Abstract
Circular RNA UVRAG (circUVRAG), a type of non‐coding RNA, is derived and cyclized by part of the exon from the UVRAG gene. However, the role of circUVRAG in bladder cancer (BLCA) has not been reported. The purpose of the present study was therefore to characterize the role of circUVRAG in BLCA. Bioinformatics analysis showed interactive relationships among circUVRAG, microRNA‐223 (miR‐223), and fibroblast growth factor receptor 2 (FGFR2). Quantitative real‐time PCR was used to detect the expression of circUVRAG in BLCA cell lines. UM‐UC‐3 cells were stably transfected with siRNA against circUVRAG, and cell proliferation and migration ability were tested using the CCK8 assay, clone formation, and Transwell assays in vitro. Tumor xenograft formation and metastasis were determined using nude mice. Fluorescence in situ hybridization was used to confirm the subcellular localization of circUVRAG, and the luciferase reporter assay was used to confirm the relationships among circUVRAG, miR‐223, and FGFR2. Results showed that circUVRAG was upregulated in BLCA cell lines. Downregulation of circUVRAG expression suppressed proliferation and metastasis both in vitro and in vivo. Downregulation of circUVRAG suppressed FGFR2 expression by “sponging” miR‐223, which was confirmed by rescue experiments and luciferase reporter assay. Overall, the results showed that downregulation of circUVRAG suppressed the aggressive biological phenotype of BLCA. Taken together, silencing circular RNA UVRAG inhibited bladder cancer growth and metastasis by targeting the miR‐223/FGFR2 axis, which may provide a potential biomarker and therapeutic target for the management of BLCA.
Keywords: bladder cancer, circular RNA UVRAG, FGFR2, metastasis, miR‐223
Abbreviations
- BLCA
bladder cancer
- circUVRAG
circular RNA UVRAG
- FGFR2
fibroblast growth factor receptor 2
- miR‐223
micro RNA‐223
- miRNA
micro RNA
- sicircUVRAG
small interfering circular RNA UVRAG
1. INTRODUCTION
Bladder cancer is a common malignant tumor of the urinary system. The 5‐year prevalence of this disorder is increasing, ranking ninth in cancer incidence worldwide and 13th as a cause of death by cancer.1, 2 Most chemotherapy clinical trials for advanced bladder carcinoma have shown poor prognoses.3 To develop an effective treatment, it is therefore necessary to first identify the potential molecular targets and novel pathways underlying tumorigenesis and development of bladder cancer.
Circular RNA (circRNA) is a novel class of competitive endogenous RNAs formed by a covalently closed loop.4, 5 Increasing evidence has shown that circRNAs are effective diagnostic biomarkers for some cancers.6, 7 Previous studies reported that abnormal expressions of circRNAs were useful to determine the progress of BLCA, and high throughput sequencing found that hsa_circ_0023642 (circUVRAG) was significantly increased in BLCA.8 However, the role of circUVRAG in the progression of BLCA is still unclear. Importantly, it has been reported that circRNAs could be targeted by endogenous miRNAs, to suppress miRNA activity, acting as miRNA sponges,9 but the regulatory roles of circUVRAG, acting as “miRNA sponges” in BLCA, are still largely unknown.
The present study sought to examine the expression of circUVRAG in BLCA, and then assess the biological roles of circUVRAG. Our results showed that circUVRAG was upregulated in BLCA, and its upregulation remarkably promoted cell proliferation and migration. Our data provide novel evidence that could form the basis for the development of novel therapeutic strategies against bladder cancer.
2. MATERIALS AND METHODS
2.1. Animal ethics statement
Four‐week‐old BALB/c nude mice (n = 20) weighing 15‐20 g (SLARC, Shanghai, China) were used in this study. All animal experiments were approved by The Ethics Committee of Huashan Hospital, Fudan University, Shanghai, China.
2.2. Cell culture and transfection
SV‐HUC and BC cell lines (T24, EJ, J82, UM‐UC‐3, TCC, and RT‐4) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured at 37°C in 5% CO2 using DMEM medium (Gibco, Gaithersburg, MD, USA) containing 10% FBS (Gibco). sicircUVRAG, miR‐223 mimics, miR‐223 inhibitors, FGFR2 overexpression vector, and their negative controls were transfected into cultured UM‐UC‐3 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. In order to further confirm the effect of circUVRAG in the in vivo experiments, lentiviral stabilized circUVRAG silenced UM‐UC‐3 cells were constructed.
2.3. Fluorescence in situ hybridization
Specific probes to the circUVRAG sequence were used for in situ hybridization as previously reported.10 In brief, FITC‐labelled probes were specific to circUVRAG. DAPI was used for cell nucleus counterstain. All procedures were conducted according to the manufacturer's protocol (Genepharma, Shanghai, China).
2.4. Bioinformatics analysis
The circRNA/miRNA target gene was predicted using the website at https://circinteractome.nia.nih.gov/. The interactive relationship between miR‐223 and FGFR2 was predicted using the website at http://www.targetscan.org/; the expression level of UVRAG and the prognoses in bladder cancer patients was predicted using the website at http://gepia.cancer-pku.cn/.
2.5. Cell proliferation assay and cloning formation assay
Cell Counting Kit‐8 assay was used to detect cell proliferation. Transfected cells were seeded into 96‐well plates at a density of 2000 cells/well in triplicate wells. Cell viability was measured by the CCK‐8 system (Gibco) at 0, 24, 48, 72, and 96 hours after seeding, according to the manufacturer's instructions.
For the colony formation assay, transfected cells were seeded into six‐well plates at a density of 2000 cells/well and maintained in DMEM medium containing 10% FBS for 10 days. The colonies were imaged and counted after they were fixed and stained.
2.6. Western blots
Protein from cells and tumor tissues were extracted and concentrated using RIPA lysis buffer containing protease inhibitors (Sigma‐Aldrich, St Louis, MO, USA) and the BCA Protein Assay kit (Vigorous Biotechnology Beijing, Beijing, China), respectively. Protein (20 μg) was resolved by SDS‐PAGE, then transferred onto nitrocellulose membranes (Millipore, Madison, WI, USA). Membranes were blocked with 5% nonfat dry milk for 2 hours before being incubated with primary antibodies at 4°C overnight. GAPDH was used as the internal control. Membranes were then incubated with HRP‐coupled secondary antibodies for 1 hour at room temperature.
2.7. RNA extraction and quantitative RT‐PCR
RNA extraction was carried out using TRIzol reagent (Invitrogen) as previously reported.4 cDNA was obtained using a pTRUEscript 1st Strand cDNA Synthesis Kit (Aidlab, Beijing, China). qRT‐PCR was carried out using the 2× SYBR Green qPCR Mix (Aidlab) on an ABI 7900HT sequence detection machine (Thermo Fisher Scientific, Waltham, MA, USA). Expression fold changes were determined using the 2−ΔΔCT method.
2.8. Migration assay
Cell migration was measured using 24‐well Transwell chambers (8 μm pore membrane; BD Biosciences, Franklin Lakes, NJ, USA). A total of 1 × 105 cells were plated into the upper chamber with 200 μL serum‐free medium while the bottom chamber was filled with 500 μL completed medium. After culturing for 24 hours, cells in the bottom chamber were fixed with 4% paraformaldehyde for 30 minutes, then stained with crystal violet for 10 minutes.
2.9. Tumor xenograft formation and metastasis assays
A total of 2 × 107 viable cells from WT or sicircUVRAG UM‐UC‐3 cells were injected into the right flanks of nude mice as in our previous study.11 Tumor sizes were measured every 5 days using a Vernier caliper, and the volume was calculated using the following formula: volume = 1/2 × length × width2. The mice were then killed for qRT‐PCR analyses 30 days after implantation.
For analysis of metastasis, UM‐UC‐3 cells were transfected with luciferase expression vectors into both WT and circUVRAG silenced cells (2 × 105), the cells were then i.v. injected into the tails of mice. After 30 days, metastasis of UM‐UC‐3 cells was analyzed by bioluminescence imaging with i.v. injection of luciferin (150 mg luciferin/kg body weight) into the mice tails.
2.10. Dual luciferase reporter assay
Reporter plasmids were obtained by inserting circUVRAG or the FGFR2 3′‐UTR sequence into the pmirGLO vector (Promega, Madison, WI, USA). For the luciferase assay, miR‐223 mimics and reporter plasmids were cotransfected into 239T cells using Lipofectamine 2000. After culturing for 48 hours, firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, Sunnyvale, CA, USA) according to the manufacturer's instructions.
2.11. Statistical analysis
Data are presented as mean ± standard deviation (SD). GraphPad Prism software, version 5.0 (GraphPad, La Jolla, CA, USA) was used to compare group differences. P ≤ 0.05 indicated a statistically significant difference.
3. RESULTS
3.1. Expression of circUVRAG was increased in BLCA cell lines
Previous studies using high‐throughput microarray assays have verified that hsa_circ_0023642 (circUVRAG) was significantly increased in BLCA when compared with normal control samples.8 The aim of the present study was therefore to confirm whether circUVRAG plays a role in the progress of BLCA. Hsa_circ_0023642 was derived and cyclized by part of the exon from the UVRAG gene. UVRAG is a UV radiation resistance‐associated gene, and its expression increases sensitivity to chemotherapy by promoting autophagy.12, 13 Bioinformatics analysis showed that high expression of UVRAG predicted a poor prognosis in bladder cancer patients (Figure 1A). We characterized the expression in BLCA cell lines and found that the expression of circUVRAG was increased in six cell lines (EJ, T24, J82, UM‐UC‐3,TCC, and RT‐4) when compared with SV‐HUC cells (normal urothelial cells). Results showed that the expression of circUVRAG in UM‐UC‐3 had the highest expression when compared with other BLCA cell lines (Figure 1B). We therefore selected UM‐UC‐3 cells to study the effect of circUVRAG in subsequent experiments. FISH assay showed that circUVRAG was predominately localized in the cytoplasm (Figure 1C). Taken together, the results suggested that circUVRAG played a role in the progression of BLCA.
Figure 1.
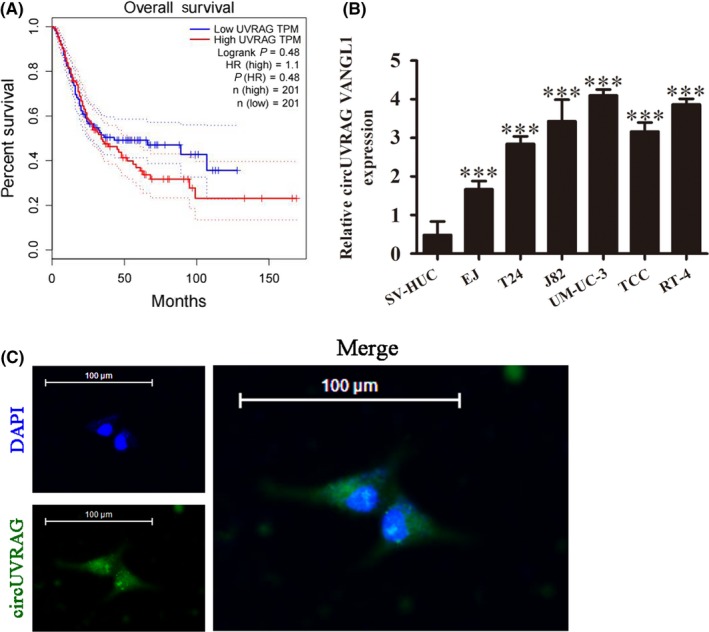
Expression of circular RNA UVRAG (circUVRAG) was increased in bladder cancer (BLCA) cell lines. A, Gepia website analysis shows survival and comparison of UVRAG high‐ and low‐expressing patients in the BLCA cohort. B, Quantitative RT‐PCR assay shows circUVRAG expression in BLCA cell lines (EJ, T24, J82, UM‐UC‐3, TCC, and RT‐4) and normal urothelial cells (SV‐HUC cells). Data are presented as mean ± SD. ***P < .001 vs SV‐HUC cells. C, FISH was carried out to determine the subcellular localization of circUVRAG. DAPI, nuclear staining (top, left); circUVRAG, green fluorescent‐tagged circUVRAG (bottom, left). Merged images shown at right
3.2. Downregulation of circUVRAG suppressed tumor formation and metastasis in nude mice xenografts
To further confirm that circUVRAG was involved in the progression of BLCA, lentiviral‐stabilized circUVRAG silenced UM‐UC‐3 cells or a negative control (NC) were used for tumor formation. Results showed that the expression of circUVRAG was significantly decreased after silencing of circUVRAG (Figure 2A). An in vivo experiment showed that downregulation of circUVRAG suppressed tumor volume and weight when compared with the NC group (Figure 2B‐D). Live imaging experiments showed that downregulation of circUVRAG suppressed metastasis of UM‐UC‐3 cells 30 days after i.v. tail injection (Figure 2E). qRT‐PCR assays showed that the expression of miR‐223 was increased after knockdown of circUVRAG in tumor tissues (Figure 2F). Western blot analyses showed that silencing of circUVRAG suppressed FGFR2 expression (Figure 2G,H). Taken together, the results suggested that downregulation of circUVRAG suppressed tumor formation and metastasis in nude mice xenografts. We also found that FGFR2 and miR‐223 were involved in the regulation of BLCA.
Figure 2.
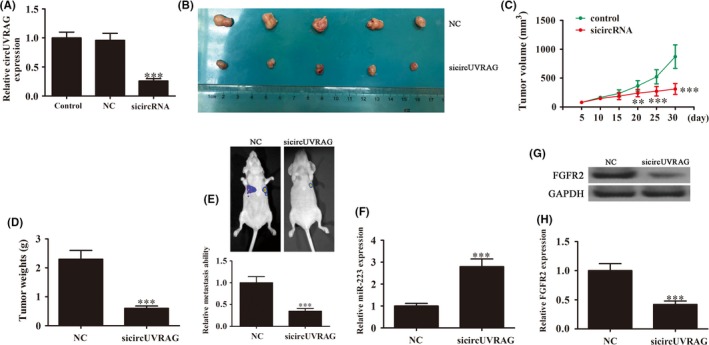
Downregulation of circular RNA UVRAG (circUVRAG) suppressed tumor formation and metastasis in nude mice xenografts. A, Quantitative RT‐PCR (qRT‐PCR) assay shows the expression of circUVRAG in adenovirus‐transfected cells and in circ‐control transfected cells. B, Representative photographs of tumor formation in xenografts of nude mice (n = 5). C, Summary of tumor volumes in mice that were measured every week. Data are presented as mean ± SD. **P < .01, ***P < .001 vs control. D, Tumor weights were measured after 30 days of injection. Data are presented as mean ± SD. ***P < .001 vs control. E, Live imaging shows the effects of circUVRAG on metastasis of UM‐UC‐3 cells 30 days after i.v. tail injection. Relative metastasis abilities were calculated. Data are presented as mean ± SD. ***P < .001 vs control. F, qRT‐PCR assay showing the expression of microRNA‐223 (miR‐223). Data are presented as mean ± SD. ***P < .001 vs control. G,H, Western blot analysis of the expression of fibroblast growth factor receptor 2 (FGFR2) in tumor tissues. Data are presented as mean ± SD. ***P < .001 vs control. NC, normal control; sicircUVRAG, small interfering circular RNA UVRAG
3.3. Silencing circUVRAG inhibited BLCA cell proliferation and migration by regulating the miR‐223/FGFR2 axis in vitro
To further confirm the relationship among circUVRAG, FGFR2, and miR‐223, UM‐UC‐3 cells were transfected with siRNA against circUVRAG, combined with or without an miR‐223 inhibitor and FGFR2 overexpression vector. qRT‐PCR assays showed that silenced circUVRAG promoted miR‐223 expression, but a miR‐223‐specific inhibitor significantly suppressed miR‐223 expression. However, FGFR2 expression had no effect on circUVRAG silence‐induced miR‐223 expression (Figure 3A). Western blot analyses showed that silenced circUVRAG suppressed FGFR2 expression, but an miR‐223‐specific inhibitor reversed the inhibition effect after circUVRAG silencing. After transfection with a FGFR2 overexpression vector, expression of FGFR2 significantly increased (Figure 3B). CCK‐8 and cloning formation assays showed that circUVRAG knockdown suppressed the proliferation of UM‐UC‐3 cells, but treatment with the miR‐223 inhibitor rescued the suppression effect of circUVRAG silencing. FGFR2 overexpression further promoted proliferation of UM‐UC‐3 cells (Figure 3C,D). To determine the effect of circUVRAG, miR‐223, and FGFR2 on metastasis, Transwell migration assays were carried out. Results showed that miR‐223 inhibitor treatment rescued circUVRAG silencing‐induced migration inhibition. FGFR2 overexpression promoted migration of UM‐UC‐3 cells (Figure 3E). Taken together, the results suggested that downregulation of circUVRAG suppressed tumor cell proliferation and metastasis by promoting miR‐223 expression but suppressing FGFR2 expression.
Figure 3.
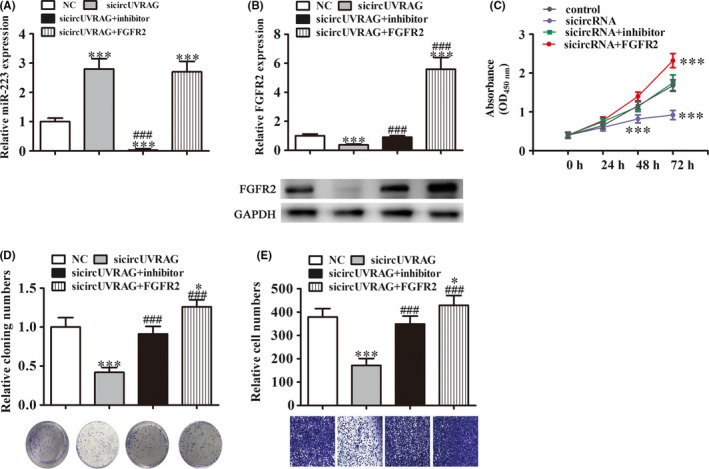
Silencing circular RNA UVRAG (circUVRAG) inhibits bladder cancer (BLCA) cell proliferation and migration by regulation of microRNA‐223 (miR‐223)/fibroblast growth factor 2 (FGFR2) axis in vitro. A, Quantitative RT‐PCR assay shows miR‐223 expression after transfection with siRNA against circUVRAG combined with or without miR‐223 inhibitor and the FGFR2 overexpression vector. Data are presented as mean ± SD. ***P < .001 vs NC. ### P < .001 vs sicircRNA. B, Western blots show FGFR2 expression. Relative protein levels were analyzed and data are presented as mean ± SD. ***P < .001 vs NC. ### P < .001 vs sicircRNA. C, CCK‐8 assays were carried out to assess cell proliferation. Data are presented as mean ± SD. ***P < .001 vs NC. D, Cloning formation assay showing cell proliferation of UM‐UC‐3. Data are presented as mean ± SD. *P < .05, ***P < .001 vs NC. ### P < .001 vs sicircRNA. E, Cell migration was determined in UM‐UC‐3 cells using Transwell assays. Data are presented as mean ± SD. *P < .05, ***P < .001 vs NC. ### P < .001 vs sicircRNA. GADPH (load control); NC, normal control; sicircUVRAG, small interfering circular RNA UVRAG
3.4. Overexpression of FGFR2 reversed miR‐223‐induced UM‐UC‐3 cell growth and migration inhibition in vitro
To further confirm the relationship between FGFR2 and miR‐223, UM‐UC‐3 cells were transfected with miR‐223 mimics combined with or without a FGFR2 overexpression vector. qRT‐PCR assays showed that the expression of miR‐223 was significantly increased after transfection with the miR‐223 mimic, but FGFR2 overexpression had no effect on miR‐223 expression (Figure 4A). Western blot analysis showed that miR‐223 expression suppressed FGFR2 expression, but FGFR2 overexpression rescued FGFR2 expression, even after transfection with the miR‐223 mimic (Figure 4B,C). CCK‐8 and cloning formation assays showed that miR‐223 overexpression suppressed the proliferation of UM‐UC‐3 cells, but FGFR2 overexpression rescued the suppressed effect of miR‐223 (Figure 4D‐F). To determine the effect of miR‐223 and FGFR2 on metastasis, Transwell migration assays showed that miR‐223 overexpression suppressed cell migration. FGFR2 overexpression promoted migration of UM‐UC‐3 cells (Figure 4G,H). Taken together, the results suggested that miR‐223 expression suppressed tumor cell proliferation and metastasis by suppressing FGFR2 expression.
Figure 4.
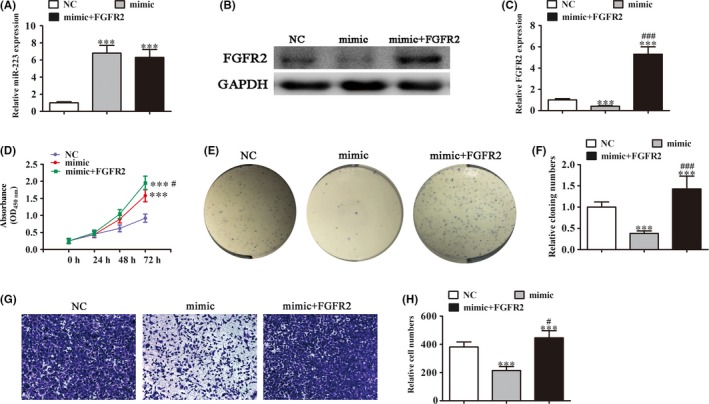
Overexpression of fibroblast growth factor receptor 2 (FGFR2) reversed microRNA‐223 (miR‐223)‐induced cell growth and migration inhibition in vitro. UM‐UC‐3 cells were transfected with miR‐223 mimics combined with or without the FGFR2 overexpression vector. A, Quantitative RT‐PCR assay showing the expression of miR‐223. Data are presented as mean ± SD. ***P < .001 vs NC. B,C, Western blot showing FGFR2 expression. Relative protein levels (C) were analyzed and data are presented as mean ± SD. ***P < .001 vs NC. ### P < .001 vs mimic. D, CCK‐8 assays were carried out to assess cell proliferation. Data are presented as mean ± SD. ***P < .001. # P < .05 vs mimic. E,F, Cloning formation assay showing cell proliferation of UM‐UC‐3. Data are presented as mean ± SD. ***P < .001. ### P < .001 vs mimic. G,H, Cell migration and invasion were determined in UM‐UC‐3 cells using Transwell assays. Data are presented as mean ± SD. ***P < .001 vs NC. # P < .05 vs mimic. NC, normal control
3.5. Interactive relationships among miR‐223, circUVRAG, and FGFR2
To confirm the relationships among miR‐223, circUVRAG, and FGFR2, bioinformatics analyses showed that circUVRAG targeted miR‐223. To further determine whether miR‐223 was a possible target of circUVRAG, a luciferase reporter analysis was used to show that miR‐223 was a downstream binding target of circUVRAG (Figure 5A). Taken together, the results showed that circUVRAG inhibited luciferase activity in WT cells but did not affect the activity in mutated cell lines (Figure 5B).
Figure 5.
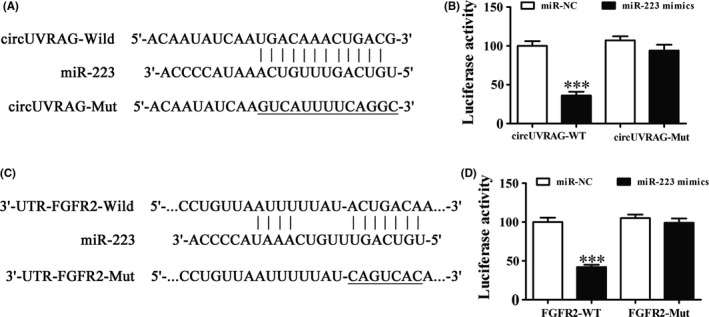
Interactive relationships among microRNA‐223 (miR‐223), circUVRAG, and fibroblast growth factor receptor 2 (FGFR2). A, Predicted binding sites of miR‐223 in circUVRAG. Mutated (Mut) version of circUVRAG is also shown. B, Relative luciferase activity was determined 48 h after transfection with miR‐223 mimic/normal control (NC) or with circUVRAG WT/Mut in HEK293T cells. Data are presented as mean ± SD. ***P < .001. C, Predicted binding sites of miR‐223 with the 3′‐UTR of FGFR2. Mutated version of the 3′‐UTR‐FGFR2 is also shown. D, Relative luciferase activity was determined 48 h after transfection with miR‐223 mimic/NC or with the 3′‐UTR‐FGFR2 WT/Mut in HEK293T cells. Data are presented as mean ± SD. ***P < .001
To further identify whether FGFR2 was a possible target of miR‐223, bioinformatics analysis was used to confirm that miR‐222 directed interactions with the 3′‐UTR of FGFR2 and suppressed FGFR2 expression at the mRNA level (Figure 5C). Luciferase reporter analysis found that miR‐223 inhibited luciferase activity in WT cells, but not in mutated cell lines (Figure 5D). The results suggested that silencing circUVRAG inhibited bladder cancer metastasis and growth by targeting the miR‐223/FGFR2 axis.
4. DISCUSSION
Studies have shown that the expression profiles of non‐coding RNAs, including miRNAs and long non‐coding RNAs (lncRNAs), are abnormal in many types of cancers, and these studies have focused on epigenetic regulation in cancer development.14, 15 Recent studies have reported that many miRNAs and some lncRNAs may serve as regulators in BLCA development.16, 17 However, whether circRNAs play a role in BLCA is unknown. In this study, we found that circUVRAG was abnormally expressed in BLCA cell lines. Downregulation of circUVRAG suppressed cell proliferation and migration in both in vivo and in vitro experiments. Recent studies have reported that UVRAG not only promotes Beclin‐1‐mediated autophagy, but it also facilitates endosome/autophagosome maturation.18, 19 Activation of autophagy can increase chemo‐ and radiotherapy resistance in bladder cancer.20, 21, 22 We also found that high expression of UVRAG predicted poor prognosis, which was consistent with the role of circUVRAG.
Results also showed that miR‐223 and FGFR2 were involved in the regulation of BLCA cell proliferation and migration. Previous studies have reported that miR‐223 acts as a suppressor and as a potential therapeutic target for glioblastoma,23 colorectal cancer,24 lung cancer,25 colon adenocarcinoma,26 and bladder cancer.27 In the present study, we also found that downregulation of circUVRAG promoted miR‐223 expression. Bioinformatics analysis found that circUVRAG interacted with several miRNAs including miR‐507, miR‐508, and miR‐653, but miR‐223 has more conservative binding sites. Suppressed miR‐223 reversed circUVRAG silencing‐induced BLCA cell proliferation and migration inhibition, suggesting that circUVRAG acted as a “miRNA sponge,” which interacted with miR‐223 and suppressed the activation of miR‐223. Luciferase reporter analysis confirmed that miR‐223 was the target of circUVRAG.
Our data also showed that miR‐223 interacted with the 3′‐UTR of FGFR and suppressed FGFR2 expression on a post‐transcriptional level. Overexpression of FGFR2 reversed miR‐223‐induced BLCA cell proliferation and migration inhibition. The FGFR2 gene at human chromosome 10q26 encodes FGFR2b and FGFR2c isoforms functioning as FGF receptors with distinct expression domains and ligand specificities. FGFR2 plays oncogenic and anti‐oncogenic roles in a context‐dependent way.28, 29 Suppression of FGFR2 shows potent antitumor activity in bladder cancer cells in both in vitro and in vivo studies.30 Taken together, these findings implicated a remarkable tumor suppressor role of the miR‐223/FGFR2 pathway in BLCA.
Collectively, our study showed for the first time that silencing circUVRAG suppressed the proliferation and metastasis abilities of BLCA cells by targeting the miR‐223/FGFR2 axis, which indicated that circUVRAG could be used as a potential biomarker for the prognosis of BLCA.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (81872102, 81802569, 81803900, 81872102, 81873853, 81874123) and Shanghai Science and Technology Commission Project (16140903600), Grant from Shanghai Healthcare Development Center (SHDC2017X10).
Yang C, Wu S, Wu X, Zhou X, Jin S, Jiang H. Silencing circular RNA UVRAG inhibits bladder cancer growth and metastasis by targeting the microRNA‐223/fibroblast growth factor receptor 2 axis. Cancer Sci. 2019;110:99–106. 10.1111/cas.13857
REFERENCES
- 1. Salim EI, Moore MA, Bener A, Habib OS, Seif‐Eldin IA, Sobue T. Cancer epidemiology in South‐West Asia ‐ past, present and future. Asian Pac J Cancer Prev. 2010;11(Suppl 2):33‐48. [PubMed] [Google Scholar]
- 2. Soerjomataram I, Lortet‐Tieulent J, Parkin DM, et al. Global burden of cancer in 2008: a systematic analysis of disability‐adjusted life‐years in 12 world regions. Lancet. 2012;380:1840‐1850. [DOI] [PubMed] [Google Scholar]
- 3. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10:396‐404. [DOI] [PubMed] [Google Scholar]
- 4. Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR‐145‐5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Y, Zeng X, He J, et al. Circular RNA as a biomarker for cancer: a systematic meta‐analysis. Oncol Lett. 2018;16:4078‐4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Z, Xie Q, He D, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bian A, Wang Y, Liu J, et al. Circular RNA Complement Factor H (CFH) Promotes Glioma Progression by Sponging miR‐149 and Regulating AKT1. Med Sci Monit. 2018;24:5704‐5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25‐miR‐103a‐3p/miR‐107‐CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li P, Yang X, Yuan W, et al. CircRNA‐Cdr1as exerts anti‐oncogenic functions in bladder cancer by sponging MicroRNA‐135a. Cell Physiol Biochem. 2018;46:1606‐1616. [DOI] [PubMed] [Google Scholar]
- 11. Yang D, Du G, Xu A, Xi X, Li D. Expression of miR‐149‐3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res. 2017;7:2209‐2219. [PMC free article] [PubMed] [Google Scholar]
- 12. Jo YK, Park NY, Shin JH, et al. Up‐regulation of UVRAG by HDAC1 inhibition attenuates 5FU‐induced Cell death in HCT116 colorectal cancer cells. Anticancer Res. 2018;38:271‐277. [DOI] [PubMed] [Google Scholar]
- 13. Wu S, He Y, Qiu X, et al. Targeting the potent Beclin 1‐UVRAG coiled‐coil interaction with designed peptides enhances autophagy and endolysosomal trafficking. Proc Natl Acad Sci U S A. 2018;115:E5669‐E5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mousa SA, Glinsky GV, Lin HY, et al. Contributions of thyroid hormone to cancer metastasis. Biomedicines. 2018;6:89‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu X, Qiu Z, Zeng J, Xiao T, Ke Z, Lyu H. A novel long non‐coding RNA, AC012456.4, as a valuable and independent prognostic biomarker of survival in oral squamous cell carcinoma. PeerJ. 2018;6:e5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kutwin P, Konecki T, Borkowska EM, Traczyk‐Borszynska M, Jablonowski Z. Urine miRNA as a potential biomarker for bladder cancer detection ‐ a meta‐analysis. Cent European J Urol. 2018;71:177‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie D, Zhang H, Shang C. Long non‐coding RNA CDKN2B antisense RNA 1 gene inhibits Gemcitabine sensitivity in bladder urothelial carcinoma. J Cancer. 2018;9:2160‐2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3‐kinase class III sub‐complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF‐1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010;316:3368‐3378. [DOI] [PubMed] [Google Scholar]
- 19. Takahashi Y, Coppola D, Matsushita N, et al. Bif‐1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mari A, D'Andrea D, Abufaraj M, Foerster B, Kimura S, Shariat SF. Genetic determinants for chemo‐ and radiotherapy resistance in bladder cancer. Transl Androl Urol. 2017;6:1081‐1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang X, Yin H, Zhang Y, et al. Hypoxia‐induced autophagy promotes gemcitabine resistance in human bladder cancer cells through hypoxia‐inducible factor 1alpha activation. Int J Oncol. 2018;53:215‐224. [DOI] [PubMed] [Google Scholar]
- 22. Lin JF, Lin YC, Tsai TF, Chen HE, Chou KY, Hwang TI. Cisplatin induces protective autophagy through activation of BECN1 in human bladder cancer cells. Drug Des Devel Ther. 2017;11:1517‐1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ding Q, Shen L, Nie X, et al. MiR‐223‐3p overexpression inhibits cell proliferation and migration by regulating inflammation‐associated cytokines in glioblastomas. Pathol Res Pract. 2018;214:1330‐1339. [DOI] [PubMed] [Google Scholar]
- 24. Ju H, Tan JY, Cao B, Song MQ, Tian ZB. Effects of miR‐223 on colorectal cancer cell proliferation and apoptosis through regulating FoxO3a/BIM. Eur Rev Med Pharmacol Sci. 2018;22:3771‐3778. [DOI] [PubMed] [Google Scholar]
- 25. Liu C, Yang Z, Deng Z, et al. Upregulated lncRNA ADAMTS9‐AS2 suppresses progression of lung cancer through inhibition of miR‐223‐3p and promotion of TGFBR3. IUBMB Life. 2018;70:536‐546. [DOI] [PubMed] [Google Scholar]
- 26. Wei LJ, Li JA, Bai DM, Song Y. miR‐223‐RhoB signaling pathway regulates the proliferation and apoptosis of colon adenocarcinoma. Chem Biol Interact. 2018;289:9‐14. [DOI] [PubMed] [Google Scholar]
- 27. Sugawara S, Yamada Y, Arai T, et al. Dual strands of the miR‐223 duplex (miR‐223‐5p and miR‐223‐3p) inhibit cancer cell aggressiveness: targeted genes are involved in bladder cancer pathogenesis. J Hum Genet. 2018;63:657‐668. [DOI] [PubMed] [Google Scholar]
- 28. Katoh Y, Katoh M. FGFR2‐related pathogenesis and FGFR2‐targeted therapeutics (Review). Int J Mol Med. 2009;23:307‐311. [DOI] [PubMed] [Google Scholar]
- 29. Wu YM, Su F, Kalyana‐Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen CH, Liu YM, Pan SL, Liu YR, Liou JP, Yen Y. Trichlorobenzene‐substituted azaaryl compounds as novel FGFR inhibitors exhibiting potent antitumor activity in bladder cancer cells in vitro and in vivo. Oncotarget. 2016;7:26374‐26387. [DOI] [PMC free article] [PubMed] [Google Scholar]


