Abstract
Next generation sequencing (NGS) has been an invaluable tool to put genomic sequencing into clinical practice. The incorporation of clinically relevant target sequences into NGS‐based gene panel tests has generated practical diagnostic tools that enable individualized cancer‐patient care. The clinical utility of gene panel testing includes investigation of the genetic basis for an individual's response to therapy, such as signaling pathways associated with a response to specific therapies, microsatellite instability and a hypermutated phenotype, and deficiency in the DNA double‐strand break repair pathway. In this review, we describe the concept of precision cancer medicine using target sequences in gene panel tests as well as the importance of the control of sample quality in routine NGS‐based genomic testing. We describe geographic and ethnic differences in cancer genomes, and discuss issues that need to be addressed in the future based on our experiences in Japan.
Keywords: DNA double‐strand break repair pathway, gene panel test, hypermutation, next generation sequencing, precision cancer medicine
1. INTRODUCTION
Large‐scale genomic studies, including The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) and the International Cancer Genome Consortium (ICGC; http://www.icgc.org/), have identified major driver‐gene mutations in various types of solid tumors,1, 2, 3 which have led to new therapeutic strategies for precision cancer medicine.4, 5, 6, 7, 8 The main platform for TCGA and ICGC projects was whole exome sequencing (WES) utilizing next generation sequencing (NGS), which has provided comprehensive gene alteration data in protein‐coding regions for all types of human cancer.3 Next generation sequencing‐based genomic sequencing has enabled the sequencing of individual cancer‐patient genomes, and has become faster and less expensive year by year, such that NGS can be used, not only in research settings, but also in clinical practice. Sequencing the whole exome, let alone the whole genome, is technically possible given today's advanced technology; however, they provide more information than what can be practically analyzed and interpreted. It is difficult to process this amount of data at a level required for clinical application, and it is too costly for individual patient diagnosis. Recently, NGS‐based gene panel tests, in which only clinically important genes are examined, have been developed to obtain genomic data in a timely and cost‐effective way based on the data learned from previous WES‐based comprehensive studies. At present, the NGS‐based gene panel test is the first choice for individual cancer‐patient care, when introducing NGS technology into daily practice.
Whole exome sequencing or other types of genome‐wide analysis have revealed geographic and ethnic diversity in the genetic alterations associated with cancer,9, 10 which indicate that the efficacy of each drug and its side‐effects might be different among populations. However, most NGS data are from North America and Europe, and there has been a paucity of data for genetic alterations in Asian cancer patients. For example, only 5.5% of participants in the TCGA cohort are from populations of Asian ethnicity (http://cancergenome.nih.gov/). It is therefore important to consider geographic and ethnic differences when looking to understand the profile of genomic alterations, and apply NGS analysis to the development of therapies for Asian patients. Recently, we studied several types of solid cancers, including colorectal, gastric, lung, and breast cancer, in Japan utilizing NGS‐based comprehensive genomic panel testing,8, 11, 12, 13, 14, 15, 16, 17, 18 and revealed some differences in genomic alterations between Asian and non‐Asian populations.
The current notion is that NGS‐based genomic sequencing will realize “precision medicine”, in which each patient receives individualized therapy based on their genetic alterations in the tumor. Through experience, we have learned that control of sample quality is crucial when applying NGS‐based genomic testing in daily practice. Moreover, we have learned that gene panel testing is a clinically useful approach to investigate the genomic mechanisms related to the therapies. These include therapy‐related signaling pathways, microsatellite instability and a hypermutated phenotype, and deficiency in the DNA double‐strand break repair pathways. In this review we therefore describe the concept of precision medicine and the utility of target sequencing using gene panel tests, based on our experience of research in a Japanese population.
2. CONCEPT OF PRECISION MEDICINE AND TARGET SEQUENCING
Precision medicine is to utilize an individual's genes, environment, and lifestyle to personalize the management of a disease.5 Today, thanks to the advancement of genomic sequencing technology, it has become possible to determine the genomic changes related to disease in each individual patient. Former US president Barack Obama proposed the “Precision Medicine Initiative” in his State of the Union address in January 2015, specifically focusing on cancer as a target disease for treatment with precision medicine. Abundant time and effort have been devoted to determining the human and cancer genome that became the foundation of Obama's precision medicine initiative (Figure 1). The Human Genome Project, which took more than 10 years to complete, revealed the entire map of a normal human genome.19 Through this comprehensive project, it was recognized that cancer is a disease of the genome, and that it was important to examine changes over the cancer genome to overcome the disease.20 Indeed, during the period of the sequencing of the human genome, cancer researchers identified the majority of important oncogenes and tumor suppressors.21 Utilizing methodology and knowledge gained from the Human Genome Project, the TCGA project has therefore been mapping the human cancer genome since 2006. The TCGA project has been undertaken to investigate not only cancer‐related DNA abnormalities, but also methylation, mRNA, and protein expression abnormalities.22, 23, 24, 25, 26, 27 Taken together, precision medicine is expected to be applied especially in the field of cancer, and aims to develop effective cancer treatment strategies by identifying genomic alterations in individual cancers. After a lengthy effort, it has been identified that target sequencing of the key genes in the cancer genome is one of the most effective ways to identify the characteristics of the disease, and to determine treatment strategies.
Figure 1.
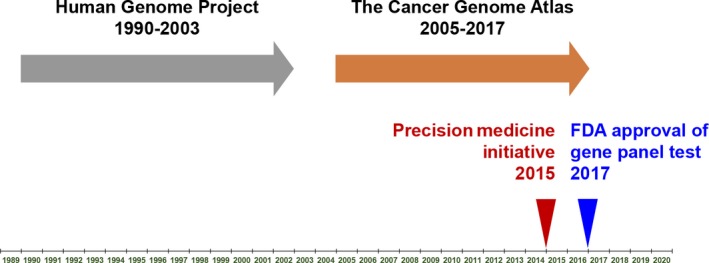
Determination of the human and cancer genomes has been a long‐term effort in the USA. The Human Genome Project followed by The Cancer Genome Atlas project were underway from 1990 to 2017. Recently, the US FDA approved next generation sequencing‐based gene panel tests as companion diagnostic tools
Although there are more than 20 000 genes in the human genome, the number of genes that are potentially related to cancer was found to be approximately 500, which includes driver‐genes of cancer. With this limited number of genes for analysis, it is possible to carry out deep sequencing, which is a method to increase accuracy by repeatedly sequencing the same site. In the USA, services for oncogenic panel testing have already begun by different enterprises and research facilities, such as FoundationOne by Foundation Medicine (Cambridge, MA, USA), Oncomine by ThermoFisher (Waltham, MA, USA), CANCERPLEX by KEW (Cambridge, MA, USA), MSK‐IMPACT by Memorial Sloan Kettering Cancer Center (New York, NY, USA), and OmniSeq Advance by Roswell Park Cancer Institute (Buffalo, NY, USA) (Table 1). In Japan, many of these genomic tests developed in the USA have been introduced and are available in the clinical research setting. In addition, original Japanese panels, such as NCC Oncopanel by Sysmex (Kobe, Japan), Todai OncoPanel by Riken genesis (Tokyo, Japan), and others, have been developed (Table 1). Some of the panel tests might have an advantage with an ability to determine tumor mutation burden (Table 1), which we describe below. In any case, it is necessary to verify the accuracy of any panel test before it is applied in clinical practice. Although some of the panel tests have been approved by the US FDA, it will be required that these panel tests be approved by the Pharmaceuticals and Medical Devices Agency in Japan for use in future clinical practice. Cancer gene panel testing allows us to analyze genetic mutations treatable with molecular‐targeted drugs, and explore the possibility of increased control over the treatment of cancer types.
Table 1.
Representative next generation sequencing‐based gene panel tests
| Panel test | No. of targeted genes | Enrichment approach | Tumor mutation burden | FDA approval | PMDA approval | References |
|---|---|---|---|---|---|---|
| Oncomine Dx Target Test | 23 genes | Amplicon | − | Yes | Yes | https://assets.thermofisher.com/TFS-Assets/LSG/brochures/oncomine-dx-target-test-flyer.pdf |
| MSK‐IMPACT | 468 genes | Capture | Yes | Yes | − | 36 |
| FoundationOne CDx | 324 genes | Capture | Yes | Yes | − | https://assets.ctfassets.net/vhribv12lmne/6Rt6csmCPuaguuqmgi2iY8/e3a9b0456ed71a55d2e4480374695d95/FoundationOne_CDx.pdf |
| NCC Oncopanel | 114 genes | Capture | − | − | − | https://www.mhlw.go.jp/file/05-Shingikai-10901000-Kenkoukyoku-Soumuka/0000179757.pdf |
| Todai OncoPanel | 464 genes | Capture | − | − | − | http://todaioncopanel.umin.jp/#sec01 |
| CANCERPLEX | 435 genes | Capture | Yes | − | − | 56 |
| OncoPrime | 223 genes | Unknown | − | − | − | 73 |
| PleSSision | 160 genes | Unknown | − | − | − | http://www.hosp.keio.ac.jp/st/cancer/info/20180529_2.pdf |
| OmniSeq Advance | 144 genes | Amplicon | Yes | − | − | 74 |
| P5 report | 52 genes | Unknown | − | − | − | http://www.okayama-u.ac.jp/user/hos/koganzai/P5report/ |
–, No data; PMDA, Pharmaceuticals and Medical Devices Agency (Japan).
3. SAMPLE QUALITY CONTROL
To utilize panel testing in daily practice, it is crucial to preserve clinical samples in a suitable quality for the analysis (Figure 2). Although the NGS‐based target sequence screening can suggest the most appropriate treatment for an individual patient, it is only possible in daily practice when commonly stored clinical samples are usable for NGS‐based analysis.28 In other words, the application of NGS‐based analysis is only possible when DNA in preserved clinical samples is maintained in appropriate conditions with the production of minimal artifacts.8 The majority of clinical samples from cancer patients, such as surgical specimens, are stored in formalin‐fixed, paraffin‐embedded (FFPE) tissue, which can be used for DNA extraction and NGS analysis if processed and preserved appropriately (Figure 2). However, the reality is that DNA in FFPE tissue is often fragmented and unusable for NGS analysis.29 DNA quality in FFPE tissue samples is often worse than expected, due to careless sample preparation and preservation.8, 30 Therefore, it is critical for clinicians to know how to prepare and preserve samples for possible future NGS analysis.8
Figure 2.
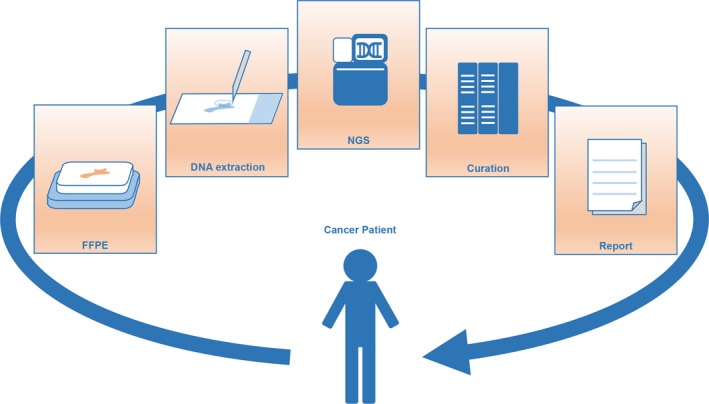
Flow of a next generation sequencing (NGS)‐based gene panel test utilizing formalin‐fixed, paraffin‐embedded (FFPE) tissue. Preparation and preservation of FFPE tissue is the first step of the panel test, followed by DNA extraction, NGS analysis, curation of the data, and report of the data with therapeutic recommendations
Previously, we confirmed that neutral buffered formalin, but not unbuffered formalin, is suitable for fixation of surgical samples to preserve DNA adequately, in agreement with a previous report.8, 31 It has been suggested that DNA damage occurs for the following reasons: formaldehyde‐induced cross‐linking; DNA fragmentation; deamination of cytosine bases leading to C–>T mutations; and the generation of basic sites.32 Furthermore, it is important for appropriate tissue preservation to fix for the appropriate time with formalin. Over‐fixation results in excessive cross‐linking, thus interfering with the extraction of nucleic acids and proteins.30 In contrast, under‐fixation causes degradation of nucleic acid and protein, or a change in gene expression in poorly formalin‐perfused areas.30 Furthermore, to avoid tissue ischemic changes, surgical specimens should be fixed with formalin immediately after removal of the specimen.30 Taken together, an appropriate preservation procedure should be carried out in daily clinical practice, following a standard protocol with neutral buffered formalin to preserve high‐quality DNA. Although DNA in FFPE tissue samples is as stable as in frozen samples, it will degrade over time.33, 34 We found that DNA extracted from FFPE tissue samples of surgical specimens older than 7 years might not be analyzed by NGS. Clinicians should consider NGS analysis for cancer patients who are at a high risk of recurrence, sooner rather than later.
Biopsy samples can be very useful for NGS analysis. For instance, genomic analysis of tissue samples from biopsies will be useful for cancer patients who are treated with neoadjuvant chemotherapy, patients whose cancer is altered or diminished, or patients with nonresectable diseases.35 Next generation sequence analysis could also provide clues for overcoming drug resistance using genomic data obtained from repeated biopsies for patients with tumor recurrence. Although the amount of tissue in an FFPE biopsy sample can be very small, we have previously reported that DNA extracted from FFPE tissue from core needle biopsies of breast cancer or endoscopic biopsies of colorectal cancer can be available for NGS‐based analysis.
In summary, utilizing FFPE samples of surgical specimens and biopsy tissue samples, NGS‐based gene panel testing can be carried out if FFPE samples are appropriately preserved. Once again, it is important for clinicians to know how to preserve these samples. Next generation sequencing‐based gene panel tests will provide useful information for cancer patients, which will be discussed in the following sections.
4. WHAT DOES AN NGS‐BASED GENE PANEL TEST TELL YOU?
An NGS‐based gene panel test can be used to identify gene alterations that are targetable by molecular‐targeted drugs (Figure 3). In the USA, a variety of NGS‐based comprehensive gene panels have been developed, and some have been approved by the FDA as companion diagnostics for multiple molecular‐targeted therapies (Table 1). Some patients have been enrolled in genomically matched clinical trials, although the rate of patients enrolled in those trials has been low.36 In Japan, clinical research has just started to reveal the value of clinical sequencing with NGS‐based gene panels following preclinical studies identifying the feasibility of clinical sequencing by several academic institutions.37 Through these experiences, we have learned what the NGS‐based gene panel test tells us for cancer patients.36
Figure 3.
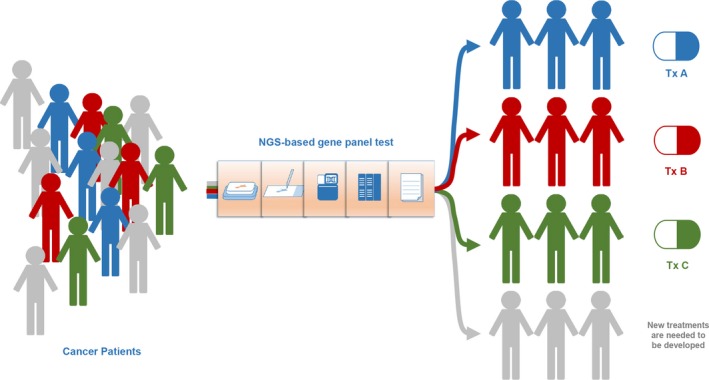
Precision cancer medicine utilizing next generation sequencing (NGS)‐based gene panel testing. Patients will be treated according to certain characteristics based on information from genomic analysis. New treatment will be needed for the patients without druggable targets
One of the most successful examples of the treatment strategies based on NGS‐based gene panel tests is that for lung adenocarcinoma, in which several genes have been shown to carry targetable driver mutations, such as EGFR, ROS1, and ALK.38 These targetable genes, such as ALK, are detectable not only in patients with lung adenocarcinoma, but also in patients with other solid cancers. Importantly, the targeted therapy will be effective not only to lung cancer patients, but also to other cancer patients with the ALK mutation. Moreover, RET is a targetable gene in non‐small‐cell lung cancer patients. Vandetanib, which targets RET rearrangements, showed clinical benefit in Japanese patients with advanced RET‐rearranged non‐small‐cell lung cancer.39 Therefore, comprehensive examination is effective for finding a specific population suitable for targeted therapy in all cancer patients.
Utilizing the gene panel test, we have also learned that we are able to detect not only the targetable driver‐genes described above, but also mutations conferring drug resistance. Moreover, gene panel testing can be used to detect hypermutation, which is expected to be a promising biomarker for immune checkpoint inhibitors. The comprehensive test further determines genetic changes in the DNA double‐strand break repair pathway, which is an emerging target for new therapies, and also closely related to hereditary genomic changes. In the following sections, we discuss the application of gene panel testing, and issues that need to be addressed.
5. TARGET GENES OR RESISTANCE‐RELATED GENES FOR MOLECULAR‐TARGETED THERAPY
In strategies involving precision medicine, selection of the most efficient treatment is based on identifying a subgroup of patients with certain characteristics in their genome. In addition to lung adenocarcinoma, another successful example of a treatment strategy involving precision medicine is anti‐human epidermal growth factor receptor 2 (HER2) therapy using trastuzumab and other molecular‐targeting drugs for breast cancer patients with tumors overexpressing HER2. Breast cancer patients with HER2 overexpression initially showed one of the worst clinical outcomes due to the biologically aggressive behavior of HER2‐positive breast cancer cells. Nevertheless, trastuzumab and other newly developed drugs that target cells overexpressing the HER2 receptor showed a favorable effect on HER2‐positive breast cancer cells and significantly improved patient survival. Of note, HER2 overexpression has been found not only in breast cancer, but also in other solid cancers such as gastric, colorectal, and lung cancer. Recently, it has been revealed that anti‐HER2 therapies are effective for patients with HER2 overexpression in various cancer types. For instance, trastuzumab is the recommended first‐line therapy for advanced gastric cancer in patients with HER2 overexpression.40 A cross‐sectional treatment applying a molecular‐targeted drug for each cancer, such as a “HER2‐oma”, accompanied by a common cancer gene alteration, can now be considered (Figure 4).
Figure 4.
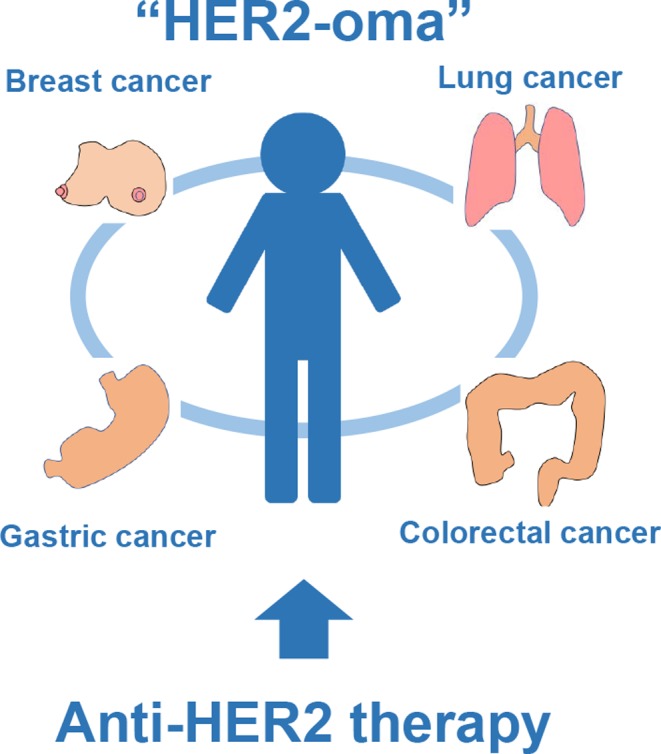
Concept of “HER2‐oma”. Not only breast cancer patients, but also lung, gastric, and colorectal cancer patients with human epidermal growth factor receptor 2 (HER2) overexpression could be potentially treated with anti‐HER2 therapies
In daily clinical practice, HER2 overexpression is determined by immunohistochemistry (IHC) and FISH. Next generation sequencing‐based gene panel testing enables detection of HER2 gene amplification. We have examined HER2 overexpression and amplification by IHC, FISH, and gene panel tests in colorectal cancer patients, and reported that gene panel testing has the same utility as IHC and FISH for detecting HER2‐positive patients that are candidates for HER2‐targeted therapy.12 Recently, it has also been revealed that the majority of HER2 somatic mutations without HER2 gene amplification in breast cancer patients are activating mutations, which can be targeted by anti‐HER2 therapies.41 Next generation sequencing‐based gene panel testing will identify patients with breast or other solid cancers with the HER2 activating mutations, who will potentially benefit from anti‐HER2 therapies.41 Moreover, gene panel tests enable simultaneous detection of gene alterations related to drug resistance. Alterations in the PI3K/mTOR pathway, MAPK pathway and other receptor tyrosine kinases (RTKs) have been considered as mechanisms of resistance to anti‐HER2 therapy.42, 43, 44 An NGS‐based gene panel test can examine these pathways in a single test, and potentially provide useful information to decide effective treatment strategies.45, 46
Specific gene alterations are significantly associated with resistance to certain molecular‐targeted therapies, such as the example of HER2 resistance as described above. Therefore, strategies to test for the presence of specific gene alterations before treatment, as a companion diagnostic test, is an effective way to determine which patients will respond to the therapy. In patients with metastatic colorectal cancer, KRAS mutations (codon 12 and 13) are indicators for resistance to anti‐epidermal growth factor receptor (EGFR) therapy.47 In addition to KRAS mutations, gene alterations in RTKs and the RAS pathway, which is downstream of EGFR, appear to indicate resistance to anti‐EGFR therapies.48 A comprehensive analysis of gene alterations in the RTKs and RAS pathways is possible with an NGS‐based gene panel test.8 Indeed, we have identified gene alterations in RTKs and RAS pathways in patients who were previously considered KRAS WT. These patients showed significantly worse outcomes compared to patients without mutations in RTKs and the RAS pathways when treated with anti‐EGFR therapies.8 Taken together, a comprehensive analysis of the gene alterations that underlie resistance to molecular‐targeted therapies is possible by gene panel testing.
6. MICROSATELLITE INSTABILITY AND HYPERMUTATION
Recently, the groundbreaking invention of immune checkpoint inhibitors has caused a paradigm shift in the field of cancer research and cancer‐patient care. Patient outcomes for advanced stage cancer treated with immune checkpoint inhibitors have shown dramatic improvement in several solid cancer types.49, 50 However, immune checkpoint inhibitors showed a significant effect in a limited population, and current methods such as IHC of programmed death‐ligand 1 cannot fully identify responders to immunotherapy. Predictive biomarkers for identifying responders are therefore highly sought after.
Recent progress in genomic analysis using NGS technology has allowed determination of the “mutation burden” in cancer. A hypermutated tumor is defined as a tumor with an increased mutation burden (a high rate of somatic mutation), which is associated with development of neoantigens and a response to immune checkpoint inhibitors.27, 51, 52, 53, 54 The hypermutated cancer cells are thought to generate numerous neoantigens, which attract cytotoxic (CD8+) T lymphocytes and activated Th1 cells to the tumor microenvironment surrounding cancer cells.55 The mutation burden was initially determined by whole exome analysis, such as TCGA.27 Later, utilizing the NGS‐based panel test, we were also able to determine the tumor mutation burden,8 which is expected to be very important in the field of immuno‐oncology and cancer treatment.
The mutation burden can be defined as the rate of peptide‐changing single‐nucleotide variants per million base pairs.56 To estimate the mutation burden, single‐nucleotide variants with a mutation allelic fraction of at least 0.1 after standard filtering were retained.56 We have previously reported that a panel test with 400 genes (roughly 1/2000th of the genome) is comparable to WES in generating mutation rates and to distinguish hypermutated and nonhypermutated tumors,8 although the average mutation rate detected by the panel test was higher than that detected by WES, reflecting the fact that the panel content includes genes that are more frequently mutated in cancer.
There can be a variety of mechanisms causing hypermutation, including exogenous mutagens, such as UV light and tobacco smoking, and endogenous mutagens, such as excessive apolipoprotein B mRNA editing enzyme, catalytic polypeptide‐like (APOBEC).57, 58, 59, 60 Among them, one of the leading causes of hypermutation is a deficiency in the DNA mismatch repair system, resulting in microsatellite instability (MSI).27 Microsatellite instability is caused by an impaired DNA mismatch repair (MMR) system, which often results from germline or somatic mutations, or promoter hypermethylation of genes involved in the DNA MMR system, such as MLH1, MLH2, MSH6, and PMS2.61 The germline mutation of MMR genes causes Lynch syndrome that often results in hereditary nonpolyposis colorectal cancer, and somatic mutations of MMR genes cause Lynch syndrome‐like tumors, both of which show hypermutated phenotypes. Microsatellite instability can be detected by investigating the 5 microsatellite loci based on the “Bethesda Guidelines”. Recently, NGS‐based analysis has been developed to detect MSI more accurately by examining a large number of MSI loci, which enables panel tests to clarify the driver and passenger mutations, as well as the presence of MSI.62, 63 Detection of MSI is now considered important for the estimation of hypermutation, especially in colorectal and gastric cancer.
We found that 17 of 201 (8%) Japanese colorectal cancers were hypermutated tumors, as identified by a gene panel test.8 Among them, we found 11 patients with MSI, and 2 patients with polymerase ε mutation, which is another cause of hypermutation.8 We also identified 32 out of 207 (15.5%) Japanese gastric cancer patients as having hypermutated tumors, which were observed to be not only an MSI subtype, but also Epstein‐Barr virus infection, which is detectable by integrative genomic analysis that makes use of NGS technologies, chromosomal instability, and genomically stable subtypes of TCGA.11 In contrast, MSI is merely observed in breast cancer. We determined tumor mutation burden in triple‐negative breast cancer patients and revealed that 3 of 51 (5.7%) patients were hypermutated. Other mechanisms rather than MSI, such as excessive APOBEC, might be involved in hypermutation in breast cancer, which needs further investigation.
7. DNA DOUBLE‐STRAND BREAK REPAIR PATHWAYS
One of the most promising targets that could be identified using the NGS‐based gene panel test are genes related to homologous recombination (HR), which is one of the major mechanisms of DNA double‐strand break repair pathways. Cancer patients with a HR deficiency (HRD), of which the most notable ones are BRCA1/2 mutations, are more sensitive to PARP inhibitors.64 Indeed, the US FDA have approved PARP inhibitors for patients with ovarian and breast cancer with a BRCA1/2 germline mutation. We examined BRCA1/2 mutations in a Japanese population and found alterations of BRCA1 and BRCA2 in 9.4% and 5.6% of Japanese TNBC patients, respectively, which is in agreement with previous reports.
There are several issues in detecting HRD in cancer patients. First, there is a difficulty in interpretation of variants with unknown significance. Second, clinical significance of somatic mutations in HR genes are still not clear.65 Somatic mutations of BRCA1/2 genes are observed in approximately 2.5% of all patients with sporadic breast cancer.65 Theoretically, it is expected that somatic BRCA1/2‐mutated breast cancers will respond to PARP inhibitors, similar to cancers with the germline mutation of those genes, however, it has not been conclusively shown that germline and somatic BRCA1/2 mutations are biologically equivalent.65 Finally, although currently BRCA1/2 mutation status is the only biomarker to identify patients for PARP inhibitors, there are other genes that, when mutated, can lead to HRD.64 However, the genes involved in HR are not yet well defined, and equal weighting might not estimate HRD appropriately.65 Although it has been possible to determine mutations in HR‐related genes utilizing NGS‐based panel tests, a system to detect HRD by panel sequencing has not been established, partially due to the difficulty in estimating the HRD based on the related genes, as described above.
Germline mutation of BRCA1/2 can be a cause of familial cancer, such as breast, ovary, prostate, and pancreatic cancer, which is known as the hereditary breast and ovarian cancer syndrome. Indeed, we found that 7 of 53 (13%) Japanese TNBC patients had possible germline BRCA1/2 alterations by additional bioinformatic analysis of solid tumor sequencing. Comprehensive genomic analysis utilizing gene panel tests revealed the frequency of BRCA1 and BRCA2 alterations to be 3.9% and 10.6% in Japanese gastric cancers, respectively.11 Among them, we confirmed 3 patients with germline mutations and a family history, which indicates that BRCA1/2 germline mutations can be a cause of gastric cancer in Japan.18 Considering the frequency of germline BRCA1/2 alterations in Japan, it appears that there is a certain population who would benefit from treatment with PARP inhibitors in Japan, not only in ovarian and breast cancer patients, but also others, potentially including gastric cancer patients.
8. GEOGRAPHIC AND ETHNIC DIFFERENCES
An EGFR mutation, which is a predictive biomarker for clinical response to EGFR tyrosine kinase inhibitors, has dramatically changed daily treatment of patients with lung adenocarcinoma. The frequency of EGFR mutation varies among different geographic and ethnic populations. There are more EGFR mutations in Asian than in Caucasian populations, and more in non‐smoking women. However, hypermutated tumors are more common in patients without EGFR or other driver mutations, who are often smokers.66 Utilizing an NGS‐based panel test, we can analyze geographic and ethnic differences between Asian and other ethnic populations.
In colorectal cancer, HER2 amplification is significantly more common in Japan than in the TCGA cohort, which indicates a possibility for anti‐HER2 therapy of colorectal cancer in patients screened by genetic testing in Japan. In breast cancer, there was significantly more MYC amplification in the TCGA cohort than in the Japanese cohort. In lung adenocarcinoma, mutations in CDKN2A, CDKN2B, and RB1, in addition to EGFR, were more common in the Japanese cohort than in the TCGA cohort. We found several differences between the Japanese cohort and the TCGA cohort or others, with the overall spectrum of genomic alterations similar among those populations. Although drug response might be different, and we need to confirm the effect of new drugs for Japanese patients, it appears that we can utilize an NGS‐based test to reveal genomic changes in cancer, in the same way as North America and European countries.
9. ISSUES TO BE ADDRESSED IN THE FUTURE
Although NGS‐based gene panel testing has the potential to change daily oncology practice, there are many issues to be addressed before it effectively saves cancer patients. One of the major issues of NGS remains the high cost.67 The NGS‐based gene panel tests cost several thousand US dollars per sample, depending on the vendor, and WES or whole genome sequencing cost more. Another major issue of NGS‐based precision medicine strategies is lack of treatment.68 The number of patients who find that their mutations are associated with a specific available treatment is limited. Clinical trials based on genetic panel tests is thought to be beneficial to the medical economy because it can narrow choice when selecting an expensive molecular‐targeted therapy, and deliver it only to those in whom it is expected to be effective. However, results from these studies and clinical trials are not as good as expected, partially due to the lack of effective treatment options compared with the number of altered genes.
It is also important to build a social system that allows genetic testing. For patients with possible BRCA1/2 germline mutations, genetic counseling should be undertaken, and additional germline tests should be examined according to the individual's needs. Not only BRCA1/2, but other genes, such as PALB2, PTEN, and TP53, have also been reported to cause hereditary carcinogenesis.69 Depending on each gene, mutation form and penetrance are different. It is urgent that genetic counseling is developed as soon as possible. Experts able to practice in the field of cancer are very limited in Japan, and training such personnel is necessary at an administrative level.
After NGS analysis, the next challenge is how to interpret data and select the most suitable therapeutic agent by linking gene alteration data with clinical information. This is currently under development, at least in Japan. There are large, publicly available genomic databases, however, the amount of genomic data that is linked with clinical treatment outcomes is as yet insufficient. Drug efficacy might be different, based not only on altered genes themselves, but also on the site and type of gene alterations. The issue of mutations of unknown significance exists, not only for BRCA1/2 mutations as described above, but also for other genes in which more clinical data will be needed for each individual driver and passenger genes. Drug efficacy might also be different based on the organ where cancer arises or metastasizes. Indeed, it has been reported that efficacy in HER2‐mutant cancers varied as a function of both tumor type and mutant allele.70 Thus, data accumulation is needed on clinical treatment outcomes following targeted therapies, based on genomic changes for each organ. Bioinformatics is indispensable for processing enormous genetic and clinical cancer patient data obtained by NGS analysis. Bioinformatics for precision oncology is a new field, and therefore human resource development is important. Widespread genetic testing and accumulation of clinical data to develop a bioinformatics knowledge base will help future patient care.
Most NGS‐based gene panel tests use tissue samples obtained by surgery or biopsy. Therefore, it is impossible for patients who have only metastasis that cannot be surgically removed or biopsied to examine their tumor by panel tests. Recently, liquid biopsy, by which NGS analysis on cancer‐derived DNA in blood is carried out, has attracted attention. There are variety of liquid biopsy technologies, which includes cell‐free DNA derived from disrupted cancer cells in the blood, and circulating tumor cells, which are cancer cells circulating in blood.71 Liquid biopsy is considered applicable to diagnosis, prediction of prognosis, recurrence monitoring, and the like. For example, if patients have metastatic breast cancer that cannot be biopsied, liquid biopsy can be used to investigate the presence or absence of estrogen susceptibility, HER2 sensitivity, or resistance of other targeted therapeutic agents. Monitoring by liquid biopsy is also expected to enable early detection of postoperative recurrence. Monitoring using microRNA is also expected as an application of liquid biopsy.72 Although these techniques are expected to make it possible to measure tumor gene mutations in patients less invasively, as described above, there are still many problems to be solved at present, such as the reproducibility and accuracy of examination. This is especially so in breast cancer, as it is thought to be more difficult than carcinomas with many driver genes, such as colon cancer and lung cancer. There are also few genetic mutations that are common among patients, and the absolute number of circulating tumor cells in blood is much smaller than that of prostate cancer. It is expected that more convenient and useful tests will be developed as technologies progress.
10. CONCLUSION
In this review, we have described the utility of NGS‐based gene panel testing and discussed issues that are a focus of future work. Cancer is a disease of the genome, and thus it is reasonable that treatment strategies should be based on genomic change. Although there are many issues that need to be addressed to fully realize precision cancer medicine, it appears that we are generally headed in the right direction to improve a cancer patient's prognosis. It is important for clinicians to understand the utility of NGS‐based genomic testing, as this will help the cancer patients of tomorrow, who are not currently able to be saved.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
Our research described in this manuscript was supported by funding from Denka Co., Ltd. M. Nagahashi is supported by the Japan Society for the Promotion of Science (JSPS) Grant‐in‐Aid for Scientific Research Grant Number JP18K19576. Y. Shimada is supported by the Japan Society for the Promotion of Science (JSPS) Grant‐in‐Aid for Scientific Research Grant Number JP18K08612. H. Ichikawa is supported by the JSPS Grant‐in‐Aid for Scientific Research Grant Number JP16K10491. H. Kameyama is supported by the Japan Society for the Promotion of Science (JSPS) Grant‐in‐Aid for Scientific Research Grant Number JP17K10624. K. Takabe is supported by NIH/NCI grant R01CA160688 and Susan G. Komen Investigator Initiated Research Grant IIR12222224. S. Okuda is supported by the JSPS Grant‐in‐Aid for Scientific Research Grant Number JP18H04123. T. Wakai is supported by the JSPS Grant‐in‐Aid for Scientific Research Grant Number JP16K15610.
Nagahashi M, Shimada Y, Ichikawa H, et al. Next generation sequencing‐based gene panel tests for the management of solid tumors. Cancer Sci. 2019;110:6–15. 10.1111/cas.13837
REFERENCES
- 1. Nakagawa H, Wardell CP, Furuta M, Taniguchi H, Fujimoto A. Cancer whole‐genome sequencing: present and future. Oncogene. 2015;34:5943‐5950. [DOI] [PubMed] [Google Scholar]
- 2. Alifrangis CC, McDermott U. Reading between the lines; understanding drug response in the post genomic era. Mol Oncol. 2014;8:1112‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17‐37. [DOI] [PubMed] [Google Scholar]
- 4. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lowy DR, Collins FS. Aiming high‐changing the trajectory for cancer. N Engl J Med. 2016;374:1901‐1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bando H, Takebe N. Perspectives on research activity in the USA on Cancer Precision Medicine. Jpn J Clin Oncol. 2016;46:106‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chamberlin MD, Bernhardt EB, Miller TW. Clinical Implementation of Novel Targeted Therapeutics in Advanced Breast Cancer. J Cell Biochem. 2016;117:2454‐2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nagahashi M, Wakai T, Shimada Y, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan DS, Mok TS, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol. 2016;34:91‐101. [DOI] [PubMed] [Google Scholar]
- 10. Dietze EC, Sistrunk C, Miranda‐Carboni G, O'Regan R, Seewaldt VL. Triple‐negative breast cancer in African‐American women: disparities versus biology. Nat Rev Cancer. 2015;15:248‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ichikawa H, Nagahashi M, Shimada Y, et al. Actionable gene‐based classification toward precision medicine in gastric cancer. Genome Med. 2017;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimada Y, Yagi R, Kameyama H, et al. Utility of comprehensive genomic sequencing for detecting HER2‐positive colorectal cancer. Hum Pathol. 2017;66:1‐9. [DOI] [PubMed] [Google Scholar]
- 13. Nagahashi M, Shimada Y, Ichikawa H, et al. Formalin‐fixed paraffin‐embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shimada Y, Kameyama H, Nagahashi M, et al. Comprehensive genomic sequencing detects important genetic differences between right‐sided and left‐sided colorectal cancer. Oncotarget. 2017;8:93567‐93579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sato S, Nagahashi M, Koike T, et al. Impact of concurrent genomic alterations detected by comprehensive genomic sequencing on clinical outcomes in east‐Asian patients with EGFR‐mutated lung adenocarcinoma. Sci Rep. 2018;8:1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okamoto T, Takada K, Sato S, et al. Clinical and genetic implications of mutation burden in squamous cell carcinoma of the lung. Ann Surg Oncol. 2018;25:1564‐1571. [DOI] [PubMed] [Google Scholar]
- 17. Nagahashi M, Ling Y, Hayashida T, et al. Actionable gene alterations in an Asian population with triple‐negative breast cancer. JCO Precis Oncol. 2018;2:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ichikawa H, Wakai T, Nagahashi M, et al. Pathogenic germline BRCA1/2 mutations and familial predisposition to gastric cancer. JCO Precis Oncol. 2018;2:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varmus H. Genomic empowerment: the importance of public databases. Nat Genet. 2003;35(Suppl 1):3. [DOI] [PubMed] [Google Scholar]
- 20. Varmus H, Stillman B. Support for the human cancer genome project. Science. 2005;310:1615. [DOI] [PubMed] [Google Scholar]
- 21. Wheeler DA, Wang L. From human genome to cancer genome: the first decade. Genome Res. 2013;23:1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202‐209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas Research Network . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061‐1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cancer Genome Atlas Network . Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next‐generation sequencing in patients with non‐small cell lung cancer. Cancer. 2015;121:631‐639. [DOI] [PubMed] [Google Scholar]
- 29. Endrullat C, Glokler J, Franke P, Frohme M. Standardization and quality management in next‐generation sequencing. Appl Transl Genom. 2016;10:2‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arreaza G, Qiu P, Pang L, et al. Pre‐analytical considerations for successful next‐generation sequencing (NGS): challenges and opportunities for formalin‐fixed and paraffin‐embedded tumor tissue (FFPE) samples. Int J Mol Sci. 2016;17:3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nuovo GJ, Silverstein SJ. Comparison of formalin, buffered formalin, and Bouin's fixation on the detection of human papillomavirus deoxyribonucleic acid from genital lesions. Lab Invest. 1988;59:720‐724. [PubMed] [Google Scholar]
- 32. Do H, Dobrovic A. Sequence artifacts in DNA from formalin‐fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64‐71. [DOI] [PubMed] [Google Scholar]
- 33. Nuovo AJ, Garofalo M, Mikhail A, Nicol AF, Vianna‐Andrade C, Nuovo GJ. The effect of aging of formalin‐fixed paraffin‐embedded tissues on the in situ hybridization and immunohistochemistry signals in cervical lesions. Diagn Mol Pathol. 2013;22:164‐173. [DOI] [PubMed] [Google Scholar]
- 34. Carrick DM, Mehaffey MG, Sachs MC, et al. Robustness of next generation sequencing on older formalin‐fixed paraffin‐embedded tissue. PLoS One. 2015;10:e0127353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramos C, Stephens P, Hoke NN, et al. Association of PIK3CA mutation with response (ExRx) to cetuximab (C) in metastatic (met) triple‐negative breast cancer (TNBC). J Clin Oncol. 2015;33:151. [Google Scholar]
- 36. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kohno T. Implementation of “clinical sequencing” in cancer genome medicine in Japan. Cancer Sci. 2018;109:507‐512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mano H. ALKoma: a cancer subtype with a shared target. Cancer Discov. 2012;2:495‐502. [DOI] [PubMed] [Google Scholar]
- 39. Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET‐rearranged advanced non‐small‐cell lung cancer (LURET): an open‐label, multicentre phase 2 trial. Lancet Respir Med. 2017;5:42‐50. [DOI] [PubMed] [Google Scholar]
- 40. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet. 2010;376:687‐697. [DOI] [PubMed] [Google Scholar]
- 41. Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395‐402. [DOI] [PubMed] [Google Scholar]
- 43. Mohd Sharial MS, Crown J, Hennessy BT. Overcoming resistance and restoring sensitivity to HER2‐targeted therapies in breast cancer. Ann Oncol. 2012;23:3007‐3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zuo Q, Liu J, Zhang J, Wu M, Guo L, Liao W. Development of trastuzumab‐resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci Rep. 2015;5:11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelly CM, Janjigian YY. The genomics and therapeutics of HER2‐positive gastric cancer‐from trastuzumab and beyond. J Gastrointest Oncol. 2016;7:750‐762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee JY, Hong M, Kim ST, et al. The impact of concomitant genomic alterations on treatment outcome for trastuzumab therapy in HER2‐positive gastric cancer. Sci Rep. 2015;5:9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoshino T, Muro K, Yamaguchi K, et al. Clinical validation of a multiplex kit for RAS mutations in colorectal cancer: results of the RASKET (RAS KEy Testing) prospective, multicenter study. EBio Med. 2015;2:317‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous‐cell non‐small‐cell lung cancer. N Engl J Med. 2015;373:123‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non‐small‐cell lung cancer. N Engl J Med. 2015;372:2018‐2028. [DOI] [PubMed] [Google Scholar]
- 52. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015;348:124‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Le DT, Uram JN, Wang H, et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med. 2015;372:2509‐2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858‐868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget. 2017;8:112103‐112115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eifert C, Pantazi A, Sun R, et al. Clinical application of a cancer genomic profiling assay to guide precision medicine decisions. Per Med. 2017;14:309‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer‐associated genes. Nature. 2013;499:214‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schlesner M, Eils R. Hypermutation takes the driver's seat. Genome Med. 2015;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alexandrov LB, Nik‐Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roberts SA, Lawrence MS, Klimczak LJ, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mensenkamp AR, Vogelaar IP, Van Zelst‐Stams WA, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch‐repair deficiency in Lynch syndrome‐like tumors. Gastroenterology. 2014;146:643‐646.e8. [DOI] [PubMed] [Google Scholar]
- 62. Kautto EA, Bonneville R, Miya J, et al. Performance evaluation for rapid detection of pan‐cancer microsatellite instability with MANTIS. Oncotarget. 2017;8:7452‐7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342‐1350. [DOI] [PubMed] [Google Scholar]
- 64. Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455‐470. [DOI] [PubMed] [Google Scholar]
- 65. den Brok WD, Schrader KA, Sun S, et al. Homologous recombination deficiency in breast cancer: A clinical review. JCO Presis Oncol. 2017;1:1‐13. [DOI] [PubMed] [Google Scholar]
- 66. Nagahashi M, Sato S, Yuza K, et al. Common driver mutations and smoking history affect tumor mutation burden in lung adenocarcinoma. J Surg Res. 2018;230:181‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fontanges Q, De Mendonca R, Salmon I, Le Mercier M, D'Haene N. Clinical application of targeted next generation sequencing for colorectal cancers. Int J Mol Sci. 2016;17:2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Peixoto R, Sousa TT, Cruz M, Maluf F, Buzaid A. Next‐generation sequencing (NGS) in metastatic breast cancer (mBC) patients: translation from sequence data into clinical practice. J Clin Oncol. 2015;33:133.25349299 [Google Scholar]
- 69. Lerner‐Ellis J, Khalouei S, Sopik V, Narod SA. Genetic risk assessment and prevention: the role of genetic testing panels in breast cancer. Expert Rev Anticancer Ther. 2015;15:1315‐1326. [DOI] [PubMed] [Google Scholar]
- 70. Hyman DM, Piha‐Paul SA, Won H, et al. HER kinase inhibition in patients with HER2‐ and HER3‐mutant cancers. Nature. 2018;554:189‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lowes LE, Bratman SV, Dittamore R, et al. Circulating tumor cells (CTC) and cell‐free DNA (cfDNA) workshop 2016: scientific opportunities and logistics for cancer clinical trial incorporation. Int J Mol Sci. 2016;17:1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shimomura A, Shiino S, Kawauchi J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kou T, Kanai M, Yamamoto Y, et al. Clinical sequencing using a next‐generation sequencing‐based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017;108:1440‐1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Conroy JM, Pabla S, Glenn ST, et al. Analytical validation of a next‐generation sequencing assay to monitor immune responses in solid tumors. J Mol Diagn. 2018;20:95‐109. [DOI] [PubMed] [Google Scholar]


