Abstract
The DNA helicase RECQL4 is known for its roles in DNA replication and repair. RECQL4 mutations cause several genetic disorders including Rothmund-Thomson syndrome (RTS), characterized by developmental defects and predisposition to osteosarcoma. Here we reprogrammed fibroblasts with a heterozygous RECQL4 mutation (c.1878 + 32_1878 + 55del24) to induced pluripotent stem cells (iPSCs). These iPSCs are pluripotent and are able to be differentiated into all three germ layers, providing a novel tool to further interrogate the role of RECQL4 DNA helicase in vitro.
| Resource Table. | |
|---|---|
| Unique stem cell line identifier | CDMLi002-A |
| Alternative name(s) of stem cell line | FCP351G |
| Institution | McGovern Medical School, The University of Texas Health Science Center Houston |
| Contact information of distributor | Dung-Fang Lee dung-fang.lee@uth.tmc.edu |
| Type of cell line | iPSCs |
| Origin | Human |
| Additional origin info | Age: 24 years Sex: F Ethnicity if known: not Hispanic, White |
| Ethnicity if known: not Hispanic, White | |
| Cell source | fibroblast |
| Clonality | clonal |
| Method of reprogramming | Sendai virus |
| Genetic modification | N/A |
| Type of modification | Germline mutation |
| Associated disease | Rothmund-Thomson syndrome |
| Gene/locus | RECQL4 (c.1878 + 32_1878 + 55del24)/ 8q24.3 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 3/31/2018 |
| Cell line repository/bank | N/A |
| Ethical approval | The Molecular Basis of Familial Cancer Predisposition Syndromes (IRB# H-7207) provide by Baylor College of Medicine |
Resource utility
We have generated an iPSC line with a heterozygous RECQL4 (c.1878 + 32_1878 + 55del24) mutation. This iPSC line will be useful to study the effects of aberrant RECQL4 protein on genomic stability.
Resource details
RECQL4, one of five human RecQ DNA helicases, is known as a guardian of the genome due to its function in multiple DNA metabolic processes, including DNA replication, recombination and repair (Van Maldergem et al., 2016; Lin et al., 2017). Compound heterozygous or homozygous mutations in the RECQL4 gene are responsible for the majority of cases of Rothmund-Thomson syndrome (RTS), an autosomal recessive disorder characterized by multiple clinical features including a poikilodermatous skin rash, small stature, skeletal dysplasias, and a striking risk for developing osteosarcoma (Van Maldergem et al., 2016; Lin et al., 2017). Here we have reprogrammed fibroblasts from an RTS heterozygous carrier of RECQL4 mutation c.1878 + 32_1878 + 55del24, a 24 base pair deletion in intron 11 of the RECQL4 gene (Fig. 1A). This mutation results in a shortened intron and has been shown to cause aberrant splicing leading to exon skipping (Wang et al., 2002; Colombo et al., 2018). The resource we describe here will be useful in characterizing the role of RECQL4 in DNA damage repair and replication and provides a valuable tool for investigating the pathogenesis of osteosarcoma.
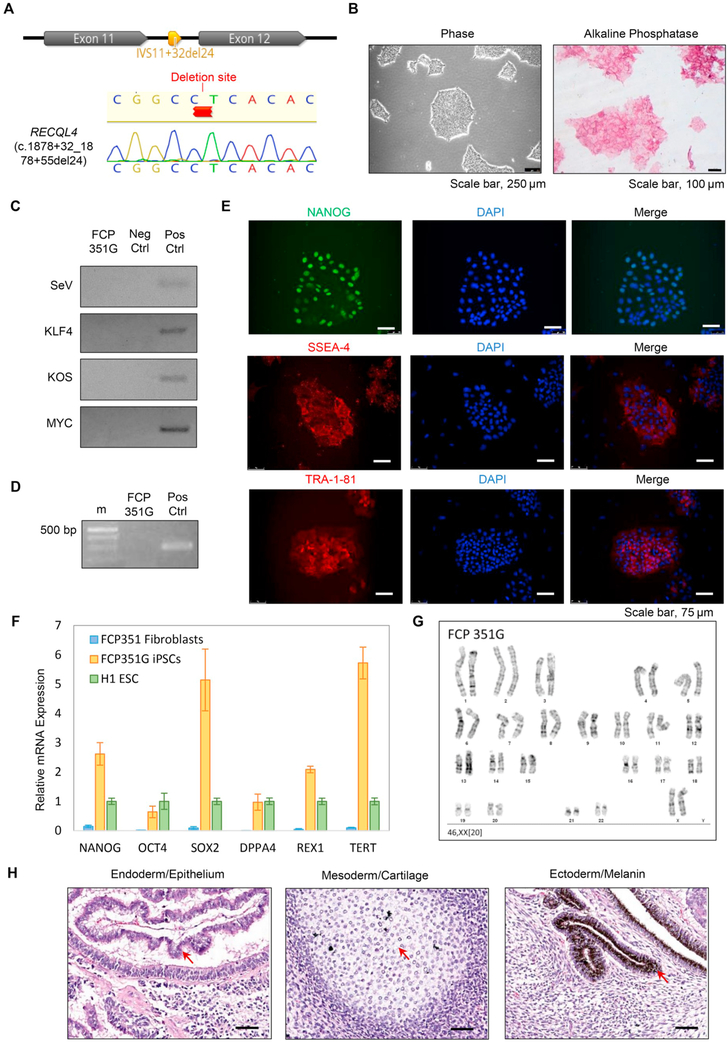
Fig. 1.
Generation and characterization of FCP351G iPSCs. (A) Sanger sequence of RECQL4 Exons 11-12 depicting the deletion found in FCP351 fibroblasts and verified by Sanger sequencing (see chromatogram) in FCP351G iPSCs. (B) Cell morphology (left panel) and alkaline phosphatase staining (right panel) of the FCP351G line. Scale bar, 250 μm (left) and 100 μm (right). (C) PCR detection of exogenous OCT4, SOX2, KLF4 and c-MYC in FCP351G iPSCs. (D) Mycoplasma PCR assay in FCP351G iPSCs.(E) Immunofluorescence staining of pluripotency factor NANOG and hESC surface markers (SSEA4 and TRA-1-81) in the FCP351G line. Scale bar, 75 μm. (F) qRT-PCR assay for the expression of endogenous pluripotency genes (NANOG, SOX2, OCT4, DPPA4, REX1, and TERT) in the FCP351G iPSCs. (G) A representative G-banded karyotype from iPSCs (FCP351G) showing 46, XX. (H) In vivo teratoma assay of the FCP351G iPSCs. Scale bar, 50 μm.
Fibroblasts were isolated from a punch biopsy taken from the arm of an individual with a heterozygous RECQL4 (c.1878 + 32_1878 + 55del24) mutation. Primary fibroblasts were reprogrammed by Yamanaka four factors OCT4, SOX2, KLF4 and MYC using CytoTune-iPS Sendai Reprogramming kit. The emerged iPS clones were picked and cultured for approximately 15 passages. The iPS clone FCP351G exhibits hESC morphology which is evident by cell clusters with a dense, domed center and well-defined borders (Fig. 1B, left panel, Scale bar, 250 μm). PCR detection of exogenous OCT4, SOX2, KLF4 and MYC in FCP351G iPSC line verified loss of Sendai virus and transgenes (Fig. 1C). PCR-based mycoplasma detection assay further validates FCP351G iPSCs are free of mycoplasma (Fig. 1D). The short tandem repeat (STR) profile of FCP351G iPSCs were identical to that of parental FCP351 fibroblasts (data available upon request). Immunofluorescence staining of FCP351G iPSCs demonstrated ubiquitous expression of pluripotency transcription factor NANOG, hESC surface markers SSEA-4 and TRA-1-81 (Fig. 1E, Scale bar, 75 μm) and alkaline phosphatase (Fig. 1B, right panel, Scale bar, 100 μ:m). Examination of pluripotent gene expression by qRT-PCR showed that FCP351G iPSCs express pluripotency gene NANOG, OCT4, SOX2, DPPA4, REX1 and TERT at a level equal to or greater than H1 hESCs, whereas FCP351 fibroblasts do not express these genes (Fig. 1F) Higher SOX2 and TERT mRNAs in FCP351G iPSCs may reult from RECQL4 mutation-induced gene up-regulation. G-band karyotype analysis confirmed the normal karyotype of the FCP351G iPSCs (Fig. 1G). Teratoma assay indicates that FCP351 iPSCs are capable of differentiation to three germ layers including epithelium (endoderm), cartilage (mesoderm), and melanin (ectoderm) (Fig. 1H), confirming their pluripotency. The characterization of the FCP351G iPSC line is summarized in Table 1.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 panel B |
| Phenotype | Qualitative analysis immunocytochemistry | Positive staining for NANOG, SSEA and TRA-1-81 | Fig. 1 panel E |
| Quantitative analysis RT-qPCR | Expression of pluripotency markers equivalent or greater than H1 embryonic stem cells | Fig. 1 panel F | |
| Genotype | Karyotype (G-banding) and resolution | 46; XX, Resolution 400 | Fig. 1 panel G |
| Identity | Microsatellite PCR (mPCR) OR | DNA Profiling not performed | N/A |
| STR analysis | 14 sites tested, all sites match | data available with authors | |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous | Fig. 1 panel A |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Negative | Fig. 1 panel D |
| Differentiation potential | Teratoma formation | Differentiation to all three germ layers confirmed by H&E Staining | Fig. 1 panel H |
| Donor screening (OPTIONAL) | N/A | N/A | N/A |
| Genotype additional info | N/A | N/A | N/A |
| (OPTIONAL) | N/A | N/A | N/A |
In summary, FCP351G iPSCs are pluripotent and demonstrate a normal karyotype. FCP351G iPSCs provide a valuable cell resource to study RECQL4 function.
Materials and methods
Cell culture
Fibroblasts were cultured in DMEM/F12 (Corning) supplemented with Glutamax (Thermo Fisher) and 10% FBS (GenDEPOT) at 37 °C with 5% CO2 after thawing until the cells were confluent. Cells were then passaged to six well plates, where they were grown to approximately 90% confluence. Cells were then reprogrammed with OCT4, SOX2, KLF4, and MYC using the CytoTune iPS 2.0 Sendai reprogramming kit (Life Technologies) according to manufacturer's instructions. Clones were picked and seeded in 96-well plates coated with Matrigel (Corning) using hESC medium supplemented with ROCK inhibitor Thiazovivin (Millipore). After one day of culture, the medium was replaced with StemMACS iPS-Brew XF media (Miltenyi Biotech). Following growth, these clones were passaged to a six well plate and cultured in the same manner for approximately 15 passages to ensure the removal of Sendai viruses.
PCR, sequencing, and STR
Genomic DNA was isolated from one 10 cm dish of 80% confluent iPSCs according to protocol using the PureLink DNA Isolation kit (Invitrogen). PCR was performed using primers listed in Table 2 and OneTaq polymerase (New England Biolabs). PCR reaction was performed according to the following protocol: 94 °C for 1 min; 35 cycles of reaction: 94 °C for 30 s, 56 °C for 45 s and 68 °C for 1 min; and 68 °C for 5 min on a Biometra TRIO Thermal Cycler (Analytik Jena). Sanger sequencing was done by Eton Bioscience (San Diego, California). STR DNA fingerprinting was done by the CCSG-funded Characterized Cell Line Core Facility at the University of Texas M.D. Anderson Cancer Center. The STRs at 14 loci (AMEL, CSF1PO, D13S317, D16S539, D18S51, D21S11, D3S1358, D5S818, D7S820, D8S1179, FGA, TH01, TPOX and vWA) were assessed in the FCP351G iPSC line and original FCP351 fibroblast.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Goat anti-NANOG | 1:500 | R and D Systems Cat# AF1997 RRID:AB_355097 |
| Pluripotency Markers | anti-SSEA4 PE-conjugated | 1:600 | R and D Systems FA1435P-025 |
| Pluripotency Markers | Mouse anti-TRA-1-85 Alexa Fluor 555-conjugated | 1:600 | R and D Systems Cat# FAB3195A RRID:AB_663789 |
| Secondary antibodies | Donkey Anti-Goat IgG (Alexa Fluor488 conjugate) | 1:500 | Jackson ImmunoResearch Labs Cat# 705–545-003 RRID:AB_2340428 |
| Primers | ||
|---|---|---|
|
| ||
| Target | Forward/reverse primer(5′-3′) | |
| PCR/Sequencing | RECQL4 Exons 11–12 | CAGATTCGGGCAGCCCAGGTAC ACCCACCTGGTCTGTGTCCCTG |
| Pluripotency Marker (qPCR) | NANOG | TTTGTGGGCCTGAAGAAAACT AGGGCTGTCCTGAATAAGCAG |
| Pluripotency Marker (qPCR) | OCT4 | AACCTGGAGTTTGTGCCAGGGTTT TGAACTTCACCTTCCCTCCAACCA |
| Pluripotency Marker (qPCR) | SOX2 | AGAAGAGGAGAGAGAAAGAAAGGGAGAGA GAGAGAGGCAAACTGGAATCAGGATCAAA |
| Pluripotency Marker (qPCR) | DPPA4 | GACCTCCACAGAGAAGTCGAG TGCCTTTTTCTTAGGGCAGAG |
| Pluripotency Marker (qPCR) | REX1 | GCCTTATGTGATGGCTATGTGT ACCCCTTATGACGCATTCTATGT |
| Pluripotency Marker (qPCR) | TERT | TGAAAGCCAAGAACGCAGGGATG TGTCGAGTCAGCTTGAGCAGGAATG |
| Housekeeping Genes (qPCR) | GAPDH | CCACTCCTCCACCTTTGAC ACCCTGTTGCTGTAGCCA |
| Sendai Virus Removal (RT-PCR) | SeV | GGATCACTAGGTGATATCGAGC ACCAGACAAGAGTTTAAGAGATATGTATC |
| Pluripotency Marker (RT-PCR) | KOS | ATGCACCGCTACGACGTGAGCGC ACCTTGACAATCCTGATGTGG |
| Pluripotency Marker (RT-PCR) | KLF4 | TTCCTGCATGCCAGAGGAGCCC AATGTATCGAAGGTGCTCAA |
| Pluripotency Marker (qPCR) | MYC | TAACTGACTAGCAGGCTTGTCG TCCACATACAGTCCTGGATGATGATG |
qRT-PCR
RNA was isolated from cells grown to 85% confluency in 6 well culture dish using Trizol (Life Technologies) according to manufacturer's instructions. 4 μg of RNA was used to make cDNA according to the reverse transcriptase protocol using iScript Reverse Transcription Supermix (BioRad). qPCR was then performed using primers described in Table 2 according to the protocol using iQ SYBR Green Supermix (Biorad) on a CFX96 machine (Bio-Rad). The reaction was performed according to the following parameters: 50 °C for 10 min, 95 °C for 5 min, 40 cycle of 95 °C for 10 s and 60 °C for 30 s, and 95 °C for 10 min. Results were calculated using the ΔΔCT method and were normalized to GAPDH expression and are displayed relative to hESC H1.
Mycoplasma testing
Mycoplasma detection was performed using PCR Mycoplasma Detection Kit (Applied Biological Materials Inc) according to the manufacturer's instructions.
Karyotyping
The cultures were harvested and G-banded using standard protocols. Clonal chromosomal changes identified by G-banding were described according to an International System for Human Cytogenetic Nomenclature ISCN (McGowan-Jordan & An, n.d.). Twenty metaphase chromosome spreads at 400 band resolution were analyzed.
Immunofluorescent staining
iPSCs were grown to 70% confluence in 12 well plates, then fixed with 4% paraformaldehyde (Affymatrix) for 10 min. Cells were then washed 2 × 500 μL in wash buffer (PBS with 1% BSA (Fisher Scientific), followed by blocking with PBS with 10% donkey serum (Jackson ImmunoResearch Labs), 1% BSA, 0.3% Triton X-100 (Sigma) for 45 min. Cells were then incubated with primary antibody diluted in dilution buffer (PBS with 1%BSA, 1% donkey serum, 0.3% Triton X-100, and 0.01% sodium azide (Sigma) at 4 °C overnight gently rocking. Cells were then washed twice. Where appropriate, secondary antibody was diluted in dilution buffer, then added to cells and incubated for 1 h at room temperature protected from light. This was followed by 2 more washes, then counterstain with DAPI diluted in dilution buffer. Cells were finally washed 2 times, then stored in PBS for visualization.
Alkaline phosphatase staining
iPSCs were grown to 70% confluence in 12 well plates, then fixed and stained using the Alkaline Phosphatase Staining Kit II (Stemgent) according to manufacturer's instructions.
Teratoma assay
Cells were grown in 10 cm dishes to 80% confluence. Cells were dissociated from the dish, then resuspended in phenol red-free Matrigel (Corning cat#354262). Immunodeficient NOD SCID mice (Charles River Laboratories) were injected subcutaneously with 2 × 107 cells in each flank and observed for palpable teratoma formation. After approximately 6 weeks, teratomas were excised then fixed in 10% formalin (Sigma). Histology was performed by Histowiz (Brooklyn, New York).
Acknowledgements
B.E.J. is supported by Tzu Chi scholarship award for excellence (The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences). R.Z. is supported by UTHealth Innovation for Cancer Prevention Research Training Program Pre-doctoral Fellowship (Cancer Prevention and Research Institute of Texas grant RP160015) and Wei Yu Family Endowed Scholarship (The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences). J.T. and Z.H. are supported by the Ke Lin Program of the First Affiliated Hospital of Sun Yat-sen University. D.W. is supported by State-sponsored Joint Ph.D. Program from China Scholarship Council (201606380093). M.-C.H. is supported by NIHR01 CA211615. L.L.W. was supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award, NIH-NICHD K08HD42136 and HD083092. D.-F.L. is the CPRIT scholar in Cancer Research and supported by NIH Pathway to Independence Award R00 CA181496 and CPRIT Award RR160019.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Colombo EA, Locatelli A, Cubells Sanchez L, Romeo S, Elcioglu NH, Maystadt I, Esteve Martinez A, Sironi A, Fontana L, Finelli P, Gervasini C, Pecile V, Larizza L, 2018. Rothmund-Thomson syndrome: insights from new patients on the genetic variability underpinning clinical presentation and cancer outcome. Int. J. Mol. Sci 19 (4). 10.3390/ijms19041103. Epub 2018/04/13. (PubMed PMID: 29642415; PMCID: PMC5979380). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Jewell BE, Gingold J, Lu L, Zhao R, Wang LL, Lee DF, 2017. Osteosarcoma: Molecular pathogenesis and iPSC modeling. Trends Mol. Med 23 (8), 737–755. 10.1016/j.molmed.2017.06.004. (PubMed PMID: 28735817; PMCID: PMC5558609). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan-Jordan JSA, An Schmid M., 2016. International System for Human Cytogenetic Nomenclature (ISCN). [Google Scholar]
- Van Maldergem L, Larizza L, Wang LL, 2016. RECQL4-related recessive conditions. In: Epstein's Inborn Errors of Development. 3. Oxford University Press, Oxford, England, pp. 1137–1144. [Google Scholar]
- Wang LL, Worley K, Gannavarapu A, Chintagumpala MM, Levy ML, Plon SE, 2002. Intron-size constraint as a mutational mechanism in Rothmund-Thomson syndrome. Am. J. Hum. Genet 71 (1), 165–167. Epub 2002/05/23. 10.1086/341234 (PubMed PMID: 12016592; PMCID: PMC384974). [DOI] [PMC free article] [PubMed] [Google Scholar]