20 cells into osteoblast lineage led to increased alkaline phosphatase activity as assessed by staining (a representative image is shown, N=4) (C), alkaline phosphatase concentration based on enzymatic activity (N=4) (D) and increased transcript levels of osteoblast lineage markers as assessed by qRTPCR analysis (N=3) (E).
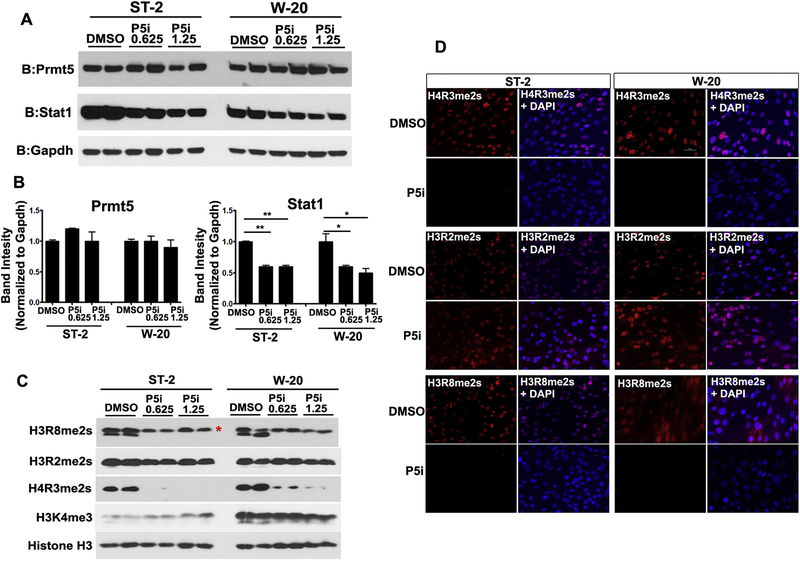
In order to delineate the effects of PRMT5 on MSC proliferation, we evaluated the protein levels of cell cycle regulators. Although we did not observe a significant change in the protein levels of p21 (data not shown), there was a downregulation of cyclin D1 upon PRMT5 inhibition in both W-20 and ST-2 cell lines (Fig S2). Prior studies in cancer cells demonstrated that PRMT5 directly upregulated cyclin D1 and inhibition of PRMT5 leads to cell cycle arrest [27, 28]. Similar to these studies, our data shows that PRMT5 is required for MSC proliferation via control of cyclin D1 levels in mesenchymal stem cells. To understand whether PRMT5 inhibition has effects on PRMT5 protein levels or function, W-20 and ST-2 cells were treated with DMSO and two different concentrations of PRMT5 inhibitor, GSK3235025 0.625 μM and 1.25 μM. GSK3235025 mediated inhibition of PRMT5 in MSCs did not alter PRMT5 protein levels (Fig 3A, 3B). Further, to understand whether inhibitor treatment altered PRMT5 function, analyses of PRMT5 catalyzed symmetrical dimethyl arginine modifications on histones were performed in the same cells. Symmetrical dimethyl arginine on histone H4 (H4R3me2s) and histone H3 (H3R8me2s) were significantly reduced in GSK3235025 treated cells whereas Histone H3 (H3R2me2s) levels were not altered compared to vehicle treated cells (Fig 3C). The differential inhibition of PRMT5 mediated methylation of arginine residues on histones H4 (H4R3) and H3 (H3R8 and H3R2) by GSK3235025 were corroborated by immunofluorescence analysis using methylation specific antibodies (Fig 3D).
Figure 3. Pharmacological inhibition of PRMT5 resulted in STAT1 downmodulation and differential blockade of histone symmetric dimethyl arginine modifications in mesenchymal stromal cells, ST-2 and W-20.
Inhibition of methyl transferase activity of PRMT5 (using GSK3235025) led to a downmodulation of STAT1 protein levels but not the levels of PRMT5 (A: western blot and B: protein band density quantitation). Decrease in global levels of symmetric dimethyl arginine marks on H4R3me2s and H3R8me2s but not H3R2me2s as assessed by western blot (C) as well as immunofluorescence (D) analyses. P5i: PRMT5 inhibitor (0.625 uM and 1.25 uM). *indicates non-specific band.
In order to delineate the dynamics of PRMT5 functions on histone symmetric arginine modifications during osteogenic differentiation, day 0 and day 14 cultures of MSCs cultured in osteogenic medium were analyzed by western blot. There was a 12% decrease in the protein levels of PRMT5 levels upon differentiation in both ST-2 and W-20 cell lines (Fig S3A). We also observed a decrease in the global levels of H4R3me2s as well as H3R8me2s during osteogenic differentiation of W-20 cells (Fig S3B). To understand the specificity of PRMT5 inhibitor, in addition to chemical inhibition of PRMT5, effects of shRNA mediated silencing of Prmt5 were evaluated in W-20 cells. Silencing of Prmt5 by shRNA reduced the levels of Prmt5 RNA but did not alter the transcript levels of Co-Pr5 (Fig S4A). Decrease in histone H4R3me2s levels were also observed in the same cells following Prmt5 silencing (Fig S4B). Similar to P5i, W-20 Prmt5 shRNA cells showed increased alkaline phosphatase staining in the presence of osteogenic medium (Fig S4C). The results from these studies substantiated the functional specificity of the small molecule inhibitor (GSK3235025) and thus we employed PRMT5 inhibitor (P5i) in all our subsequent analyses.
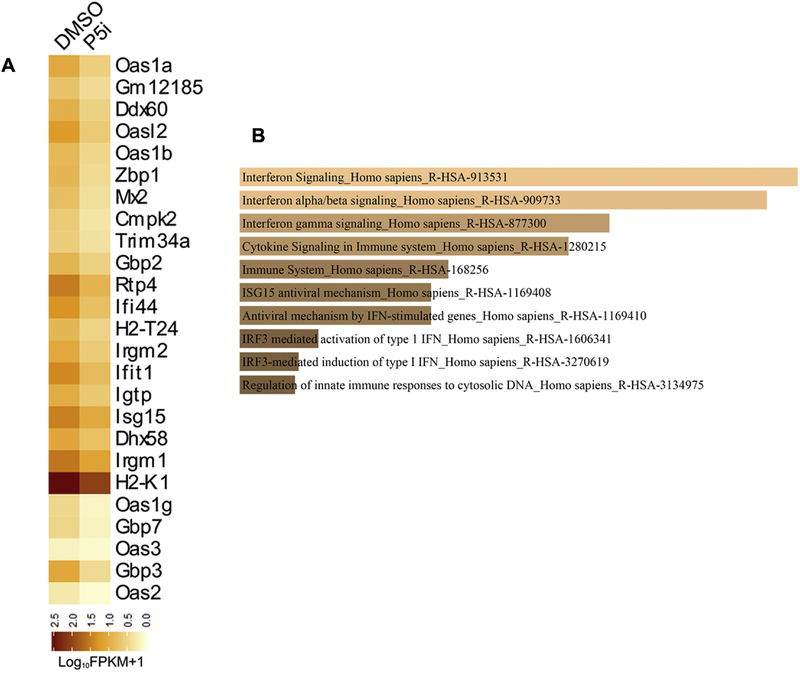
To understand the genome-wide molecular changes that occur upon PRMT5 inhibition during osteoblast differentiation, RNA sequencing was performed on total RNA isolated from W −20 cells following osteoblast differentiation (Day 10) and treated with either DMSO or GSK3235025. Principal component and dendrogram analyses of RNA sequencing data revealed separation of samples predominantly based on biological variation (Fig S5). Transcripts levels that were altered more than 1.5-fold up or down (log2FC, q<0.05) upon PRMT5 inhibition were deduced by differential gene expression analysis using Cuffdiff. Interestingly, PRMT5 inhibition down modulated a set of genes regulated by basal interferon signaling during osteogenic differentiation (Fig 4A). Gene set enrichment analysis using reactome pathway further revealed alteration in type I interferon pathway genes as significant enriched term in the downregulated gene list following PRMT5 blockade (Fig 4B). Similar analysis of genes upregulated 1.5-fold or more (log2FC) upon PRMT5 inhibition (Fig S6) revealed that extracellular matrix organization (Mfap4, Ctsk, Col7a1, Itgb3) and genes involved in elastic fiber formation were significantly enriched terms for upregulated gene set (Fig S7).
Figure 4. RNA-sequencing analysis of W-20 MSCs undergoing osteoblast differentiation.
PRMT5 enzymatic activity was inhibited using GSK3235025 (1.25 uM) in W-20 cells during osteogenic differentiation. Total RNA was collected upon treatment with vehicle control (DMSO) and PRMT5 inhibitor (P5i) at Day 10 and RNA sequencing was performed. (A) Heatmap of genes downregulated 1.5-fold or more (log2FC, q<0.05) upon Prmt5 inhibition in W-20 cells following differentiation into osteoblasts. (B) Reactome pathway term analysis(p<0.01) of RNA-seq data revealed that interferon pathway gene set to be primarily modulated by Prmt5 inhibition in MSCs during osteogenic differentiation.
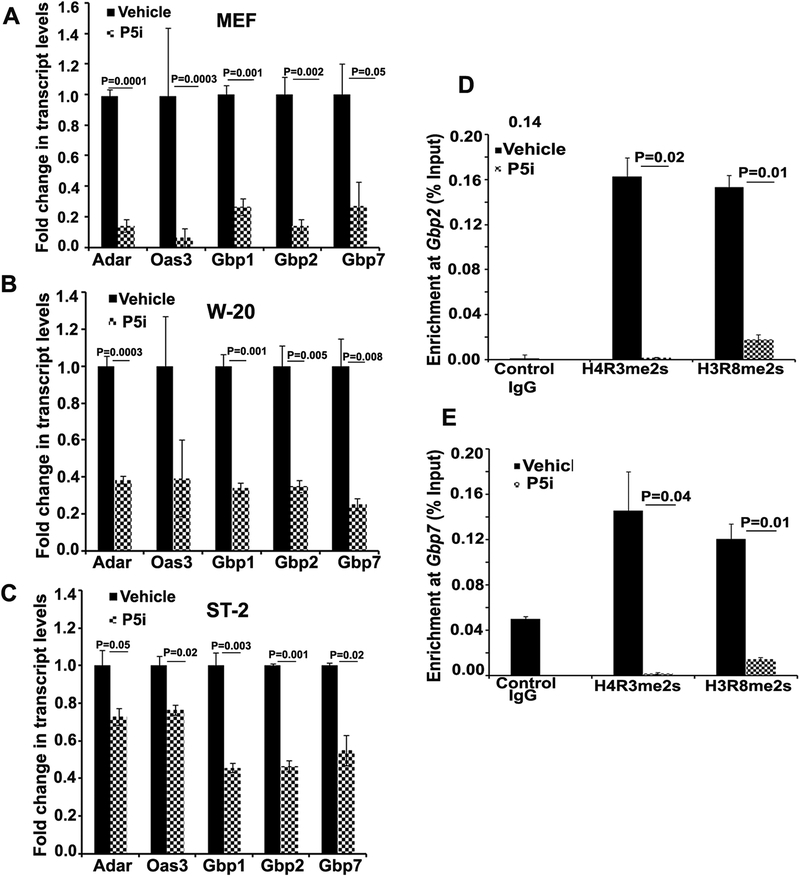
To validate the candidates obtained in RNA-sequencing, RT-qPCR analysis was performed on randomly chosen set of genes that were downregulated more than 1.5-fold or more upon PRMT5 inhibition and with a statistical significance of <0.05. These genes included interferon-stimulated genes including Gbp family proteins (Gbp1, Gbp2, Gbp7) as well as Adar and Oas3. Expression analyses of these genes performed in mouse embryonic fibroblasts (MEFs) (Fig 5A) as well as in W-20 (Fig 5B) and ST-2 (Fig 5C) cells undergoing osteogenic differentiation revealed significant decrease in the transcript levels of these genes upon PRMT5 inhibition. Guanylate binding protein (Gbp) family proteins are classical interferon stimulated genes, and are members of intrinsic IFN signaling pathway. Silencing of Gbp1 has also been shown to alter osteogenic differentiation of MSCs [29]. Interestingly, significant increase in the transcript levels of interferon stimulated gene (ISGs) and interferon signaling pathway genes including members of Gbp family genes was observed during osteogenic differentiation (Fig S8A and S8B). This increase in the expression of ISGs during differentiation is consistent with prior studies indicating the occurrence of enhanced, differential, cell-type specific IFN signaling response in differentiated cells in comparison to intrinsic signaling that occur in pluri- and multi-potent stem cells [30].
Figure 5. Prmt5 plays a role in the regulation of basal interferon pathway gene expression.
Quantitative RT-PCR analysis of interferon stimulated gene (ISG) expression upon PRMT5 blockade (P5i at a concentration of 1.25 uM) in (A) mouse embryonic fibroblasts (MEFs) and in MSCs undergoing osteoblast differentiation (Day 10) (B) W-20 and (C) ST-2 cells. Chromatin Immunoprecipitation (ChIP) analyses of the promoter regions of two interferon pathway genes Gbp2 and Gbp7 revealed decreased enrichment of arginine methylation marks on H4R3 and H3R8 upon PRMT5 inhibition (P5i −1.25 uM) in both ST-2 (D) and W-20 (E). PRMT5 inhibitor (1.25 uM). (N=3)
Further, we tested if loss of PRMT5 mediated methylation of arginine residues on histones H4 (H4R3me2s) and histone H3 (H3R8me2s) were linked to the downregulation of basal interferon-stimulated gene expression using locus specific chromatin immunoprecipitation analysis. PRMT5 inhibition resulted in decreased enrichment of H4R3me2s and H3R8me2s marks on Gbp2 (Fig 5D) and Gbp7 (Fig 5E), two genes whose transcript levels were significantly down regulated by Prmt5 inhibition in both ST-2 and W-20 cell lines during osteogenesis. Thus, Prmt5 mediated histone arginine methylation sustains basal levels of interferon signaling in MSCs and in osteoblast lineage cells. We also analyzed enrichment of H4R3me2s and H3R8me2s on the promoters of Gbp2 and Gbp7 during osteogenic differentiation. Both these histone modification marks were less pronounced in differentiated cells compared to MSCs (Fig S8C and S8D). These findings suggest that besides transcriptional regulation, some of the functions of PRMT5 on ISG expression might be post transcriptional, affecting splicing and or RNA stability. Consistent with this notion, blockade of PRMT5 in undifferentiated cells (Fig 5) as well as continued blockade of PRMT5 during the process of osteogenic differentiation (Fig S9), led to significant decline in ISG transcript levels.
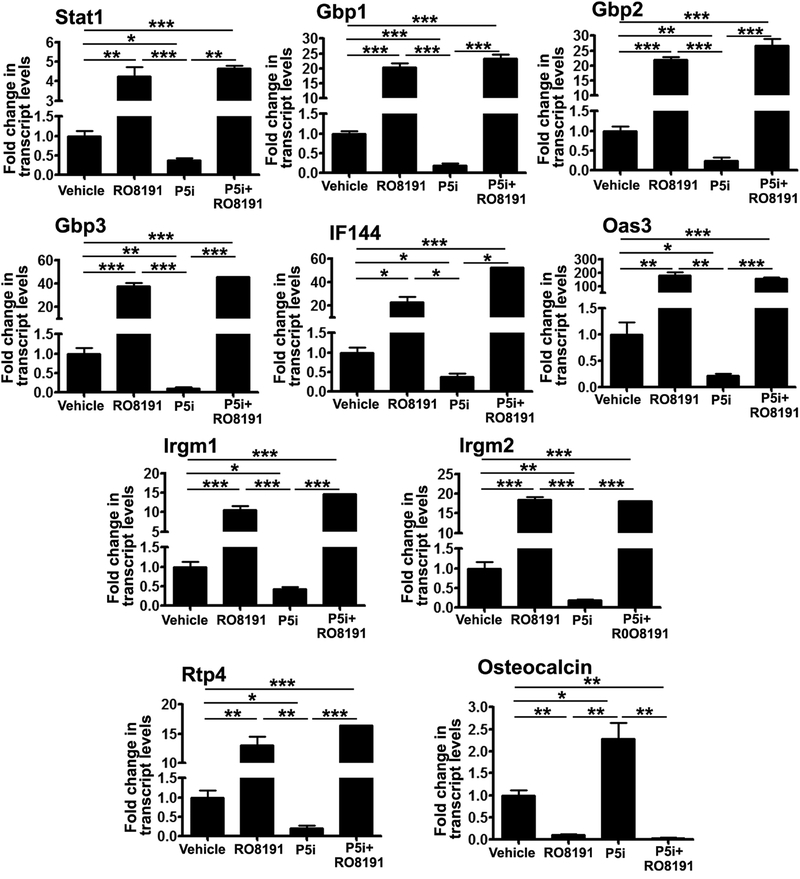
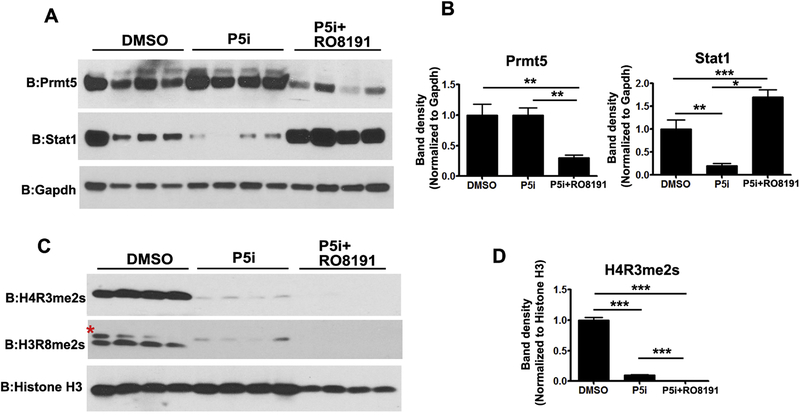
To gain further insights into whether exogenous activation of type I IFN signaling has effects on osteogenic differentiation and also counter the effects of PRMT5 inhibition, we employed a potent type I IFN receptor agonist, RO8191. Addition of RO8191 to osteogenic medium led to decreased alkaline phosphatase and alizarin red staining and was sufficient to block the increased differentiation that was seen with Prmt5 inhibition, corroborating the inhibitory effect of interferon signaling activation on osteogenic differentiation potential (Fig S10). Prmt5 inhibition alone led to a decrease in transcript levels of multiple ISGs (Fig 6). Stat1 levels were also decreased both at transcript and protein levels in MSCs upon PRMT5 inhibition during OB differentiation (Fig 6, Fig 7A and B). Co-treatment with RO8191 significantly increased the transcript levels of many IFN pathway genes including ISGs and Stat1 and rescued the effects of PRMT5 inhibition under the same conditions (Fig 6). RO8191 treatment did not alter histone symmetric arginine modifications mediated by PRMT5 (Fig 7C and D). Previously, Kim et al [31] have shown that STAT1 interferes with nuclear localization of RUNX2 a key transcriptional regulator of osteoblast differentiation. To test whether downregulation of Stat1 levels (Fig 6 and Fig 7A) seen with PRMT5 inhibition had effects on RUNX2 levels and localization, WB and immunofluorescence analyses were performed on W-20 cells undergoing osteogenic differentiation (day 12) in the presence of P5i and P5i+RO8191. Western blot analysis revealed increased RUNX2 protein levels upon PRMT5 inhibition were abrogated by R08191 (Fig S11A and S11B). RUNX2 protein is predominantly nuclear localized in DMSO as well as PRMT5 inhibitor treated cells in contrast to cytoplasmic RUNX2 that was evident only upon treatment with type IFN agonist R08191 (Figure S11C).
Figure 6. Overriding effects of type I IFN receptor agonist RO8191 over PRMT5 inhibition during OB differentiation.
Continued blockade of PRMT5 during OB differentiation led to decreased expression of IFN signaling pathway genes but was rescued by type I IFN receptor agonist RO8191 (50 uM). PRMT5 blockade led to increased expression of osteocalcin and RO8191 treatment did not overcome effects of PRMT5 inhibition. P5i: PRMT5 inhibitor (1.25 uM). (N=3)
Figure 7. Analysis of protein changes upon cotreatment with type I IFN receptor agonist RO8191 and PRMT5 inhibitor.
(A and B) PRMT5 inhibition alone did not alter the protein levels of PRMT5 but co-treatment with RO8191 led to a decrease in PRMT5 levels. PRMT5 inhibition mediated down modulation of STAT1 protein levels was reversed by cotreatment with RO8191. (C). However, cotreatment with RO8191 (50 uM) did not rescue the decrease in histone arginine metylation marks, H4R3me2s and H3R8me2s mediated by the blockade of PRMT5. PRMT5 inhibitor (1.25 uM).
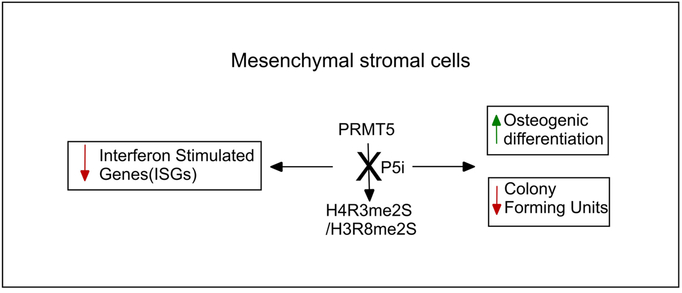
Interferon response is required for antiviral responses and recent evidence suggests the involvement of the intrinsic interferon signaling in stem cell functions and distinctly active ISG expressions in stem cells and terminally differentiated cells [30]. Our study has revealed a role for PRMT5 in the homeostatic regulation of basal interferon signaling in MSCs and during their differentiation into osteoblasts. Involvement of the immune system and cytokines including interferons in bone homeostasis, specifically the blockade of RANKL signaling and osteoclastogenesis by type I and type II interferons, have been studied in detail [32]. Interferon alpha and beta receptor subunit 1 (IFNAR1) null mice did not display deficiency in osteoblast formation in vivo [33]. However, recent studies have revealed a role for basal interferon signaling in osteoblast biology. STAT1, a critical transcription factor involved in IFN alpha/ beta signaling that also act as bridge between type I and type II interferon signaling pathways, [34] plays a critical role in osteoblast differentiation. Stat1 knockout mice displayed increased bone formation rate and Stat1 deletion favored osteoblast formation in in vitro differentiation studies [31]. Similarly, siRNA mediated inhibition of Gbp1, an interferon inducible gene enhanced osteogenic differentiation of human bone marrow derived MSCs [29]. Overall, the above studies reveal a negative correlation between IFN signaling and osteogenic differentiation. Our study has identified PRMT5 as a key player in the cell intrinsic mechanisms regulating basal interferon pathway gene expression in MSCs and during their differentiation into osteoblasts, in turn serves as an internal brake against osteogenic lineage differentiation (Fig 8).
Figure 8.
Model depicting the PRMT5 functions in gene regulation in MSCs and during osteogenic differentiation.
Treatment of human pre-osteoblasts with IFN beta inhibited osteoblast mediated mineralization [35]. Exploring the role of 1 alpha, 25- Dihydroxyvitamin D3 on bone mineralization, the existence of basal interferon signaling in human osteoblasts and expression of interferon beta itself by these cells were recently reported [36]. Treatment of human pre-osteoblasts with vitamin D3 resulted in down-modulation of interferon-stimulated genes with a concomitant up-regulation of extracellular matrix genes. Thus, there appears to be a direct negative feedback loop between interferon signaling and expression of extracellular matrix genes by osteoblasts. Interestingly, PRMT5 has been shown to repress 1, 25-dihydroxy vitamin D3 (1,25(OH)2D3 ) catabolism by negatively regulating Cyp24a1, a 25-hydroxyvitamin D3 24-hydroxylase [37]. Upon PRMT5 inhibition, levels of Cyp39a1, 24S-hydroxycholesterol 7-alpha-hydroxylase, an enzyme involved in Vitamin D precursor metabolism were increased (0.9 fold, log2FC). Potential roles of PRMT5 in vitamin D3 dependent calcium homeostasis and VDR mediated gene expression in vivo require further exploration.
Together, our studies have revealed that PRMT5 acts as an endogenous block towards MSC osteogenic differentiation. PRMT5 has multifunctional roles in the regulation of gene expression at the transcriptional level via histone methylation as well as during RNA splicing via sm protein methylation. H4R3 methylation at multiple loci has been associated with transcriptional repression [38, 39] whereas H3R8 methylation is associated with gene activation including during skeletal muscle stem cell differentiation by mediating myogenin transcription via recruitment of chromatin remodeling machinery [9]. During myogenic differentiation, PRMT5 mediated enrichment of H4R3me2s was observed on osteoblast specific Runx2 promoter element, but it was not an essential epigenetic modification for Runx2 gene repression [40]. In our study, blockade of methyl transferase activity of PRMT5 led to global loss of both H3R8me2s and H4R3me2s as well as at specific interferon stimulated gene promoters. As the osteogenic differentiation specific increase in the levels of subset of ISGs was susceptible to PRMT5 functional inhibition that is independent of promoter H4R3me2s levels, we surmise that additional factors including RNA metabolism and cell cycle regulation mediated by PRMT5 enzymatic activity may also play a role in determining the outcome of gene regulation at distinct genes. It remains to be investigated if the increased expression of extracellular matrix genes upon PRMT5 inhibition results from either direct genomic and / or non-genomic actions of PRMT5 or as a result of relief from negative feedback loop from IFN signaling. Recently, Dong Y et al [41] demonstrated that in vivo administration of PRMT5 inhibitor prevented ovariectomy-induced bone loss by blocking osteoclast differentiation and increased bone volume. Since bone remodeling is a multi-cellular process involving osteoblasts and osteocytes that also modulate osteoclast formation and function, it is important to understand the effects of PRMT5 inhibition on mesenchymal precursor cells and osteoblast lineage cells in vivo during development as well as ageing induced bone loss. Thus, our current study has implications in gaining a comprehensive understanding of the effect of PRMT5 inhibition on bone biology in vivo as symmetric histone arginine methylation catalyzed by PRMT5 has distinct effects on various cell types involved in bone formation and metabolism.
Supplementary Material
Figure S1. PRMT5 inhibition enhanced differentiation of ST-2 MSCs into osteogenic lineage. ST-2 cells were differentiated in the presence of OM and stained for alkaline phosphatase activity (A representative image is shown, N=4) (A). RT-qPCR analysis revealed increased expression of Col1a1 and Ocn, markers of osteogenic lineage upon PRMT5 inhbition (1.25 uM) (N=3) (B).
Figure S2. PRMT5 inhibition (P5i-0.625 uM and 1.25 uM) led to decrease in protein levels of Cyclin D1 in both ST-2 and W-20 cells. (A) western blot (B) ImageJ based quantitation of protein band densities.
Figure S3. Analysis of PRMT5 and associated histone symmetric arginine methylation changes during differentiation of MSCs into osteoblasts. During differentiation of W-20 MSCs in osteogenic medium, there was a 12% decrease in the protein levels of PRMT5 in differentiated cells compared to Day 0 (A) and there was a decrease in the global protein levels of PRMT5 mediated symmetric histone arginine methylation marks, H4R3me2s and H3R8me2s (46 and 57% respectively compared to Day 0) (B). Immunofluorescence analysis further corroborated maintenance of H4R3me2s marks during osteogenic differentiation (C).
Figure S4. Effects of short hairpin RNA mediated silencing of PRMT5 mirrored that of PRMT5 inhibitor, GSK3235025 in W-20 MSC cells. PRMT5 specific shRNA led to a decrease in the transcript levels of PRMT5 and not that of Co-Pr5 (A), led to a decrease in the symmetric histone arginine methylation marks, H4R3me2s and H3R2me2s (B) in undifferentiated W-20 cells. When cultured in osteogenic media, Prmt5 RNAi increased differentiation of W-20 MSCs as assessed by alkaline phosphatase staining (C).
Figure S5. (A) Principal component analysis of RNA sequencing data for W-20 cells differentiating in osteoblast medium in the presence of DMSO (DM1,DM2) and P5i (1.25 uM) (P51,P52). 86.73% and 10.82% variance are explained by principal component 1 (PC1) and principal component 2(PC2). Red dots are P5i treated samples and blue dots are DMSO treated samples. First principal component, PC1 separated samples based expectedly on biological variation (B). Dendrogram obtained from VST transformed count data clustered replicates and separated based on biological variation.
Figure S6. Heatmap of upregulated genes following PRMT5 inhibition. RNA-seq analysis was performed on W-20 cells undergoing differentiation (Day 10), either treated with vehicle (DMSO) or PRMT5 inhibitor (P5i-1.25 uM) and log fold change values were plotted as a heatmap using cummerbund for upregulated genes showing fold change of 1.5 or more (log2FC, q<0.05).
Figure S7. Reactome pathway analysis of upregulated genes upon PRMT5 inhibition.
Figure S8. Increased expression of IFN pathway genes during osteoblast differentiation. W-20 cells were differentiated in OB medium and the expression kinetics of IFN pathway genes (A) Gbp family genes (B) IFN genes were analyzed by qRT-PCR analyses. ChIP analyses for H4R3me2s and HR8me2s at two distinct loci, viz Gbp2 and Gbp7 (C and D).
Figure S9. PRMT5 blockade (P5i-1.25 um) during osteogenic differentiation led to a decrease in the enrichment of symmetric histone arginine methylation mark, H4R3me2s in several gene loci. Similar to undifferentiated cells, PRMT5 blockade during osteogenic differentiation led to a decrease in H4R3me2s enrichment in several gene loci viz. Stat1, Runx2, Gbp2, Gbp7, Col1a1 and Ocn.
Figure S10. PRMT5 inhibition (P5i-1.25 uM) led to increased differentiation (A) and mineralization (B), which was abrogated by exogenous activation of type I IFN signaling via RO8191 (50 uM).
Figure S11. Osteogenic differentiation in the presence of PRMT5 inhibitor led to increased RUNX2 protein levels and that increase is countered in presence of RO8191 (A and B) in W-20 cells. (C). Immunofluorescence analyses of Runx2 protein in the presence of PRMT5 inhibitor (P5i-1.25 uM) and RO8191 (50 uM).
Supplementary table 1. List of genes modulated 1.5 fold up or down (log2FC) following inhibition of PRMT5 as deduced from RNA-sequencing analysis.
Supplementary table 2. Sequences of PCR primers used in the qRT-PCR analysis.
Highlights.
Osteogenic differentiation is regulated by interferon signaling
Prmt5 inhibition leads to decreased basal/intrinsic ISG (interferon stimulated gene) expression both in MSCs and during differentiation
Prmt5 is an endogenous block towards MSC osteogenic differentiation
Acknowldgements
This work was aided by grants K01AR069197 (to S.K.K from National Institute of Arthritis and Musculoskeletal and Skin diseases), R21AG047412 (to S.B.K from National Institutes of Aging).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bedford MT, Arginine methylation at a glance, J Cell Sci 120(Pt 24) (2007) 4243–6. [DOI] [PubMed] [Google Scholar]
- [2].Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U, Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln, Curr Biol 11(24) (2001) 1990–4. [DOI] [PubMed] [Google Scholar]
- [3].Blanc RS, Richard S, Arginine Methylation: The Coming of Age, Mol Cell 65(1) (2017) 8–24. [DOI] [PubMed] [Google Scholar]
- [4].Bedford MT, Clarke SG, Protein arginine methylation in mammals: who, what, and why, Mol Cell 33(1) (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lacroix M, El Messaoudi S, Rodier G, Le Cam A, Sardet C, Fabbrizio E, The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5, EMBO Rep 9(5) (2008) 452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA, Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency, Genes Dev 24(24) (2010) 2772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang T, Gunther S, Looso M, Kunne C, Kruger M, Kim J, Zhou Y, Braun T, Prmt5 is a regulator of muscle stem cell expansion in adult mice, Nat Commun 6 (2015) 7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dacwag CS, Bedford MT, Sif S, Imbalzano AN, Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation, Mol Cell Biol 29(7) (2009) 1909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN, The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling, Mol Cell Biol 27(1) (2007) 384–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].LeBlanc SE, Konda S, Wu Q, Hu YJ, Oslowski CM, Sif S, Imbalzano AN, Protein arginine methyltransferase 5 (Prmt5) promotes gene expression of peroxisome proliferator-activated receptor gamma2 (PPARgamma2) and its target genes during adipogenesis, Mol Endocrinol 26(4) (2012) 583–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu F, Cheng G, Hamard PJ, Greenblatt S, Wang L, Man N, Perna F, Xu H, Tadi M, Luciani L, Nimer SD, Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis, J Clin Invest 125(9) (2015) 3532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Norrie JL, Li Q, Co S, Huang BL, Ding D, Uy JC, Ji Z, Mackem S, Bedford MT, Galli A, Ji H, Vokes SA, PRMT5 is essential for the maintenance of chondrogenic progenitor cells in the limb bud, Development 143(24) (2016) 4608–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Girardot M, Hirasawa R, Kacem S, Fritsch L, Pontis J, Kota SK, Filipponi D, Fabbrizio E, Sardet C, Lohmann F, Kadam S, Ait-Si-Ali S, Feil R, PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome, Nucleic Acids Res 42(1) (2014) 235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E, Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery, Genes Dev 27(17) (2013) 1903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S, Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes, Mol Cell Biol 24(21) (2004) 9630–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang L, Pal S, Sif S, Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells, Mol Cell Biol 28(20) (2008) 6262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ, Jin L, Jacques SL, West KA, Lingaraj T, Stickland K, Ribich SA, Raimondi A, Scott MP, Waters NJ, Pollock RM, Smith JJ, Barbash O, Pappalardi M, Ho TF, Nurse K, Oza KP, Gallagher KT, Kruger R, Moyer MP, Copeland RA, Chesworth R, Duncan KW, A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models, Nat Chem Biol 11(6) (2015) 432–7. [DOI] [PubMed] [Google Scholar]
- [18].Thies RS, Bauduy M, Ashton BA, Kurtzberg L, Wozney JM, Rosen V, Recombinant human bone morphogenetic protein-2 induces osteoblastic differentiation in W-20–17 stromal cells, Endocrinology 130(3) (1992) 1318–24. [DOI] [PubMed] [Google Scholar]
- [19].Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L, Improving RNA-Seq expression estimates by correcting for fragment bias, Genome Biol 12(3) (2011) R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat Protoc 7(3) (2012) 562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L, Differential analysis of gene regulation at transcript resolution with RNA-seq, Nat Biotechnol 31(1) (2013) 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A, Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool, BMC Bioinformatics 14 (2013) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P, The Reactome Pathway Knowledgebase, Nucleic Acids Res 46(D1) (2018) D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Valcu M, Valcu CM, Data transformation practices in biomedical sciences, Nat Methods 8(2) (2011) 104–5. [DOI] [PubMed] [Google Scholar]
- [25].Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S, PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine, J Biol Chem 280(5) (2005) 3656–64. [DOI] [PubMed] [Google Scholar]
- [26].Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG, Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues, J Biol Chem 287(11) (2012) 7859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW, Agostinelli C, Pileri S, Denis GV, Baiocchi RA, Sif S, Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing, J Biol Chem 288(49) (2013) 35534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, Liu KL, Yu CT, Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade, Cancer Sci 103(9) (2012) 1640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bai S, Mu Z, Huang Y, Ji P, Guanylate Binding Protein 1 Inhibits Osteogenic Differentiation of Human Mesenchymal Stromal Cells Derived from Bone Marrow, Sci Rep 8(1) (2018) 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM, Intrinsic Immunity Shapes Viral Resistance of Stem Cells, Cell 172(3) (2018) 423–438 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H, Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation, Genes Dev 17(16) (2003) 1979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Takayanagi H, Sato K, Takaoka A, Taniguchi T, Interplay between interferon and other cytokine systems in bone metabolism, Immunol Rev 208 (2005) 181–93. [DOI] [PubMed] [Google Scholar]
- [33].Takayanagi H, Kim S, Matsuo K, Suzuki H, Suzuki T, Sato K, Yokochi T, Oda H, Nakamura K, Ida N, Wagner EF, Taniguchi T, RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta, Nature 416(6882) (2002) 744–9. [DOI] [PubMed] [Google Scholar]
- [34].Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP, Trapani JA, Levy DE, Hertzog PJ, Clarke CJ, Johnstone RW, Functional crosstalk between type I and II interferon through the regulated expression of STAT1, PLoS Biol 8(4) (2010) e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Woeckel VJ, Koedam M, van de Peppel J, Chiba H, van der Eerden BC, van Leeuwen JP, Evidence of vitamin D and interferon-beta cross-talk in human osteoblasts with 1alpha,25-dihydroxyvitamin D3 being dominant over interferon-beta in stimulating mineralization, J Cell Physiol 227(9) (2012) 3258–66. [DOI] [PubMed] [Google Scholar]
- [36].Woeckel VJ, Eijken M, van de Peppel J, Chiba H, van der Eerden BC, van Leeuwen JP, IFNbeta impairs extracellular matrix formation leading to inhibition of mineralization by effects in the early stage of human osteoblast differentiation, J Cell Physiol 227(6) (2012) 2668–76. [DOI] [PubMed] [Google Scholar]
- [37].Seth-Vollenweider T, Joshi S, Dhawan P, Sif S, Christakos S, Novel mechanism of negative regulation of 1,25-dihydroxyvitamin D3-induced 25-hydroxyvitamin D3 24-hydroxylase (Cyp24a1) Transcription: epigenetic modification involving cross-talk between protein-arginine methyltransferase 5 and the SWI/SNF complex, J Biol Chem 289(49) (2014) 33958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM, PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing, Nat Struct Mol Biol 16(3) (2009) 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C, Negative regulation of transcription by the type II arginine methyltransferase PRMT5, EMBO Rep 3(7) (2002) 641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, van Wijnen AJ, van Zundert B, Allende ML, Montecino M, Epigenetic Control of the Bone-master Runx2 Gene during Osteoblast-lineage Commitment by the Histone Demethylase JARID1B/KDM5B, J Biol Chem 290(47) (2015) 28329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dong Y, Song C, Wang Y, Lei Z, Xu F, Guan H, Chen A, Li F, Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2, Cell Signal 34 (2017) 55–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PRMT5 inhibition enhanced differentiation of ST-2 MSCs into osteogenic lineage. ST-2 cells were differentiated in the presence of OM and stained for alkaline phosphatase activity (A representative image is shown, N=4) (A). RT-qPCR analysis revealed increased expression of Col1a1 and Ocn, markers of osteogenic lineage upon PRMT5 inhbition (1.25 uM) (N=3) (B).
Figure S2. PRMT5 inhibition (P5i-0.625 uM and 1.25 uM) led to decrease in protein levels of Cyclin D1 in both ST-2 and W-20 cells. (A) western blot (B) ImageJ based quantitation of protein band densities.
Figure S3. Analysis of PRMT5 and associated histone symmetric arginine methylation changes during differentiation of MSCs into osteoblasts. During differentiation of W-20 MSCs in osteogenic medium, there was a 12% decrease in the protein levels of PRMT5 in differentiated cells compared to Day 0 (A) and there was a decrease in the global protein levels of PRMT5 mediated symmetric histone arginine methylation marks, H4R3me2s and H3R8me2s (46 and 57% respectively compared to Day 0) (B). Immunofluorescence analysis further corroborated maintenance of H4R3me2s marks during osteogenic differentiation (C).
Figure S4. Effects of short hairpin RNA mediated silencing of PRMT5 mirrored that of PRMT5 inhibitor, GSK3235025 in W-20 MSC cells. PRMT5 specific shRNA led to a decrease in the transcript levels of PRMT5 and not that of Co-Pr5 (A), led to a decrease in the symmetric histone arginine methylation marks, H4R3me2s and H3R2me2s (B) in undifferentiated W-20 cells. When cultured in osteogenic media, Prmt5 RNAi increased differentiation of W-20 MSCs as assessed by alkaline phosphatase staining (C).
Figure S5. (A) Principal component analysis of RNA sequencing data for W-20 cells differentiating in osteoblast medium in the presence of DMSO (DM1,DM2) and P5i (1.25 uM) (P51,P52). 86.73% and 10.82% variance are explained by principal component 1 (PC1) and principal component 2(PC2). Red dots are P5i treated samples and blue dots are DMSO treated samples. First principal component, PC1 separated samples based expectedly on biological variation (B). Dendrogram obtained from VST transformed count data clustered replicates and separated based on biological variation.
Figure S6. Heatmap of upregulated genes following PRMT5 inhibition. RNA-seq analysis was performed on W-20 cells undergoing differentiation (Day 10), either treated with vehicle (DMSO) or PRMT5 inhibitor (P5i-1.25 uM) and log fold change values were plotted as a heatmap using cummerbund for upregulated genes showing fold change of 1.5 or more (log2FC, q<0.05).
Figure S7. Reactome pathway analysis of upregulated genes upon PRMT5 inhibition.
Figure S8. Increased expression of IFN pathway genes during osteoblast differentiation. W-20 cells were differentiated in OB medium and the expression kinetics of IFN pathway genes (A) Gbp family genes (B) IFN genes were analyzed by qRT-PCR analyses. ChIP analyses for H4R3me2s and HR8me2s at two distinct loci, viz Gbp2 and Gbp7 (C and D).
Figure S9. PRMT5 blockade (P5i-1.25 um) during osteogenic differentiation led to a decrease in the enrichment of symmetric histone arginine methylation mark, H4R3me2s in several gene loci. Similar to undifferentiated cells, PRMT5 blockade during osteogenic differentiation led to a decrease in H4R3me2s enrichment in several gene loci viz. Stat1, Runx2, Gbp2, Gbp7, Col1a1 and Ocn.
Figure S10. PRMT5 inhibition (P5i-1.25 uM) led to increased differentiation (A) and mineralization (B), which was abrogated by exogenous activation of type I IFN signaling via RO8191 (50 uM).
Figure S11. Osteogenic differentiation in the presence of PRMT5 inhibitor led to increased RUNX2 protein levels and that increase is countered in presence of RO8191 (A and B) in W-20 cells. (C). Immunofluorescence analyses of Runx2 protein in the presence of PRMT5 inhibitor (P5i-1.25 uM) and RO8191 (50 uM).
Supplementary table 1. List of genes modulated 1.5 fold up or down (log2FC) following inhibition of PRMT5 as deduced from RNA-sequencing analysis.
Supplementary table 2. Sequences of PCR primers used in the qRT-PCR analysis.