Transmembrane protein 106B (TMEM106B; NM_001134232) was recently identified as a gene responsible for a form of hypomyelinating leukodystrophy (HLD).1,2 All 5 cases identified to date carry the identical c.754 G > A, (p.Asp252Asn) mutation.1,2 Although the exact function is unknown,3 studies of TMEM106B in the context of frontotemporal lobar degeneration with 43-kD TAR DNA-binding protein (TDP-43) pathology (FTLD-TDP) indicate that TMEM106B likely acts as a lysosomal regulator and can modify risk for FTLD-TDP.4 However, the molecular effects of the (p.Asp252Asn) substitution have not yet been reported for TMEM106B-associated HLD. The HLDs are heterogeneous conditions, with the known disease genes playing roles in myelin sheath structure (e.g., PLP1) and other cellular functions that are not oligodendrocyte specific, including protein translation, molecular chaperoning, and cytoskeletal regulation.5 We set out to assess if this recurrent TMEM106B substitution was affecting lysosome biology or had an alternate role underlying the HLD pathogenesis. Implication of lysosome biology in HLD provides exciting new advances in our understanding of the molecular underpinnings of this condition and the complexities of neurodevelopment.
Functional analysis
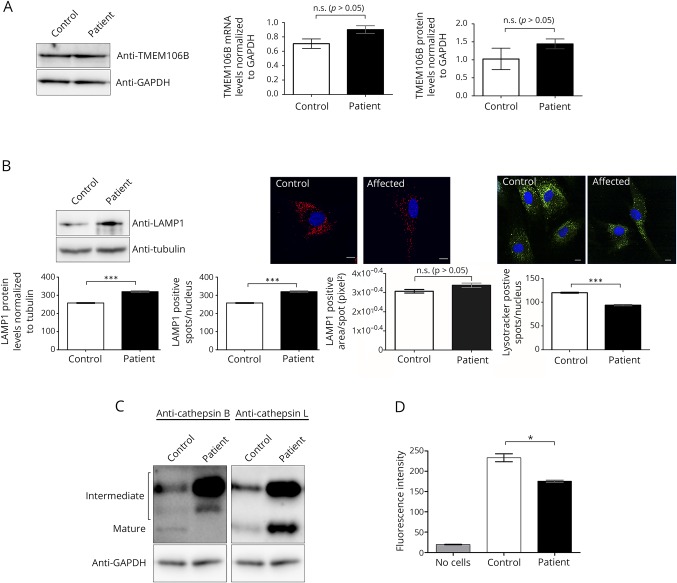
Using patient-derived fibroblasts (patient 4),1 we assessed TMEM106B messenger RNA (mRNA) and protein levels and found that these were unaltered in patient cells compared with controls (figure, A). TMEM106B has been shown to affect lysosome number, morphology, and acidification.4,6 LAMP1 staining, which marks lysosomes, showed an increase in the number of lysosomes in patient fibroblasts compared with controls, although the average size of the lysosome remained unchanged (figure, B). Staining with a pH-sensitive fluorescent dye showed that despite the increased number of lysosomes, a substantial decrease in the number of lysotracker-positive foci was observed, indicating that patient lysosomes show impaired acidification (figure, B).
Figure. Lysosome dysfunction observed in patient-derived fibroblasts with the TMEM106B p.Asp252Asn substitution.
(A) Control and patient fibroblasts had comparable levels of both mRNA and TMEM106B protein when examined by real-time quantitative PCR and western blot analysis, respectively (p > 0.05, 2-tailed Student's t-test). (B) An increase in the lysosome-specific marker LAMP1 was observed in patient cells by both Western blot analysis and immunofluorescent staining (p < 0.001, 2-tailed Student's t-test). (A substantial decrease was observed in the number of lysotracker-positive spots in the patient cells by immunofluorescence; p < 0.001, 2-tailed Student's t-test.) For immunofluorescent studies, fibroblast images were collected from 10 wells per sample from 3 independent experiments using the Opera High Content Screening System (Perkin Elmer) and analyzed using Columbus software. Scale bars represent 10 μm. Three control fibroblast cell lines were used for these studies. (C) Western blot analyses showed an accumulation of the intermediate forms of both cathepsin B and cathepsin L, but accumulation of only the mature form of cathepsin L in patient fibroblasts. The mature form of cathepsin B is not detectable by Western blot analysis in patient cells. (D) A fluorescence-based enzyme assay shows reduction in the activity of cathepsin B in affected fibroblasts (p < 0.05, 1-way analysis of variance, Bonferroni post-test).
Impairment in lysosomal acidification can affect the processing and function of lysosomal enzymes. Therefore, we next examined the levels of cathepsin B, cathepsin L, and dipeptidyl peptidase VII (DPP7) as these are lysosomal proteases previously shown to be decreased in Tmem106b null mice.6 The (p.Asp252Asn) variant results in a decrease in the mature form of cathepsin B protein and, importantly, a concomitant decrease in the activity levels of this enzyme in patient cells (figure, C and D). This reduction is predicted to not only affect the degradation of cathepsin B protein substrates but also the regulation of the T-cell transcription factor (TFEB).7 As cathepsin B suppresses lysosomal number in a TFEB-dependent manner,7 it is likely that the reduction in cathepsin B activity contributes to the increased lysosome number observed. Patient cells also displayed an accumulation of both intermediate and mature forms of cathepsin L and no change in DPP7 levels (figure, C, data not shown). Taken together, these results show that the (p.Asp252Asn) substitution affects multiple aspects of lysosome biology.
Discussion
TMEM106B is a structural component of the lysosomal membrane and, importantly, plays a role in lysosome acidification.6 The acidity of the lysosome is important to mediate multiple aspects of lysosomal function, including maintaining active lysosome enzymes, directing the maturation of endosomes, and maintaining intralysosomal calcium levels.8 Therefore, impaired acidification in our patient cells is predicted to have noteworthy implications for lysosome function. Lysosomes are required for the generation of myelin during the development of the CNS. Notably, this includes trafficking of PLP1 protein, the main component of myelin, from the late endosome/lysosome to the cell membrane for exocytosis in oligodendrocytes.8 In addition, disruption of endosome-lysosome biogenesis caused by mutations in another lysosomal-associated gene, vacuolar protein sorting 11, causes another form of HLD.5 Tmem106b null mice are phenotypically normal, and numerous loss-of-function variants in TMEM106B are reported in the gnomAD database, supporting a non-haploinsufficiency mechanism underlying TMEM106B-related HLD. Further studies are required to elucidate the specific dominant negative or gain-of-function effect of the (p.Asp252Asn) mutation that results in decreased lysosomal acidification. Also, potential effects on other HLD-related proteins, such as PLP1, which would have direct consequences on myelin formation need to be examined. We appreciate that these experiments are based on a sample from a single individual and that additional studies from other (p.Asp252Asn)-positive individuals are necessary to confirm these findings. Nevertheless, this report provides an important first step in defining the role of TMEM106B in lysosome function in HLD. TMEM106B now joins the catalogue of essential lysosomal proteins implicated in human neurologic disease.
Acknowledgments
The authors would like to thank the patient and family; without whom this work would not be possible. The authors also wish to acknowledge Dr. Wendy Mears for her tissue culture expertise. This work was supported by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Génome Québec, and the Children's Hospital of Eastern Ontario Foundation.
Author contributions
Y. Ito: study concept and design, acquisition, analysis, and interpretation of data, and manuscript preparation. T. Hartley: critical revision of manuscript for intellectual content. S. Baird: acquisition of data and critical revision of manuscript for intellectual content. S. Venkateswaran: critical revision of manuscript for intellectual content. C. Simons: critical revision of manuscript for intellectual content. N.I. Wolf: critical revision of manuscript for intellectual content. K.M. Boycott: critical revision of manuscript for intellectual content. D.A. Dyment: analysis and interpretation of data, manuscript preparation, and study supervision. K.D. Kernohan: analysis and interpretation of data, manuscript preparation, and study supervision.
Study funding
This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Génome Québec, and Children's Hospital of Eastern Ontario Foundation.
Disclosure
Y. Ito, T. Hartley, S. Baird, S. Venkateswaran, and C. Simons report no disclosures. N.I Wolf has served on scientific advisory boards for European Leukodystrophy Association (ELA) and Mission Massimo Foundation (both without compensation); has received travel funding from ELA; has served on the editorial boards of Neuropediatrics and Neurology; has received research support from Hersenstichting, Metakids, the Stofwisselkracht Foundation, the M.O. Knip Foundation, and Yasho's Leukodystrophy Foundation; and holds stock/stock options in Aer Beatha. K.M. Boycott, D.A. Dyment, and K.D. Kernohan report no disclosures. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Simons C, Dyment D, Bent SJ, et al. . A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain 2017;140:3105–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Kubisiak T, Ji H, Xiao J, Wang J, Burmeister M. The recurrent mutation in TMEM106B also causes hypomyelinating leukodystrophy in China and is a CpG hot spot. Brain 2018;141:e36. [DOI] [PubMed] [Google Scholar]
- 3.Nicholson AM, Zhou X, Perkerson RB, et al. . Loss of Tmem106b is unable to ameliorate frontotemporal dementia-like phenotypes in an AAV mouse model of C9ORF72 -repeat induced toxicity. Acta Neuropathol 2018;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholson AM, Rademakers R. What we know about TMEM106B in neurodegeneration. Acta Neuropathol 2016;132:639–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pouwels PJW, Vanderver A, Bernard G, et al. . Hypomyelinating leukodystrophies: translational research progress and prospects. Ann Neurol 2014;76:5–19. [DOI] [PubMed] [Google Scholar]
- 6.Klein ZA, Takahashi H, Ma M, et al. . Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 2017;95:281–296.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Man SM, Kanneganti TD. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy 2016;12:2504–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Yuan Y, Su W, Gu Y, Chen G. The roles of lysosomal exocytosis in regulated myelination. J Neurol Neuromed 2016;1:4–8. [Google Scholar]