The gut microbiome, a term that describes the entire intestinal habitat including resident microorganisms (the microbiota) and their metabolic byproducts, has a profound impact on systemic health ranging from immune development, metabolism, and protection from infection1–4. The innovative use of the 16S ribosomal RNA gene for phylogenic analysis, paired with the decreasing cost of sequencing paved the way for numerous surveys describing the composition of human bacterial microbiota5,6. The ability to characterize bacterial membership at various body sites beyond our limited capacity to culture organisms has enabled us to find associations between the microbiota and many human diseases.
The role of the microbiome in cardiovascular disease (CVD) has only been investigated in a small number of human studies that have primarily linked differences in the composition of microbiota to various disease processes associated with CVD7. For example, taxa abundant in atherosclerotic plaques were also increase in patient oral cavities8. Studies in both animal models and humans have demonstrated a link between microbial metabolism of diet-derived metabolites and increased levels of trimethylamine N-oxide9. Together these studies point tantalizingly toward the promise of modulation of microbiome for treatment of CVD.
In this issue of Circulation Research, Malik et al. sought to move microbiome studies from observation to intervention by examining whether Lactobacillus plantarum 229v altered vascular endothelial function in men with stable coronary artery disease10. To completely meet these goals the following questions must be asked: Did the intervention alter the gut microbiome? Did the intervention alter measures of vascular disease? Are the vascular measures a reliable clinical surrogate?
The study used G3 PhyloChip to assess the effect of oral L. plantarum 229v on the microbiota though PhyloChip does not, as stated, rely on “16S rDNA sequencing” but rather is a microarray-based approach. To the best of our knowledge, no recent papers have benchmarked G3 PhyloChip relative to Illumina 16S rRNA gene sequencing so it is difficult to compare the two methods. The microarray based approach enables detection of taxa from both Bacteria and Archaea11. Archaea are an understudied minority member of the gut microbiota which have been proposed as a potential means to decrease trimethylamine levels in the gut12. The authors reported that, L. plantarum 229v did not result in significant differences in the overall microbiota, including the number of observed species (richness) but they noted significant differences in the relative abundance of 70 operational taxonomic units between the pre and post-treatment samples with an enrichment of Lactobacillus spp. in the post-treatment samples. However, these findings were not significant after correcting for multiple comparisons. Perhaps other analytical methods like machine learning based models might be better able to parse the differences between samples13,14. Overall the findings from this study are in line with other work, which have seen minimal effects of probiotic supplementation on community structure15,16. Plasma levels of acetic, propionic, and butyric acid, which are derivatives of microbial derived short chain fatty acids, were measured as a surrogate to determine if L. plantarum 229v supplementation altered the microbiome17. Following supplementation, levels of acetic acid significantly decreased while levels of propionic acid increased. This result begs the question if the changes reflect L. plantarum 229v metabolism or if they reflect changes in metabolism of other members of the microbiota from the dietary supplementation. Others have found that while no significant alterations in the community structure were observed during probiotic therapy, the gut microbial transcriptional program was significantly changed15. This suggests that future studies seeking to determine the effects of an intervention on the microbiota should move beyond metrics of community structure (“who is there”) and instead focus on what the microbes are doing.
Following the probiotic supplementation the outcome measures included change in brachial artery flow mediated dilation (FMD) from 3.55+/−1.96 to 4.73+/−2.32 with no change in resting vessel diameter, hyperemic sheer stress, or vascular smooth muscle mediated vasodilation. They then took plasma from 4 subjects and tested the net/bulk effects on adipose arterioles. There were reductions in IL-8 and IL-12 and again, effects were limited to the endothelium as the effects were smooth muscle independent and blocked by L-NAME. Direct measures of bioavailable nitric oxide (NO) are feasible but were not made18. Following a washout period, a subset of patients also took oral vancomycin as the authors had previously seen a therapeutic effect in animal models19. Vancomycin did not result in a significant change in brachial artery FMD post treatment. It is tempting to speculate that this result lends support for more targeted microbial therapeutics, however, the value of vancomycin as an intervention control for the specificity of the probiotic therapy is yet to be determined.
The role of NO as the key factor in the maintenance of endothelial function, overall vascular health, and arterial tone is an established paradigm and central theme of translational vascular biology. Though brachial artery FMD is technically challenging, it does provide the best non-invasive assessment and is a frequently used accepted “biomarker” of the state of NO bioavailability in the endothelium. Changes in FMD suggest that the responsive of the endothelium can be inversely related to disease progression, although even in this space BAR serves more as a marker for prediction of risk and its role is less clear as a response to therapy.
It is quite likely that the gut microbiome can influence the balance of vascular homeostasis (Figure 1) in critical features like NO production, inflammation, and thrombosis. Variations in baseline and alterations within the microbiome should be considered as an important and influential factor. Studies of the microbiome within and outside cardiovascular medicine are likely to and indeed should increase. In parallel, we will see an increase in the complexity of the analytic tools needed to fully establish the absolute and relative importance of this area on vascular health.
Figure 1.
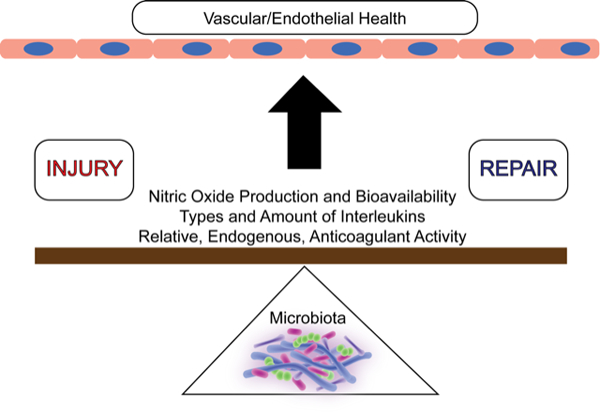
The gut microbiome (bottom) can serve as a fulcrum in the ongoing battle of vascular injury and repair that determines overall endothelial health.
Acknowledgments
Sources of Funding: Dr. Leslie received support from T32 AI 7496–23. Dr. Annex is supported by RO1HL16455, HL121635, and HL101200.
Footnotes
Disclosures: No relevant conflicts of interest.
References
- 1.Marchesi JR, Ravel J. The vocabulary of microbiome research: A proposal. Microbiome. 2015;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leslie JL, Young VB. The rest of the story: The microbiome and gastrointestinal infections. Current opinion in microbiology. 2015;0:121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: Metabolism of nutrients and other food components. European journal of nutrition. 2018;57:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proceedings of the National Academy of Sciences. 1977;74:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Human Microbiome Project C, Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fåk F, Tremaroli V, Bergström G, Bäckhed F. Oral microbiota in patients with atherosclerosis. Atherosclerosis. 2015;243:573–578 [DOI] [PubMed] [Google Scholar]
- 8.Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences. 2011;108:4592–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik M, Suboc TM, Tyagi S, Salzman N, Wang J, Ying R, Tanner MJ, Kakarla M, Baker JE, Widlansky ME. Lactobacillus plantarum 299v supplementation improves vascular endothelial function and reduces inflammatory biomarkers in men with stable coronary artery disease. Circulation Research. 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. mBio. 2015;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brugère J-F, Borrel G, Gaci N, Tottey W, O’Toole PW, Malpuech-Brugère C. Archaebiotics. Gut Microbes. 2014;5:5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxter NT, Ruffin MT, Rogers MAM, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Medicine. 2016;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, Ravel J, Haverkamp M, Fiorino A-M, Botelho C, Andreyeva I, Hibberd PL, Fraser CM. Functional dynamics of the gut microbiome in elderly people during probiotic consumption. mBio. 2015;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Medicine. 2016;8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen JD, Miller EM, Schwark E, Robbins JL, Duscha BD, Annex BH. Plasma nitrite response and arterial reactivity differentiate vascular health and performance. Nitric oxide : biology and chemistry. 2009;20:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam V, Su J, Hsu A, Gross GJ, Salzman NH, Baker JE. Intestinal microbial metabolites are linked to severity of myocardial infarction in rats. PLoS One. 2016;11:e0160840. [DOI] [PMC free article] [PubMed] [Google Scholar]


