Abstract
KIF15, the vertebrate kinesin-12, is best known as a mitotic motor protein, but continues to be expressed in neurons. Like KIF11 (the vertebrate kinesin-5), KIF15 interacts with microtubules in the axon to limit their sliding relative to one another. Unlike KIF11, KIF15 also regulates interactions between microtubules and actin filaments at sites of axonal branch formation and in growth cones. Our original work on these motors was done on cultured rat neurons, but we are now using zebrafish to extent these studies to an in vivo model. We previously studied kif15 in zebrafish by injecting splice-blocking morpholinos injected into embryos. Consistent with the cell culture work, these studies demonstrated that axons grow faster and longer when KIF15 levels are reduced. In the present study, we applied CRISPR/Cas9-based knockout technology to create kif15 mutants and labeled neurons with Tg(mnx1:GFP) transgene or transient expression of elavl3:EGFP-alpha tubulin. We then compared by live imaging the homozygotic, heterozygotic mutants to their wildtype siblings to ascertain the effects of depletion of kif15 during Caudal primary (CaP) motor neuron and Rohon-Beard (R-B) sensory neuron development. The results showed, compared to the kif15 wildtype, the number of branches was reduced while axon outgrowth was accelerated in kif15 homozygotic and heterozygotic mutants. In R-B sensory neurons, after laser irradiation, injured axons with loss of kif15 displayed significantly greater regenerative velocity. Given these results and the fact that kif15 drugs are currently under development, we posit kif15 as a novel target for therapeutically augmenting regeneration of injured axons.
Keywords: kif15 (kinesin-12), axon, regeneration, zebrafish, laser injury
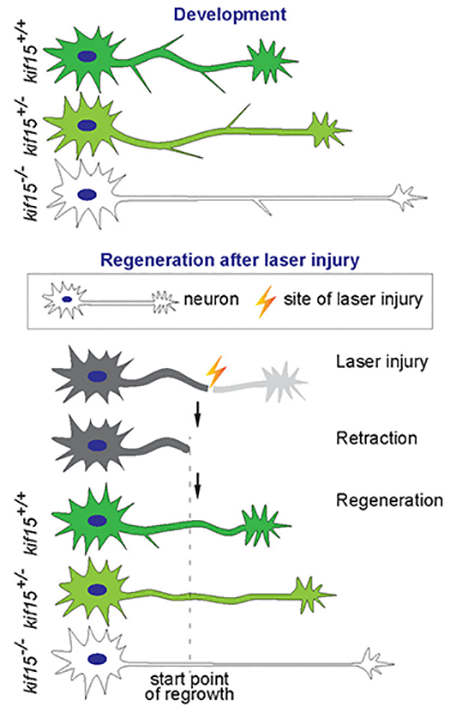
Graphical Abstract
1. Introduction
The microtubule cytoskeleton is a cell component essential to neural activities, including neurogenesis during development1,2 and axonal regeneration3. Normal microtubule function is required for neurogenesis, neuronal migration and polarization, which are key to embryonic neural development2,4–8. In addition, after spinal cord injury, microtubule-based interventions have been shown to enhance axon regeneration3. Members of the superfamily of kinesins, which are microtubule-based ATPase motor proteins that transport cargos along microtubules and also regulate the organization and movements of microtubules in the mitotic spindle, have been found to be active players in neuronal development and regeneration after injury of the adult nervous system9–11. In terminally differentiated neurons, microtubule motors were originally considered to function mainly in vesicle transport, but our work suggests that certain kinesins usually considered in the context of mitosis may play a role in coordinating and reconstructing the microtubule network by generating forces that regulate sliding of microtubules in the growth cone and axonal shaft9–11. KIF15 (Kinesin-12, KSNL7 or HKLP2), mainly studied in recent years as a potential target for cancer therapy12,13, was found in rats to be highly expressed in both brain and peripheral ganglia during embryonic development, with gradually diminished expression thereafter11. In cultured rat neurons, KIF15 was found to participate in axonal growth, navigation, branching11, dendrite morphology9 and neuronal migration13.
We are pursuing KIF15 as a target for therapy for nerve injury. Previously, we observed kif15 morphant zebrafish influenced axonal growth with faster extension and less branching14. However, morpholino oligonucleotide-based knockdown technology has a few shortcomings, including mosaicism and attenuation. It is practical to study axon regeneration by laser injury, but in a mosaic individual, the downregulation of the expression of the gene of interest in a certain neuron is not guaranteed, preventing us from collecting reliable data with that approach. In order to overcome these shortcomings to make it possible to observe axon growth during post-injury regeneration in neurons with confirmed reduction in gene expression, we employed CRISPR/Cas9-based knockout technology to create kif15 mutants. Not only does this approach avoid the shortcomings of the previous approach, but also enables us to feasibly analyze the dosage function of kif15 gene by comparing the homozygotic and heterozygotic mutants. Thus we were able to compare the mutants to their wildtype siblings to ascertain the effect of depletion of kif15 during axon regeneration following laser injury.
2. Results
2.1. Generation of kif15 mutant zebrafish
We utilized CRISPR/Cas9 mediated mutagenesis to knock out kif15 in zebrafish. The zebrafish KIF15 protein is encoded by kif15 located in chromosome 25. The reference transcript is 4466 base pairs (bp) (XM_002666923) and encodes a protein of 1376 amino acids (aa) (XP_002666969). KIF15 contains a Kinesin motor domain (aa 18–367) and a Hyaluronan-mediated motility receptor, C-terminal (HMMR_C) domain (aa 1262–1372). The kinesin motor domain coding sequence lies from the 3’ part of exon 1 to the 5’ part of exon 11. Based on the genomic structure of kif15, we searched for potential target sites (proto-spacers) for CRISPR/Cas9 mediated mutagenesis in exon 3–10, mainly focusing on exon 7–10 (Table 1). In this case CRISPR/Cas9 is sufficient in inducing mutation at any of these sites, a resulting allele, if still able to express, should code for a deformed protein with only an incomplete kinesin motor domain and lacking all other parts.
Table 1.
design of proto-spacers in zebrafish kif15
| Proto-spacer | Proto-spacer + PAM* seq (5’−3’) | Strand | Location | Activity** |
|---|---|---|---|---|
| k1 | tacactgtaaaccagagccacgg | sense | Exon 3 | N |
| k2 | cttctttctcatcaatcgagagg | sense | Exon 6 | N |
| k3 | ctatgaacgagcacttgcagagg | antisense | Exon 7 | N |
| k4 | ctacgggaggacatcaagagagg | sense | Exon 7 | N |
| k5 | tgaatcttctcgttcacatgcgg | sense | Exon 8 | N |
| k6 | agaagattcacggttcatagagg | antisense | Exon 8 | N |
| k7 | ggggtggcggaacaggcgagtgg | sense | Exon 8 | Y |
| k8 | ggagtccaaggagaccggacagg | sense | Exon 8 | Y |
| k9 | gacaggaagtggtgaacattagg | sense | Exon 8 | N |
| k10 | gaccgagacacatcagagagcgg | antisense | Exon 9 | N |
| k11 | tcttatcaggactctctcggcgg | sense | Exon 10 | N |
| k12 | ggctctttgtgcaaactgcaggg | antisense | Exon 10 | N |
: PAM, proto-spacer adjacent motif.
: In the Activity column, a “Y” indicates the capability to induce a mutation ratio greater than 10% detected 24 h after injection while an “N” stands for not-detectable.
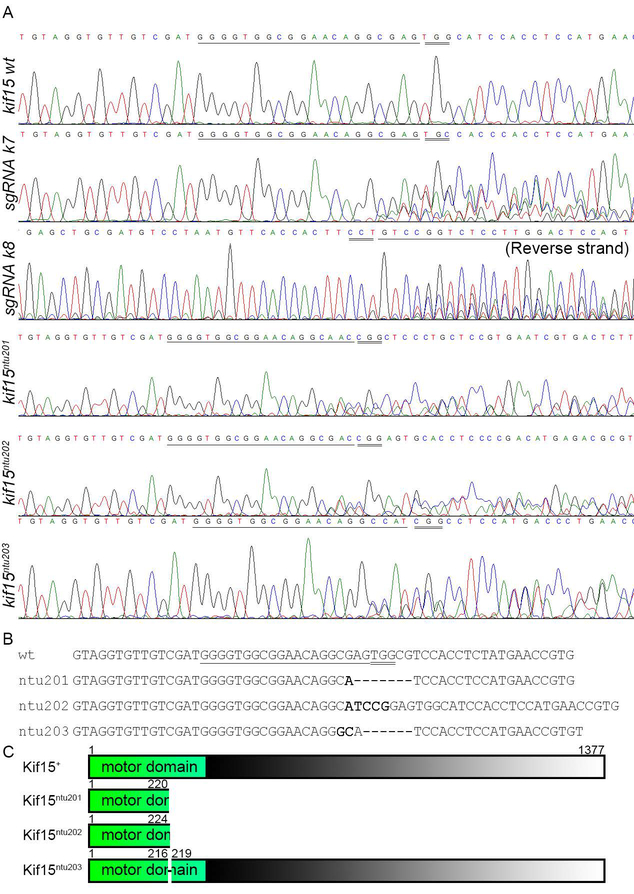
Twelve sgRNAs (k1–k12, detailed in Table 1) corresponding to 12 target sites were examined by microinjection and PCR-sequencing. Two sgRNAs, k7 and k8, both targeted in exon 8, were found able to induce mutation with Cas9 (Fig.1A). They were then used to produce kif15 mutants. About 500 1-cell embryos were injected with a mixture of Cas9 mRNA, k7 sgRNA and k8 sgRNA. When reaching sexual maturity, they were mated with wildtype ones. F1 fish were examined for kif15 genotype by taking fin clips, genomic DNA PCR and sequencing of PCR amplicons. By sequencing, we found heterozygotic mutants harboring three sorts of mutated kif15 alleles, namely ntu201 (−7 bp), ntu202 (+5 bp) and ntu203 (−6 bp) (Fig.1), among which the first two alleles were frameshift mutations. The F1 kif15ntu201 mutants were then selected for following studies. When the F1 mutants with kif15ntu201 allele reached sexual maturity, they were mated with Tg(mnx1:GFP) transgenic fish to breed the F2 generation to be visible of the interested neurons by means of the fluorescence microscope. The F3 generation was generated by F2 in-cross followed by fluorescence selection and PCR genotyping.
Figure 1.
Generation of Kif15 mutant alleles using CRISPR/Cas9 in AB line zebrafish.
(A) Target site k7 and k8 were shown to be effectively cleaved in AB line zebrafish, forming chimera, and various heritable mutations were found in the F1 generation. Underlined sequences are proto-spacers, while Double underlined triplets are PAM.
(B) Sequence alignment of wildtype and inherited mutated alleles of kif15. Underlined fragment is the proto-spacer identified by sgRNA k7. Double underlined triplets are PAM recognized by Cas9.
(C) Deduced translation products showed that mutated kif15 alleles code for disrupted Kif15 proteins lacking part of the kinesin motor domain and the entire tail. Numbers indicate peptide length. The numbers 216 and 219 indicate loss of residues 217 and 218 in mutated allele ntu203.
In the genotype identification of the F1 individuals generated by crossing mutant Founders with Tg(mnx1:GFP) with GFP-labeled CaP motor neurons15, we observed the overall morphological changes during zebrafish development and alteration of reproductive capacity in homozygotic and heterozygotic kif15ntu201 mutants. Compared with their wildtype siblings, the animals either lacking one or both of kif15 alleles were able to finish embryonic development and hatching; and the larvae harboring mutated kif15 alleles were morphologically healthy and were able to swim and feed normally (data not shown). This result matched our hypothesis that KIF15 mainly plays the role in nervous system development and its function of spindle organization and separation may be redundant by KIF11 motor protein14,16,17.
2.2. Loss of KIF15 affected axonal morphology in CaP motor neurons
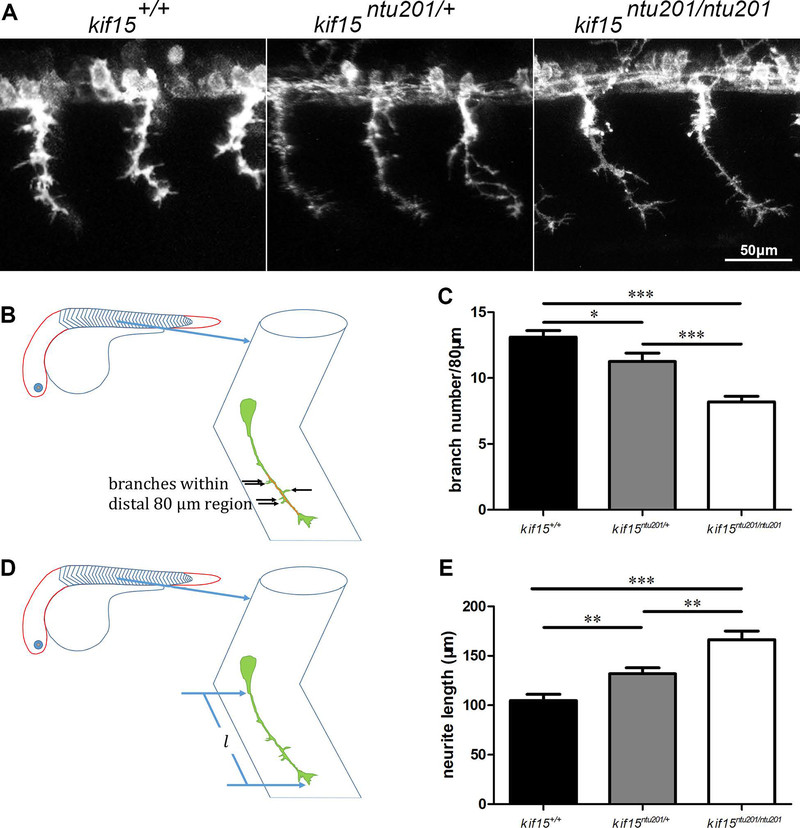
We then investigated the neural morphological alterations in kif15ntu201 mutant zebrafish on a Tg(mnx1:GFP) background. Taking advantage of mnx1 promotor reporter system which specifically induced GFP protein expression in CaP neurons, we found that the axonal shaft of CaP motor neurons was smoother and slender (Shown in Fig.2A) in zebrafish lacking KIF15 during development from 29 hpf (hours post fertilization) to 31 hpf. Therefore, we quantified the axonal branch number and neurite length. As described and shown in Fig.2B, compared with the kif15 wildtype, the number of branches was reduced by 37% and 14% in kif15 homozygotic and heterozygotic mutants, at 31 hpf, respectively (Fig.2C). Homozygotic loss of KIF15 resulted in significant increase of neurite length (166.4 μm) compared to wildtype (104.9 μm), and an increase was shown in heterozygotic mutants (131.9 μm) at 31 hpf (Fig.2DE). The different results between homozygotic and heterozygotic mutants revealed a dosage effect of kif15 gene function on the axonal morphology.
Figure 2.
Changes in CaP motor neuron morphology in mutants of kif15.
(A) CaP neuron morphology in kif15 heterozygotic and homozygotic mutants compared to wildtype siblings at 31 hpf. GFP signals are shown in white pseudo color.
(B) Schematic diagram of branch counting, showing examination of branch numbers in the most distal 80 μm axon of a GFP-labeled CaP neuron in the somite area (V-shaped structures shown in the zebrafish embryo and enlarged) at 31 hpf.
(C) Statistical results of branch numbers of CaP axon in kif15+/+, kif15ntu201/+ and kif15ntu201/ntu201 zebrafish embryos. The columns and bars represent data in mean±S.E.M. (standard error of the mean). The variance was tested using one-way ANOVA, with F=21.63 and P<0.0001, * indicates a p < 0.05 in Student’s t-test, and *** indicates a p < 0.001 (n=20, data were collected from 10 individuals of each genotype measuring lengths of 2 axons in somite 16 and 17 of each fish, and calculated by averaging measurement of total 20 neurons for one genotype).
(D) Schematic diagram of axon length, showing examination of axon length of a GFP-labeled CaP neuron in the somite area at 31 hpf.
(E) Statistical results of axon length of CaP neurons in kif15+/+, kif15ntu201/+ and kif15ntu201/ntu201 zebrafish embryos. The data were presented as mean±S.E.M. The variance was tested using one-way ANOVA, with F=19.14 and P<0.0001. ** indicates a p < 0.01 in Student’s t-test, and *** indicates a p < 0.001 (n=20, data were collected from 10 individuals of each genotype measuring lengths of 2 axons in somite 16 and 17 of each fish, and calculated by averaging measurement of total 20 neurons for one genotype).
2.3. Loss of KIF15 caused swifter axonal extension during development
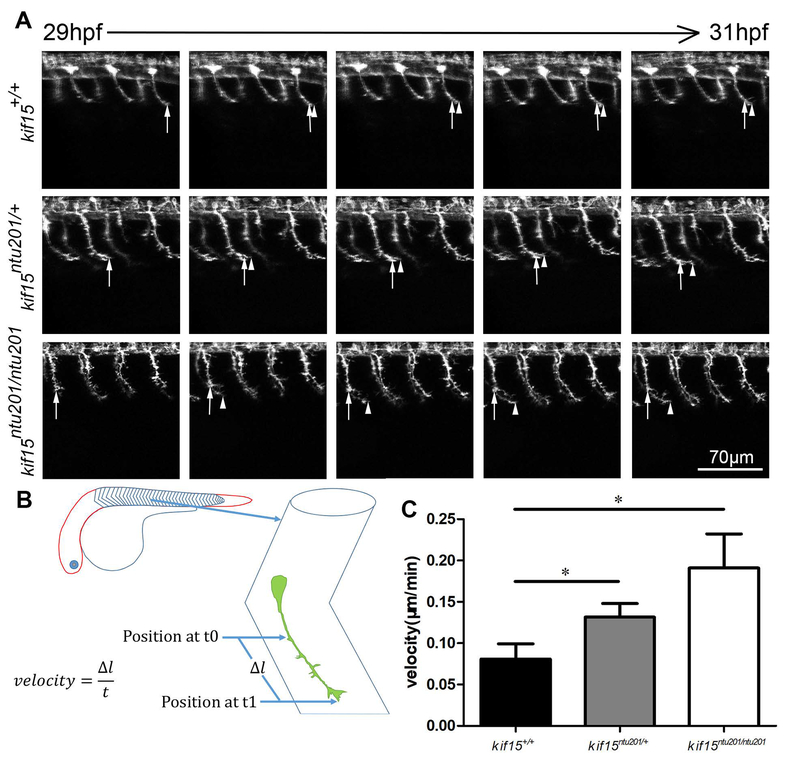
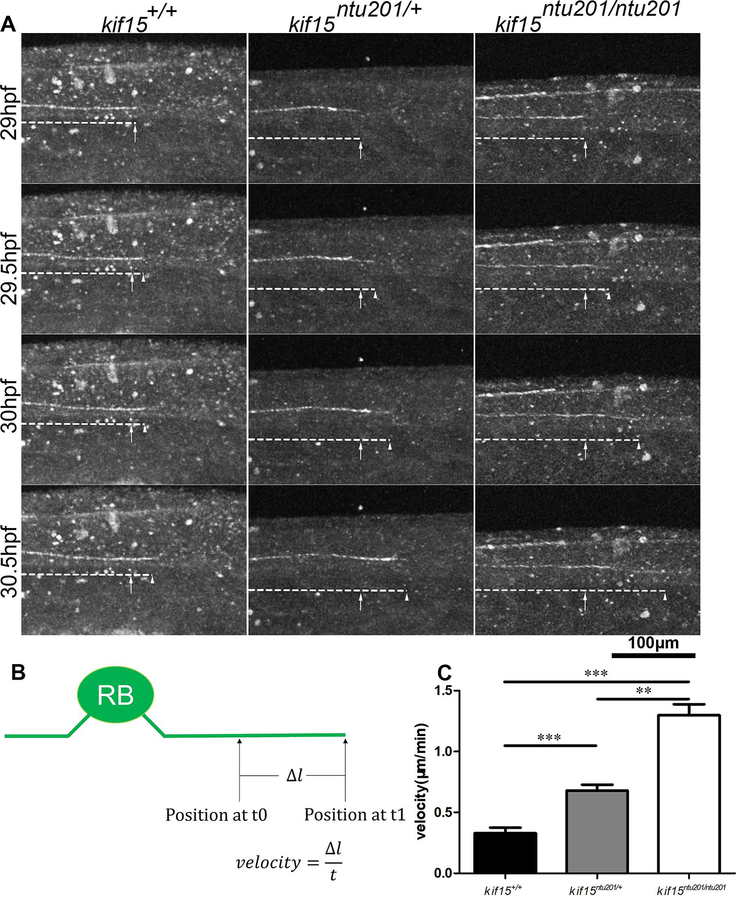
The effect of loss of KIF15 was observed on Tg(mnx1:GFP) background or with injected transiently expressing elavl3:EGFP-alpha tubulin, respectively. When the zebrafish embryos reached 29 hpf, they were anesthetized and embedded in 1% low melt point agarose. Axon length was measured for GFP labeled CaP neurons around the 16th somite at 31 hpf. We calculated the velocity by measuring the distance of axonal tip outgrowth of a CaP neuron between somite 16–22 during 29 hpf to 31 hpf (Fig.3, Supplementary Movie 1), or that of an R-B neuron during 29 hpf to 30.5 hpf (Fig.4, Supplementary Movie 2), respectively. The results showed from 29 hpf to 31 hpf, in mnx1 expressing CaP motor neurons, kif15 mutants exhibited accelerated axon extension. Homozygotic mutants showed approximately 130% faster (0.19 μm/min) axonal growth compared to wildtype (0.08 μm/min), whereas heterozygotes had approximately a 60% increase (0.13 μm/min) (Fig.3, Supplementary Movie 1). In elavl3:EGFP-alpha tubulin transient expression mutants, we focused on labelled R-B neurons. Similarly, from 29 hpf to 30.5 hpf, homozygotic mutants showed 294% faster (1.3 μm/min) axonal growth compared to wildtype (0.33 μm/min), whereas heterozygotes 106% faster (0.68 μm/min) in R-B neurons (Fig.4, Supplementary Movie 2).
Figure 3.
Elimination of Kif15 resulted in accelerated axonal extension in developing CaP motor neurons.
(A) Axon extension in developing CaP neurons in kif15 heterozygotic and homozygotic mutants compared to wildtype siblings. Arrows show growth cone position at 29 hpf. Arrow heads show real time growth cone positions. GFP signals are shown in white pseudo color.
(B) Schematic diagram of measurement of axon extension.
(C) Statistical results of axon extension velocity of CaP neurons in kif15+/+, kif15ntu201/+ and kif15ntu201/ntu201 zebrafish embryos. The data were presented as mean±S.E.M. The variance was tested using one-way ANOVA, with F=4.408 and P=0.0287. * indicates a p < 0.05 in Student’s t-test (n=20, data were collected from 10 individuals of each genotype measuring lengths of 2 axons in somite 16 and 17 of each fish, and calculated by averaging measurement of total 20 neurons for one genotype).
Figure 4.
Elimination of Kif15 resulted in accelerated axonal extension in developing Rohon-Beard sensory neurons.
(A) Axon extension in developing R-B neurons in kif15 heterozygotic and homozygotic mutants compared to wildtype siblings. Arrows show growth cone position at 29 hpf. Arrow heads show real time growth cone positions. GFP signals are shown in white pseudo color.
(B) Schematic diagram of measurement of axon extension.
(C) Statistical results of axon extension velocity of R-B neurons in kif15+/+, kif15ntu201/+ and kif15ntu201/ntu201 zebrafish embryos. The data were presented as mean±S.E.M. The variance was tested using one-way ANOVA, with F=57.32 and P<0.0001. ** indicates a p < 0.01 and *** indicates a p < 0.001 in Student’s t-test (n=9, data were collected from 9 individuals of each genotype, where one axon was measured for each animal).
The effects of kif15 gene on axonal outgrowth in both CaP or R-B neurons in homozygotic kif15ntu201 mutants were significantly stronger than that in heterozygotic kif15ntu201 mutants; in this case, the velocity of axonal outgrowth is minimal in kif15 wildtype. Interestingly, in mnx1 expressing CaP motor neurons, loss of KIF15 impacted axon growth more significantly than in R-B neurons. We observed during development the axonal tips in CaP neuron moved forward in a swinging way, while that of in R-B neurons ran ahead straightly (Supplementary Movies 1 and 2).
2.4. Loss of KIF15 accelerated axon regeneration after laser injury
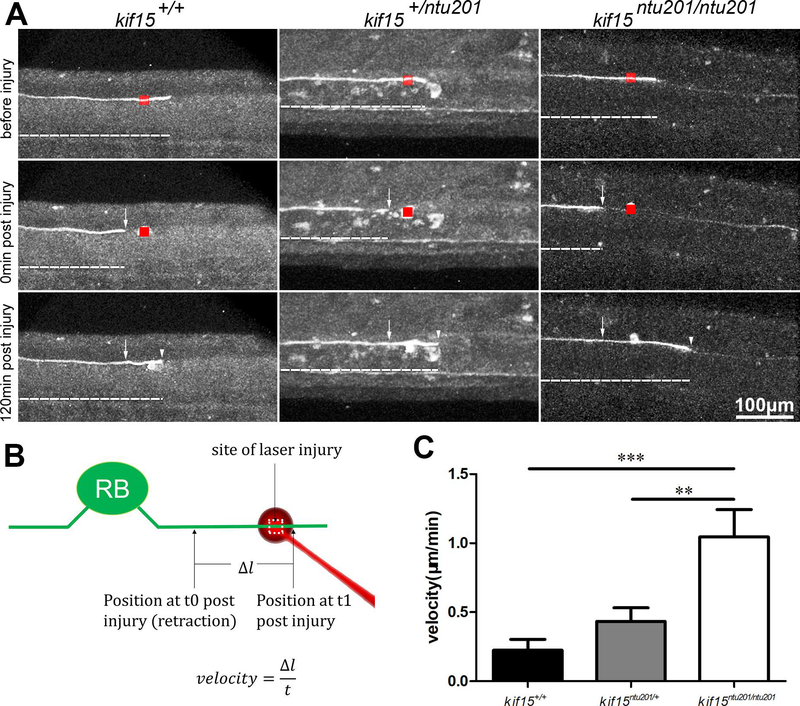
Finally, we investigated the effect of loss of kif15 in regenerating axons after laser injury. Axons expressing fluorescent proteins were cauterized at 26 hpf using 800 nm laser and observed afterwards. Both CaP neurons and R-B neurons were able to regenerate in the injured axon, during which process the loss of kif15 had an effect. However, we were not able to assess the regenerative growth of CaP axon because of the complex morphology and variable extension direction (data not shown). In R-B sensory neurons, we clearly evaluated the regenerative velocity of the injured axons. As shown in Fig.5, after laser irradiation, injured axons retracted for a short distance followed by regenerating (Supplementary Movie 3). We measured the axon length of regenerating for 120 min after injury. The results showed that loss of kif15 promoted axonal regenerative velocity in R-B neurons. Homozygotic mutants showed approximately 377% faster axonal growth at a speed of (1.05 μm/min) compared to wildtype (0.22 μm/min), whereas heterozygotes 95% faster at a speed of (0.43 μm/min) during injured axon regeneration.
Figure 5.
Elimination of Kif15 resulted in accelerated axonal extension in regenerating Rohon-Beard (R-B) sensory neurons after laser injury.
(A) Axon growth in regenerating R-B neurons in kif15 heterozygotic and homozygotic mutants compared to wildtype siblings. Arrow heads show real time growth cone positions. Arrows show retracted axon after injury. Red boxes show laser irradiated area. GFP signals are shown in white pseudo color.
(B) Schematic diagram of measurement of axon growth after laser injury. Arrows show growth cone position at 29 hpf. Arrows show real time growth cone positions.
(C) Statistical results of axon regeneration velocity of R-B neurons in kif15+/+, kif15ntu201/+ and kif15ntu201/ntu201 zebrafish embryos. The data were presented as mean±S.E.M. The variance was tested using one-way ANOVA, with F=9.880 and P=0.0006. ** indicates a p < 0.01, and *** indicates a p<0.001 in Student’s t-test (n=10).
3. Discussion
Kinesin-12 is a microtubule motor protein expressed in all cells in vertebrate organisms, including mitotic cells and post-mitotic neurons18, and is generally regarded to regulate the bipolar microtubule spindle apparatus in dividing cells by generating forces that regulate the sliding of microtubules12,17,19,20. Our previous data showed that kif15 is strongly expressed in developing nervous system in zebrafish with the expression gradually decreasing as the neurons mature14, which is similar to the case in rodent neurons11. Depletion of KIF15 from cultured rodent neurons results in longer axons with fewer branches11, while its depletion from cultured astrocytes results in increased migration21. In the present study, we applied CRISPR/Cas9-based knockout technology to create kif15 mutants, and labeled neurons by hybridizing with Tg(mnx1:GFP) zebrafish or by injecting plasmid pDestTol2-elavl3:EGFP-alpha tubulin. Taking potential off-target effects into consideration, axon development and regeneration were observed in the F3 generation generated from heterozygotic F2 mutants, as any potential off-target mutation would have been diluted to a frequency low enough so as not to obstruct our judgement of the relevance between kif15 targeted mutation and altered axon morphology and growth. In a simple, visual method by live imaging under a fluorescence microscope, we compared the homozygotic, heterozygotic mutants to their wildtype siblings to ascertain the effects of depleting one or both of kif15 alleles on axon outgrowth during neuron development. The results showed, compared to the kif15 wildtype, the number of branches and axonal diameter of CaP motor neurons were reduced in kif15 homozygotic and heterozygotic mutants. The difference of the homozygotic and heterozygotic mutants revealed an obvious dosage effect of kif15 gene function on the axonal morphology. We further measured the velocity of axonal outgrowth of CaP and R-B neurons, and found that homozygotic kif15ntu201 mutants had a significantly greater axonal growth phenotype than heterozygotic kif15ntu201 mutants.
Interestingly, although animals with homozygotic or heterozygotic mutated kif15 had altered neuron morphology during development, the whole individuals appeared normal. Mutants were able to swim and feed just as well as their wildtype siblings, suggesting their neural systems were functional and probably healthy. Also, the generally normal morphology and behavior of mutants suggest that one, the mitosis during development was not strongly affected and two, the morphological changes in neurons were not secondary results from developmental delay or any systemic dysfunction. We interpret these findings as consistent with sufficient redundancy, typical in vertebrates, as well as compensatory mechanisms, that absence or reduction of one key player is tolerated by the system. However, that is not to say the system is the same, and as indicated by our studies on regeneration, which indicate that when challenged, a KIF15-reduced nervous system is better able to regenerate. For this, we evaluated the effect of loss of kif15 gene in regenerating axons after laser injury. Compared to the mechanical methods such as surgery which is operated manually, the two-photon laser injury approach was performed under the identical preset parameters. The consistency of damage to each individual is guaranteed. We used a two-photon laser-based laser injury. In this strategy, the axons of target neurons were damaged by heat generated by laser radiation instead of undergoing fluorescent protein laser bleaching. Both CaP neurons and R-B neurons were able to regenerate in the injured axon, during which process the depletion of kif15 seemed to influence. However, we were not able to assess the regenerative growth of CaP axon because of the complex morphology and variable extension direction (data not shown). As we observed during neural development, the axonal tips in CaP neuron moved forward in a swinging way, while that of in R-B neurons ran ahead straightly. Therefore we focus on observing axonal regeneration of the Rohon-Beard (R-B) neuron, since the primary sensory neurons present during the embryonic and early larval stages, with axon outgrowth straightforward, and many research used it as an object to study the neural functions22,23. We found after laser irradiation, injured axons with depletion of kif15 had significantly improved axonal regenerative velocity. To our best knowledge, this study provides the first in vivo regeneration time-lapse data of KIF15’s role in axonal regeneration.
Why does kif15 express in the nervous system if axons grow better without it? Our previous studies indicate that both KIF15 and KIF11 act as brakes on axonal microtubule transport, and that these brakes are responsive to signaling cascades that regulate features of axonal growth and navigation. Axons presumably grow faster when either of these motors is inhibited because of greater mobility of short microtubules and also less compressive force on the longer microtubules24. A potential issue with KIF11 inhibition as therapy for nerve injury arises from studies indicating that axon navigation is impaired when KIF11 is suppressed25, because of KIF11’s role in regulating the amount of microtubule sliding into the growth cone. It was reported that KIF15 works as a tetramer12,26, similar to KIF1127, and is believed to be capable of cross-linking microtubules and switching microtubule tracks, while other work suggests that it acts as a dimer28. The KIF15 tetramer may behave as a brake via its cross-linking function, with our data showing accelerated axon growth explicable on the basis of releasing the KIF15 brake. Studies to date indicate that KIF15 can do the same sorts of things as KIF11, in terms of regulating microtubule sliding, but can also regulate microtubule-actin interactions, via KIF15’s unique ability to interact with Myosin-IIB, a property not shared by KIF1121,29. We believe the braking effect of KIF15 is at least partly the effect of KIF15-Myosin-IIB complex. The process of an axon generating collateral branches requires the interplay between microtubules and actin filaments, which is a factor impacting the diameter of the stalk of an axon and is also the machinery of the Kif15 brake hypothesis. Our present results demonstrate the effects of KIF15, thus opening the door for therapeutically control not only the window of time to optimally inhibit KIF15, but also the level of inhibition most conducive to both axonal growth and navigation. Drugs could potentially be developed that attenuate KIF15’s microtubule-microtubule functions while not affecting its microtubule-actin functions, or vice versa. Finally, it is relevant to note that KIF15 inhibition may have beneficial effects on other cell types relevant to nerve regeneration, such as the cells that form the glial scar21. The cancer community is already at work developing drugs against KIF15, and hence the time is ripe for the nerve regeneration community to participate.
4. Materials and methods
4.1. Zebrafish husbandry
Zebrafish were provided by the Zebrafish Center at Nantong University. Zebrafish embryos of AB and Tg(mnx1:GFP) (also known as Tg(hb9:eGFP) or Tg(mnx1:GFP)ml2)15 were obtained through natural mating and maintained at 28.5°C. Embryos after 24 hours post fertilization (hpf) were treated with 0.2 mM 1-phenyl-2-thio-urea (PTU, a tyrosinase inhibitor commonly used to block pigmentation and aid visualization of zebrafish development).
4.2. Bioinformatics
The zebrafish kif15 genomic information was obtained from Genbank (Gene ID: 573988, mRNA XM_002666923.4, protein XP_002666969.1). Conserved domains of the kinesin-12 proteins were localized by the Pfam database (http://pfam.sanger.ac.uk/). kif15 sequences of wildtype and mutated alleles were aligned by Vector NTI software (http://www.thermofisher.com/). Primers for PCR and sequencing were designed by Primer Premier 5 software (http://www.premierbiosoft.com/).
4.3. CRISPR/Cas9 design and preparation
Target sites of 23 bp including a 20 bp proto-spacer and a 3 bp proto-spacer adjacent motif (PAM) to the 3’ direction of proto-spacer were manually selected with two stringencies: one, the 1st or 2nd base of the proto-spacer is purine (R, A/G) to assure T7 promoter driven transcription and two, the PAM is NGG. sgRNA templates were then prepared by PCR with a forward primer composed of the first 17 bases of minimum T7 promoter followed by 20 bases identical to target proto-spacer and 20 bases identical to the first 20 bases of sgRNA scaffold, a reverse primer complementary to the last 25 bases of sgRNA scaffold (Table 2) and a template plasmid pT7-gRNA kindly provided by Prof. Bo Zhang at Peking University. The PCR products were used as templates for in vitro transcription using MAXIscript T7 Kit (Ambion, USA) to obtain sgRNAs. Capped Cas9 mRNA was prepared by in vitro transcription using mMessage mMachine T7 Kit with a zebrafish optimized Cas9 template plasmid pGH-T7-zCas930 kindly provided by Prof. Bo Zhang at Peking University. Cas9 mRNA and sgRNA were mixed and adjusted to a final concentration of 300ng/μl:100ng/μl.
Table 2.
PCR primers for gRNA template preparation and mutant genotyping
| Primer | sequence (5’−3’) |
|---|---|
| gRNA-template-F | taatacgactcactata + proto spacer + gttttagagctagaaatagc |
| gRNA-template-R | aaaaaaagcaccgactcggtgccac |
| kif15-4F3 | agggatcggtggagaaa |
| kif15-4R5 | cttgcccagagtgtattgt |
| kif15-seqR59 | ctgtccggtctccttggact |
4.4. Microinjection
Each embryo received about 1 nl of the mixed RNA solution using borosilicate glass capillaries (world precision Inc. WPI) with PV830 Pneumatic picopump (WPI). Injected embryos were grown until sexual maturity, and mated with Tg(mnx1:GFP) transgenic zebrafish for CaP neuron imaging. Heterozygotic mutants were in-crossed and resulting embryos were injected with pDestTol2-elavl3:EGFP-alpha tubulin (prepared using Golden Gate cloning with Tol2kit #394 pDestTol2pA2, Addgene #72640 p5E-elavl3 from Joshua Bonkowsky, pME-EGFP-alpha tubulin in which the EGFP-alpha tubulin coding sequence was subcloned from Addgene #12298 from Patricia Wadsworth, and Tol2Kit #302 p3E-polyA) construct for R-B neuron imaging.
4.5. Genotyping
CRISPR/Cas9 mutagenesis efficiency was examined by PCR-sequencing. Genomic DNA was isolated with YSY Trace DNA buffer (YSY biotech, Nanjing, China) from 24hpf embryos that had received injection. PCR amplicons around the target region for kif15 mutation (primers for PCR and sequencing are listed in Table 2). The resulting F1 generation were genotyped by fluorescence microscopy for the transgenes and by PCR-sequencing as described above. Genomic DNA was isolated from fin clips with YSY Trace DNA buffer.
4.6. Imaging
For confocal imaging of neuronal development in elavl3:EGFP-alpha tubulin transient expression or Tg(mnx1:GFP) zebrafish, embryos were anesthetized with egg water/0.16 mg/mL tricaine/1% 1-phenyl-2-thiourea (Sigma) and embedded in 1% agarose. Using a 20x objective, confocal stack images of the trunk region were obtained in time intervals of 10 minutes. Images were taken using a Leica TCS SP5 LSM confocal microscope (Leica, Wetzlar, Germany). Analysis was performed using Imaris (http://www.bitplane.com/).
4.7. Laser injury
We generally translated a protocol using Zeiss confocal microscope for laser injury31,32 and a method based on Olympus FV1000 MPE two-photon laser scanning microscope33 to Leica TCS SP5 LSM equipped with two-photon laser, but with minor alterations. In brief, the embedded embryos were observed under a 20x water lens and the selected axons were placed in the center of the field of vision. The display was zoomed in to 64x, and 800nm two-photon laser was used to perform a single frame xy scan at 50% intensity.
4.8. Statistics
Neurite length between was measured by Image J software (https://imagej.nih.gov/ij/). The branch numbers within the distal 80 μm of axon length were calculated. All data analysis, statistical comparisons, and graphs were generated using GraphPad Prism 5 (http://www.graphpad.com/scientific-software/prism/). Data are expressed as mean ± S.E.M. (standard error of the mean). Statistical analysis was performed using one-way ANOVA for the analysis of variation among genotype groups follow by Student’s t-test for comparison between each two groups. The final figure processing was performed with Adobe Illustrator CS6.
Supplementary Material
Supplementary Movie 1
(Associated with Fig. 3)
CaP motor neuron growth in developing zebrafish embryos with or without KIF15 from 29 hpf to 34 hpf. CaP motor neurons are labelled with GFP in a Tg(mnx1:GFP) background.
Supplementary Movie 2
(Associated with Fig. 4)
Rohon-Beard sensory neuron growth in developing zebrafish embryos with or without KIF15 from 28.5 hpf to 30.5 hpf. R-B neurons are labelled with GFP with transient expressing elavl3:EGFP-alpha tubulin.
Supplementary Movie 3
(Associated with Fig. 5)
Rohon-Beard sensory neuron growth after laser injury in developing zebrafish embryos with or without KIF15. R-B neurons are labelled with transient expressing elavl3:EGFP-alpha tubulin.
Synopsis.
KIF15, a microtubule-associated motor protein, was reported previously impacting axonal growth, navigation and branching in cultured rat neurons. To elucidate kif15 function in vivo, we employed CRISPR/Cas9-based knockout technology to create kif15 mutants in zebrafish. By comparing the homozygotic and heterozygotic mutants with their wildtype siblings, the effects of kif15 depletion were ascertained during axon development and regeneration. That is, a disuse of kif15 renders axons fast-growing, smooth and slender, and also with greater regenerative velocity after axonal laser injury.
Acknowledgments
This study was supported by grants to the Nantong University group from the National Natural Science Foundation of China (31171007; 31371078; 31500965; 31701049), Natural Science Foundation of Jiangsu Province (BK20150404, BK20171253), the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions and financial support from the program of China Scholarships Council (No. 201808320097) to Z.D.. Additional support was from a grant to the Drexel group from the National Institutes of Health (R01 NS28785). The authors thank Prof. Yanqin Shen at Jiangnan University for technical help in the axonal laser injury. The authors declare no conflict of interest.
References
- 1.Dong B, Deng W, Jiang D. Distinct cytoskeleton populations and extensive crosstalk control Ciona notochord tubulogenesis. Development. 2011;138(8):1631–1641. [DOI] [PubMed] [Google Scholar]
- 2.Pramparo T, Libiger O, Jain S, et al. Global Developmental Gene Expression and Pathway Analysis of Normal Brain Development and Mouse Models of Human Neuronal Migration Defects. PLoS Genet. 2011;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellal F, Hurtado A, Ruschel J, et al. Microtubule Stabilization Reduces Scarring and Causes Axon Regeneration After Spinal Cord Injury. Science. 2011;331(6019):928–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuckowree JA, Vickers JC. Cytoskeletal and morphological alterations underlying axonal sprouting after localized transection of cortical neuron axons in vitro. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23(9):3715–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geraldo S, Gordon-Weeks PR. Cytoskeletal dynamics in growth-cone steering. J Cell Sci. 2009;122(20):3595–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci. 2008;30(1–3):36–46. [DOI] [PubMed] [Google Scholar]
- 7.Poulain FE, Sobel A. The microtubule network and neuronal morphogenesis: Dynamic and coordinated orchestration through multiple players. Mol Cell Neurosci. 2010;43(1):15–32. [DOI] [PubMed] [Google Scholar]
- 8.Sakakibara A, Ando R, Sapir T, Tanaka T. Microtubule dynamics in neuronal morphogenesis. Open Biol. 2013;3(7):130061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S, Liu M, Mozgova OI, Yu W, Baas PW. Mitotic motors coregulate microtubule patterns in axons and dendrites. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32(40):14033–14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S, Liu M, Son Y-J, et al. Inhibition of Kinesin-5, a Microtubule-Based Motor Protein, As a Strategy for Enhancing Regeneration of Adult Axons. Traffic. 2011;12(3):269–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Nadar VC, Kozielski F, Kozlowska M, Yu W, Baas PW. Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(44):14896–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife. 2014;3:e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klejnot M, Falnikar A, Ulaganathan V, Cross RA, Baas PW, Kozielski F. The crystal structure and biochemical characterization of Kif15: a bifunctional molecular motor involved in bipolar spindle formation and neuronal development. Acta Crystallographica Section D: Biological Crystallography. 2014;70(Pt 1):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu M, Liu D, Dong Z, et al. Kinesin-12 influences axonal growth during zebrafish neural development. Cytoskeleton. 2014;71(10):555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan-Steet H, Fox MA, Meyer D, Sanes JR. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132(20):4471–4481. [DOI] [PubMed] [Google Scholar]
- 16.Sturgill EG, Norris SR, Guo Y, Ohi R. Kinesin-5 inhibitor resistance is driven by kinesin-12. The Journal of cell biology. 2016;213(2):213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Current biology : CB. 2009;19(20):1703–1711. [DOI] [PubMed] [Google Scholar]
- 18.Buster DW, Baird DH, Yu W, et al. Expression of the mitotic kinesin Kif15 in postmitotic neurons: implications for neuronal migration and development. J Neurocytol. 2003;32(1):79–96. [DOI] [PubMed] [Google Scholar]
- 19.Vanneste D, Takagi M, Imamoto N, Vernos I. The Role of Hklp2 in the Stabilization and Maintenance of Spindle Bipolarity. Curr Biol. 2009;19(20):1712–1717. [DOI] [PubMed] [Google Scholar]
- 20.Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell cycle. 2011;10(20):3533–3544. [DOI] [PubMed] [Google Scholar]
- 21.Feng J, Hu Z, Chen H, et al. Depletion of kinesin-12, a myosin-IIB interacting protein, promotes migration of cortical astrocytes. J Cell Sci. 2016;129(12):2438–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menelaou E, Husbands EE, Pollet RG, Coutts CA, Ali DW, Svoboda KR. Embryonic motor activity and implications for regulating motoneuron axonal pathfinding in zebrafish. Eur J Neurosci. 2008;28(6):1080–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi CC, Kaji T, Artinger KB. Transcriptional control of Rohon-Beard sensory neuron development at the neural plate border. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238(4):931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahn OI, Baas PW. Microtubules and Growth Cones: Motors Drive the Turn. Trends Neurosci. 2016;39(7):433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baas PW, Matamoros AJ. Inhibition of kinesin-5 improves regeneration of injured axons by a novel microtubule-based mechanism. Neural regeneration research. 2015;10(6):845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drechsler H, McAinsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci U S A. 2016;113(12):E1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. [DOI] [PubMed] [Google Scholar]
- 28.Reinemann DN, Sturgill EG, Das DK, et al. Collective Force Regulation in Anti-parallel Microtubule Gliding by Dimeric Kif15 Kinesin Motors. Curr Biol. 2017;27(18):2810.-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, Feng J, Bo W, et al. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Molecular biology of the cell. 2017;28(4):545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Wang Z, Xiao A, et al. Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. Journal of genetics and genomics = Yi chuan xue bao. 2014;41(1):43–46. [DOI] [PubMed] [Google Scholar]
- 31.O’Brien GS, Rieger S, Martin SM, Cavanaugh AM, Portera-Cailliau C, Sagasti A. Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. Journal of visualized experiments : JoVE. 2009(24):e1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Donnell KC, Vargas ME, Sagasti A. WldS and PGC-1alpha regulate mitochondrial transport and oxidation state after axonal injury. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(37):14778–14790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahu S, Zhang Z, Li R, et al. A Small Organic Compound Mimicking the L1 Cell Adhesion Molecule Promotes Functional Recovery after Spinal Cord Injury in Zebrafish. Mol Neurobiol. 2018;55(1):859–878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie 1
(Associated with Fig. 3)
CaP motor neuron growth in developing zebrafish embryos with or without KIF15 from 29 hpf to 34 hpf. CaP motor neurons are labelled with GFP in a Tg(mnx1:GFP) background.
Supplementary Movie 2
(Associated with Fig. 4)
Rohon-Beard sensory neuron growth in developing zebrafish embryos with or without KIF15 from 28.5 hpf to 30.5 hpf. R-B neurons are labelled with GFP with transient expressing elavl3:EGFP-alpha tubulin.
Supplementary Movie 3
(Associated with Fig. 5)
Rohon-Beard sensory neuron growth after laser injury in developing zebrafish embryos with or without KIF15. R-B neurons are labelled with transient expressing elavl3:EGFP-alpha tubulin.