Abstract
DNA nanostructures largely rely on pairing DNA bases; thus, sequence designing is required. Here we demonstrate a sequence-independent strategy to fabricate DNA nanogel (NG) inspired by cisplatin, a chemotherapeutic drug that acts as a DNA cross-linker. A simple heating and cooling of the genomic DNA extracts and cisplatin produces DNA NG with a size controlled by the heating time. Furthermore, the drug loaded NG has been formulated by spontaneous mixing DNA segments, cisplatin and doxorubicin. The in vitro cell studies demonstrate the doxorubicin loaded NG alters the drug distribution in cells while its cytotoxic potential is well maintained. This chemotherapeutics-inspired method provides a facile one-pot and cost effective strategy to fabricate size-controllable DNA NG that potentially acts as drug carrier.
Keywords: DNA, DNA nanotechnology, Cisplatin, Nanogel, drug delivery
1. Introduction
Due to the biocompatibility, biological recognition ability, predictable conformation and programmability of DNA sequences, numerous DNA nanostructures have been constructed and intensively explored as multifunctional biomaterials.[1–6] DNA molecules self-assemble into predefined structures in one, two and three dimensions following the strict rules of Watson-Crick base pairing, such as DNA origami and tiles, etc.[7–10] In addition to the fundamental base pairing method, enzyme mediated DNA polymerization and chemical crosslinking methods have been introduced to create different DNA materials at both nano and bulk scales.[11–14] The maturation of these construction strategies allows for the building of complex and predictable DNA architectural structures. New applications with novel DNA nanostructures have been reported. Alternatively, DNA, as the inherent genetic material in all organisms, has been widely used in gene and nucleic acid based therapies.[15] However, DNA itself is vulnerable toward enzymatic degradation and is incapable of penetrating the cell membrane; consequently, carriers such liposomes and cationic polymers are commonly used to ferry DNA into cells.[15]
Interestingly, DNA can not only be a drug by itself, but also a carrier for active therapeutic agents after forming nanostructures. Unlike free DNA strands, DNA nanostructures are more resistant to nucleases and can be internalized through caveolin or receptor mediated endocytosis.[16–18] The targeted delivery of drugs to specific disease sites have been realized in vivo by attaching a targeting ligand, such as aptamers, or tuning the morphology of DNA nanostructures through active and negative targeting mechanisms.[3, 19] To date, the typical drug that were integrated within DNA nanostructures were those with the ability to bind or intercalate with DNA. Among the list of drugs used in these nanostructures, Doxorubicin (Dox) is the most prevalent.[6, 20] Dox is a chemotherapeutic drug widely used in the treatment of multiple cancers that functions in binding DNA and inhibiting DNA-associated enzymes.[21] Dox is able to preferentially intercalate between GC bases of DNA. In most cases GC-rich sequences were intentionally added into DNA strands to load more Dox into DNA nanostructures.[20, 22, 23] Inspired by the loading of drugs into DNA nanostructures based on their interactions with DNA, we sought to exploit DNA-drug interactions in order to build new DNA nanocarriers. Compared to the base pairing strategy in DNA nanotechnology, the developed DNA-drug interaction strategy is less reliant on DNA sequences to build the nanostructures.
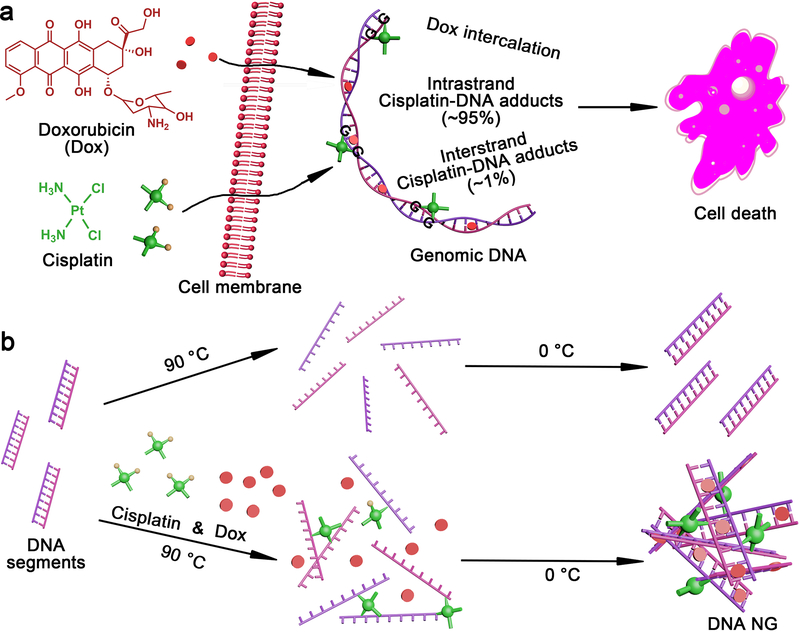
Among the DNA targeting drugs,[24] DNA crosslinkers have the potential to hold different DNA strands together into hydrogels. Cisplatin (Cis), a well-known chemotherapeutic drug, binds to DNA preferably at the N7 position of guanine nucleoside and blocks DNA replication and transcription, causing cell death (Figure 1a).[25] Among the cisplatin-DNA adducts, most are 1,2-d(GpG) intrastrand crosslinks (~95%), while a small portion of the GG interstrand crosslink (<1%) was also found.[25–27] Intrigued by this interstrand crosslinking capability, we proposed that cisplatin could act as a crosslinker to mediate the formation of the DNA nanogel in a sequence-independent manner. Using genomic DNA segments as an example, we verified the formation of a spherical DNA nanogel (NG) with a tailored size by simply mixing DNA and cisplatin under a heating and cooling process (Figure 1b). To demonstrate the possible application of this DNA NG, Dox was loaded into DNA NG and their drug delivery performance was evaluated in cells.
Figure 1.
Synthesis of DNA NG inspired by DNA as therapeutic target. a, DNA as the therapeutic target in cancer therapy. b, Synthesis of DNA NGs. Cisplatin as crosslinker in mediating the formation of DNA NG. Dox could be loaded in the DNA NG.
2. Results and Discussion
Genomic DNA extracts from Herring sperm (≤ 50 bp), the DNA segments widely used in interaction between DNA and binding agent studies,[28, 29] were chosen as model DNA to fabricate DNA NG. The DNA and cisplatin (4:1 w/w) was simply mixed in pure water, heated to 90 °C, cooled on ice and then purified by dialysis in water to remove free cisplatin (Figure 1b). The heat was introduced in this process for the following considerations: Firstly, the genomic DNA segments used here were shorter than the persistence length (150 bp), thus the dsDNA would be rigid[30] which is not favorable in the formation of NG. The heating process denatured the DNA double strands into a single stranded DNA (ssDNA), which is more flexible. Secondly, the high temperature could accelerate the rate of cisplatin crosslinking[27] by denatuting the DNA and increasing the probability of crosslinking two ssDNA. During the on ice cooling phase, DNA strands could re-anneal and/or brought together by the cisplatin crosslinker (Figure 1b). In addition, the decrease of physical distance between different DNA strands mediated by cisplatin crosslinking may also force the mismatches of base pairs and further facilitate NG formation.
To study the heating effects, NGs were prepared with 5, 10 and 15 minutes of 90 °C heating, named DNA NG-1, 2 and 3, respectively. In the presence of cisplatin, the solution’s opaqueness increased as the heating time was elongated (Figure 2a), implying the formation of NGs which enhanced light scattering. The dynamic light scattering (DLS) analysis proved the presence of negatively charged DNA NG varying from 198 nm, 320 nm and 392 nm for NG-1, 2 and 3, respectively (Figure S1). The cisplatin encapsulated in DNA NGs was only around 2.6 – 3.2 ng per μg of DNA (Table S1), which was in the similar range of cisplatin binding to calf thymus DNA with an incubation of 12h at 37°C.[31] The residual cisplatin and rising size of DNA NG was dramatically different from what was observed in cisplatin crosslinked hyaluronan (HA),[32, 33] which was possibly due to the difference between cisplatin’s crosslinking efficiency towards DNA bases and HA carboxyl groups.
Figure 2.
Characterization of DNA NGs in comparison to free DNA. a, Photos of aqueous solution of DNA and DNA NGs. b, UV-Vis absorption spectra. c, Agarose gel electrophoresis. d, Morphology of DNA NGs observed by TEM.
The formation of NG was also evidenced by the red shift absorption peak from 260 to ~270 nm (Figure 2b). As revealed by the structural biology, cisplatin binding bent and unwound DNA strands,[26, 34] which led to a different absorption profile of DNA. Agarose gel electrophoresis was conducted to confirm the formation of NGs as shown in Figure 2c. When the heating time was increased, more DNA retarded in the loading well because of their increased size.
The stability of DNA nanostructures is one of the main concerns for their application in cell environments. To test its stability, DNA NG-2, for example, was co-incubated with fetal bovine serum (FBS) at 37 °C up to 3 days. After its incubation, the NG integrity was evaluated by electrophoresis. While the band of free DNA disappeared after 24 hours of incubation, the main band for DNA NG-2 remained in the loading well even after 72 hours of incubation with FBS (Figure S2), thus suggesting that DNA NGs are more resistant to enzymatic degradation. The formation of NG must have restricted the accessibility of DNA to nucleases. In addition, the binding of cisplatin potentially provides extra steric hindrance for enzyme recognition.
Cisplatin possesses a high electron density, thus the morphology of DNA NGs observed from the transmission electron microscopy (TEM) appeared as a black sphere structure with a clear size increase (Figure 2d). The TEM size of the three DNA NGs was 148, 300 and 397 nm (Figure S3), well corroborating the size distribution determined by DLS (Figure S1a). In the free DNA control, as expected, no specific structure could be detected by TEM.
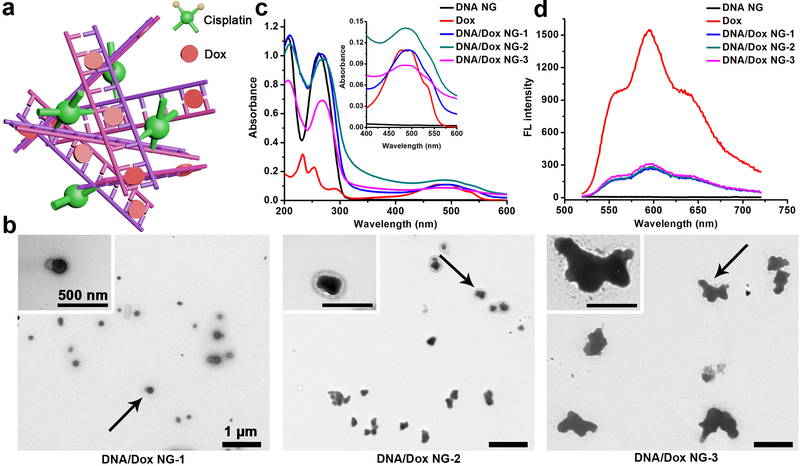
To explore the potential of current DNA NG as a drug carrier (Figure 3a), Dox was introduced during the mixing of DNA and cisplatin; the final products were referred to DNA/Dox NG-1, 2, 3 with a heating time of 5, 10 and 15 minutes, respectively. Because of the intrinsic absorbance and fluorescence of Dox, the formulated DOX containing DNA nanostructures can be analyzed conveniently. Similar to the drug free DNA NG, the incubated mixtures exhibited strong Dox color, while demonstrating a decreased transparency as the heating time increased, implying a successful withholding of Dox as well as a NG formation (Figure S4a). The DLS analysis further confirmed the size change, with DNA/Dox NG-1, 2 and 3 ranged from 150 nm, 245 nm to 416 nm (Figure S4a). When the size of DNA/Dox NG increased, the zeta potential gradually dropped as shown in Figure S4b. In the UV-Vis absorption spectra, DNA/Dox NG demonstrates a clear Dox absorption band compared with DNA NG (Figure 3c). After being incorporated into NGs, the Dox fluorescence was severely quenched (Figure 3d). This quenching effect was reported when Dox intercalated between the DNA bases.[35] Interestingly, the morphology of DNA/Dox NG turned more irregular in contrast to spherical structure of DNA NG from the TEM observations (Figure 3b). The intercalation of Dox induced a conformational change in the DNA, which could partialy explain the irregular morphology of DNA/Dox NGs.[36] The TEM size of DNA/Dox NG-3, specically, grew rapidly and inhomogenously which was also corroborated by the precipitates spotted at the bottom of the preparation solution (Figure S5 and S4a). The cisplatin in DNA/Dox NG was very low (~3 ng per μg of DNA), with an amount similar to that in DNA NG, while the Dox amount was around 100 ng per μg of DNA (Table S2). This Dox value was equal to one Dox binding for every 7–8 base pairs of DNA. Considering the similar Dox content in all DNA/Dox NGs together with the fact that a nanoparticle size of below 200 nm demonstrated a better tumor accumulation in vivo,[37] the DNA/Dox NG-1 was selected as the Dox delivery vehicle in the cell study.
Figure 3.
Characterization of DNA/Dox NG. a, Schematic illustration of DNA/Dox NG. The number of cisplatin and Dox in the cartoon doesn’t represent the real drug content in NG (Table S2). b, TEM observation. c, Absorption spectra of free Dox, DNA and DNA/Dox NG. d, Fluorescent spectra. Free Dox and DNA NG were set as controls.
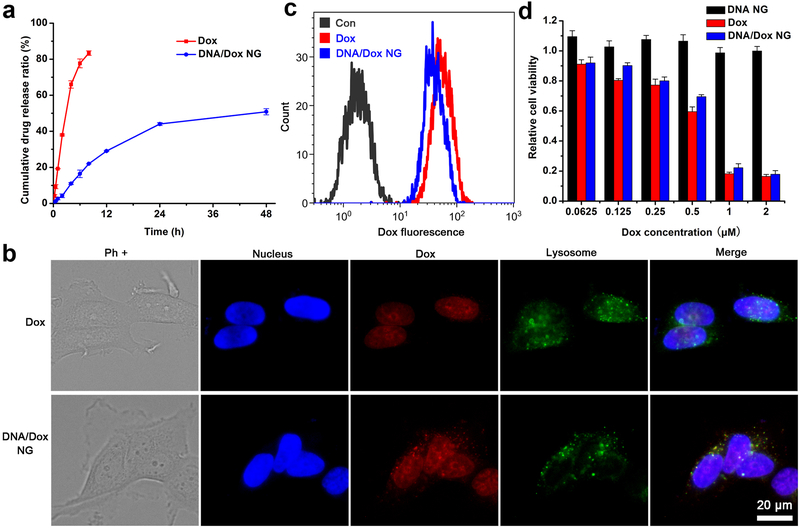
The controlled drug release played a pivotal role in the drug release system; as such, the release of Dox from DNA/Dox NG-1 was followed in PBS at 37 °C. The free Dox, which freely diffused throughout the dialysis membrane, was released rapidly; while the DNA/Dox NG-1 demonstrated a slow release behavior, and reached a plateau after 24 hours of incubation (Figure 4a). The DNA/Dox NG were slightly swelled in PBS after 2 days of incubation (Figure S6), possibly due to the partial loss of Dox. Around 50% of Dox had been released into the solution. Additionally, the loss of a crosslinker presumably also contributed to this swelling since conjugated cisplatin could be freed in a chloride-containing environment.[32] However, no significant aggregation of DNA/Dox NG was observed, which indicated the NG still continued to behave as nanoparticles even after being introduced into a biological environment.
Figure 4.
Dox delivery mediated by DNA/Dox NG-1. a, Release of Dox in PBS at 37 °C. b, Intracellular localization of Dox in MCF-7 cells after 3 hours incubation. The Dox and cisplatin content in DNA/Dox NG group was set at 1 μM and 0.05 μM. Nucleus and lysosomes were stained by Hoechst and Lysotracker green, respectively. c, Cellular uptake of free Dox and DNA/Dox NG determined by FACS. d, Cell viability determined by MTS assay. The DNA content in DNA/NG was the same with that in DNA/Dox NG.
With the intrinsic fluorescence of Dox, the cellular uptake and localization of DNA/Dox NG was evaluated in a breast cancer cell line MCF-7. Free Dox diffused through the cell membrane and localized in the nucleus with its DNA binding ability (Figure 4b). But for the DNA/Dox NG treated cells, besides nuclear localization, spotty Dox fluorescence was also seen within the lysosomes. Resembling most of other DNA nanostructures, which were internalized through endocytosis and trafficked to the lysosomes,[38, 39] DNA/Dox NG carried Dox into the lysosomes following its internalization. The Dox located within nucleus, for DNA/Dox NG, could be ascribed to the Dox released either extracellularly or intracellularly during the co-incubation process. As quantified by flow cytometry (FACS), the intracellular Dox delivered by DNA/Dox NG was comparable to that of free Dox, which could freely diffuse though the cell membrane and bind the cell’s nucleus (Figure 4c). This indicated that DNA/Dox NG mediated a similar cytotoxicity in vitro compared with free Dox (Figure 4d). To exclude the possible cytotoxicity contributed by the cisplatin retained in DNA/Dox NG, Dox plus equal amount of free cisplatin was also tested. It turned out the addition of free cisplatin to Dox didn’t cause a difference in toxicity, as the amount of the cisplatin was low (≤ 0.1 μM) (Figure S7). In addition, the empty DNA NG itself caused no cytotoxic effects under the tested concentrations. Due to the extremely low cisplatin crosslinker within DNA NG, the cytotoxicity of cisplatin could be negligible. Therefore, the final cytotoxicity could be mainly contributed to Dox in DNA/Dox NG. Although the drugs were encapsulated in the DNA NG, the biodegradability of DNA might be a beneficial factor for efficient intracellular drug release.
3. Conclusions
In summary, a facile one pot strategy to construct drug-loaded DNA NG, inspired by DNA as therapeutic target, has been demonstrated. In comparison to current prevalent methods of preparing DNA nanostructures, our unconventional strategy neither requires sequence design nor the assistance of enzyme reactions and its size could be easily tuned by varing the heating times. Similar to its action mechanism in cells, cisplatin could crosslink its DNA target and quickly form well-organized NGs within minutes. The scaffolding DNA segments in the current study contained a myriad of short DNA strands which may also be replaced by DNA with specific functions combined with specific sequence designs. Importantly, the contained drugs could be released efficiently in cells, without losing their therapeutic effects. Finally, the present strategy may be extended to the use of other molecules with actions similar to cisplatin since many drugs have been identified with ability to form interstrand crosslinks with DNA.
Supplementary Material
Acknowledgements:
This study was financially supported in part by NIH GM094880.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author
References
- [1].Chen Y-J, Groves B, Muscat RA, Seelig G, Nat. Nanotechnol 2015, 10, 748. [DOI] [PubMed] [Google Scholar]
- [2].Zhang F, Nangreave J, Liu Y, Yan H, J. Am. Chem. Soc 2014, 136, 11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li J, Fan C, Pei H, Shi J, Huang Q, Adv. Mater 2013, 25, 4386. [DOI] [PubMed] [Google Scholar]
- [4].Chao J, Liu H, Su S, Wang L, Huang W, Fan C, Small 2014, 10, 4626. [DOI] [PubMed] [Google Scholar]
- [5].Yang D, Hartman MR, Derrien TL, Hamada S, An D, Yancey KG, Cheng R, Ma M, Luo D, Acc. Chem. Res 2014, 47, 1902. [DOI] [PubMed] [Google Scholar]
- [6].Jiang D, England CG, Cai W, J. Controlled Release 2016, 239, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rothemund PWK, Nature 2006, 440, 297. [DOI] [PubMed] [Google Scholar]
- [8].Yin P, Hariadi RF, Sahu S, Choi HMT, Park SH, LaBean TH, Reif JH, Science 2008, 321, 824. [DOI] [PubMed] [Google Scholar]
- [9].Wei B, Dai M, Yin P, Nature 2012, 485, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ke Y, Ong LL, Shih WM, Yin P, Science 2012, 338, 1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Um SH, Lee JB, Park N, Kwon SY, Umbach CC, Luo D, Nat. Mater 2006, 5, 797. [DOI] [PubMed] [Google Scholar]
- [12].Zhang L, Zhu G, Mei L, Wu C, Qiu L, Cui C, Liu Y, Teng IT, Tan W, ACS Appl. Mater. Interfaces 2015, 7, 24069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sun W, Ji W, Hu Q, Yu J, Wang C, Qian C, Hochu G, Gu Z, Biomaterials 2016, 96, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun W, Jiang T, Lu Y, Reiff M, Mo R, Gu Z, J. Am. Chem. Soc 2014, 136, 14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guo X, Huang L, Acc. Chem. Res 2012, 45, 971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Conway JW, McLaughlin CK, Castor KJ, Sleiman H, Chem. Commun 2013, 49, 1172. [DOI] [PubMed] [Google Scholar]
- [17].Liang L, Li J, Li Q, Huang Q, Shi J, Yan H, Fan C, Angew. Chem., Int. Ed 2014, 53, 7745. [DOI] [PubMed] [Google Scholar]
- [18].Surana S, Bhat JM, Koushika SP, Krishnan Y, Nat. Commun 2011, 2, 340. [DOI] [PubMed] [Google Scholar]
- [19].Zhang Q, Jiang Q, Li N, Dai L, Liu Q, Song L, Wang J, Li Y, Tian J, Ding B, ACS nano 2014, 8, 6633. [DOI] [PubMed] [Google Scholar]
- [20].Kumar V, Palazzolo S, Bayda S, Corona G, Toffoli G, Rizzolio F, Theranostics 2016, 6, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tacar O, Sriamornsak P, Dass CR, J. Pharm. Pharmacol 2013, 65, 157. [DOI] [PubMed] [Google Scholar]
- [22].Zhu G, Niu G, Chen X, Bioconjugate Chem 2015, 26, 2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu J, Wei T, Zhao J, Huang Y, Deng H, Kumar A, Wang C, Liang Z, Ma X, Liang X-J, Biomaterials 2016, 91, 44. [DOI] [PubMed] [Google Scholar]
- [24].Hurley LH, Nat. Rev. Cancer 2002, 2, 188. [DOI] [PubMed] [Google Scholar]
- [25].Johnstone TC, Suntharalingam K, Lippard SJ, Chem. Rev 2016, 116, 3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kartalou M, Essigmann JM, Mutat. Res 2001, 478, 1. [DOI] [PubMed] [Google Scholar]
- [27].Fichtinger-Schepman AMJ, Van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J, Biochemistry 1985, 24, 707. [DOI] [PubMed] [Google Scholar]
- [28].Dautzenberg H, Zintchenko A, Konák C, Reschel T, Šubr V, Ulbrich K, Langmuir 2001, 17, 3096. [Google Scholar]
- [29].Li Y, Jiang Y, Yan X-P, Anal. Chem 2006, 78, 6115. [DOI] [PubMed] [Google Scholar]
- [30].Roh YH, Ruiz RCH, Peng S, Lee JB, Luo D, Chem. Soc. Rev 2011, 40, 5730. [DOI] [PubMed] [Google Scholar]
- [31].Liedert B, Pluim D, Schellens J, Thomale J, Nucleic Acids Res 2006, 34, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang W, Tung C-H, ACS Appl. Mater. Interfaces 2017, 15, 8547. [DOI] [PubMed] [Google Scholar]
- [33].Zhang W, Zhang Z, Tung C-H, Chem. Commun 2017, 53, 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang H, Zhu L, Reid BR, Drobny GP, Hopkins PB, Science 1995, 270, 1842. [DOI] [PubMed] [Google Scholar]
- [35].Wu C, Han D, Chen T, Peng L, Zhu G, You M, Qiu L, Sefah K, Zhang X, Tan W, J. Am. Chem. Soc 2013, 135, 18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Agudelo D, Bourassa P, Bérubé G, Tajmir-Riahi H-A, Int. J. Biol. Macromol 2014, 66, 144. [DOI] [PubMed] [Google Scholar]
- [37].Albanese A, Tang PS, Chan WCW, Annu. Rev. Biomed. Eng 2012, 14, 1. [DOI] [PubMed] [Google Scholar]
- [38].Lee DS, Qian H, Tay CY, Leong DT, Chem. Soc. rev 2016. 45, 4199. [DOI] [PubMed] [Google Scholar]
- [39].Shen X, Jiang Q, Wang J, Dai L, Zou G, Wang Z-G, Chen W-Q, Jiang W, Ding B, Chem. Commun 2012, 48, 11301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.