Abstract
Background:
Convolutional neural networks (CNNs) are advanced artificial intelligence algorithms well suited to image classification tasks with variable features. These have been used to great effect in various real-world applications including handwriting recognition, face detection, image search, and fraud prevention. We sought to retrain a robust CNN with coronal computed tomography (CT) images to classify osteomeatal complex (OMC) occlusion and assess the performance of this technology with rhinologic data.
Methods:
The Google Inception-V3 CNN trained with 1.28 million images was used as the base model. Preoperative coronal sections through the OMC were obtained from 239 patients enrolled in 2 prospective CRS outcomes studies, labelled according to OMC status, and mirrored to obtain a set of 956 images. Using this data, the classification layer of Inception-V3 was retrained in Python using a transfer learning method to adapt the CNN to the task of interpreting sinonasal CT images.
Results:
The retrained neural network achieved 85% classification accuracy for OMC occlusion, with a 95% confidence interval for algorithm accuracy of 78–92%. Receiver operating curve analysis on the test set confirmed good classification ability of the CNN with an area-under-the-curve of 0.87, significantly different than both random guessing and a dominant classifier that predicts the most common class. (p<0.0001).
Conclusions:
Current state-of-the-art CNNs may be able to learn clinically relevant information from 2-dimensional sinonasal CT images with minimal supervision. Future work will extend this approach to 3-dimensional images in order to further refine this technology for possible clinical applications.
Keywords: Sinusitis, Chronic Disease, Machine Learning, Neural Network
INTRODUCTION
Chronic rhinosinusitis (CRS) is a heterogeneous group of diseases characterized by persistent inflammation of the paranasal sinuses. While symptoms are essential for accurate diagnosis, clinical testing tools such as computed tomography (CT) and nasal endoscopy help otolaryngologists confirm their suspicions and appropriately select treatment options. As such, CT imaging represents an integral part of the workup and management of chronic sinusitis. Most patients electing to proceed with sinus surgery receive a preoperative CT scan1, and a recent ESS appropriateness study defined the presence of inflammation on CT as a minimum criterion for surgical intervention2. Due to the high rate of recurrent disease and need for revision3, some patients may also receive multiple CT scans of the sinuses over their lifetime. Thanks to modern data archiving, these images represent a permanent record of the inflammatory state of these refractory CRS patients and can be used to potentially observe changes over time and understand disease evolution in this subset of patients. Unfortunately, our ability to capture clinically meaningful information from these scans for research purposes is limited by the need for highly trained observers to document features on CT scans in a labor-intensive way. The Lund-Mackay score4 and similar CT scoring systems5,6 were developed to reduce this burden, but their semi-quantitative nature may neglect important information7. As a result, the literature regarding correlation of these CT scoring systems and patient-reported outcomes has been mixed8. Interestingly, more recent studies using semi-automated volumetric analysis of CT scans have shown a higher degree of association with symptoms9,10, suggesting that more comprehensive methods of CT image analysis may be important for linking symptoms with objective measures of inflammation. These methods, however, are time-consuming and may not be practical for routine clinical use. The rise of artificial intelligence (AI) and deep learning11 however, provides an intriguing method to possibly solve this problem and leverage the information within these scans for the purposes of improving our understanding of CRS.
Convolutional neural networks (CNNs) represent a particular form of AI that is well suited to the task of computer vision12,13. Briefly, a CNN is a biologically-inspired computer algorithm that uses thousands of simple units, modeled after neurons, that take an input from one layer of units, perform a mathematical transformation, and feed the output to the next set of neurons. Similar to their biological counterparts, while each individual neuron is simple to understand, structured networks of them have demonstrated complicated, emergent properties that are useful for pattern-recognition tasks9,10. These algorithms have been used to great effect in multiple applications11, most notably the ImageNet challenge, an annual international computer vision competition that asks computer scientists to develop algorithms to classify a given set of images into over 1,000 categories. For the past several years, CNN-based AIs have been at the top of the ImageNet rankings14. As this technology matures, its role in medical applications is being investigated in areas that may benefit from automated classification of images. One promising study demonstrated that the performance of a CNN in diagnosing benign and malignant skin lesions from photographs alone was equivalent and, in some cases, superior to a group of board-certified dermatologists15. Another was able to correctly classify bacteria seen on gram-stain slides with 94% accuracy16. Within otolaryngology, two pilot studies have used CNNs to detect cancer cells17 and endolymphatic hydrops in images from mice18. In the present study, we sought to use clinical data from two prospective CRS outcomes studies to assess the performance of CNNs in a rhinologic application, establish a proof-of-concept, and chart possible avenues of future research.
MATERIALS AND METHODS
Study Design and Inclusion Criteria
Findings from both prospective clinical investigations pooled for this study have been previously published19–23. Subjects in both studies were adults 18 years of age or older with CRS diagnosed by fellowship trained rhinologists at Vanderbilt University Medical Center (VUMC) and Oregon Health and Science University (OHSU) using current American Academy of Otolaryngology guidelines24 and the International Consensus Statement on Allergy and Rhinology25. Patients at OHSU were enrolled between March 2011 and March 2015 while patients at VUMC were enrolled between March 2014 and November 2017. The Institutional Review Board (IRB) at each enrollment site provided study authorization, annual review, and continual safety monitoring. Informed written consent was obtained in English per good clinical practice guidelines outlined by the International Conference on Harmonization26
Patients in both studies were eligible for enrollment if they had previously completed, at a minimum, one course (14 days) of culture-directed or empiric antibiotics, either corticosteroid nasal sprays (21 days) or systemic corticosteroid therapy (5 days), and daily saline solution irrigation (~240ml). For the purposes of the present study, only patients undergoing endoscopic sinus surgery for management of CRS were included.
Exclusion criteria
Patients at VUMC were excluded from enrollment if they had a history of autoimmune disease, cystic fibrosis, steroid dependency, or oral steroid use within 4 weeks of surgery. A similar limitation was not a part of the study protocol at OHSU; however, as the present study does not seek to predict outcomes of treatment, all patients enrolled at OHSU with archived preoperative computed tomography were included.
Computed Tomography data
Each patient received a fine-cut CT scan of the sinuses with <1 mm slices performed in the axial plane within 3 months of surgery. Due to limitations with current generation CNNs, a representative 2-dimensional coronal section through the osteomeatal complex (OMC) was recorded for each patient’s CT. To standardize across patients and imaging techniques as much as possible, the plane of interest was defined as the first coronal section through the natural maxillary os immediately posterior to the nasolacrimal duct. OMC occlusion was selected as the clinical feature of interest due to its ease of visualization on coronal CT as well as its potential role in the pathogenesis and severity of CRS27,28.
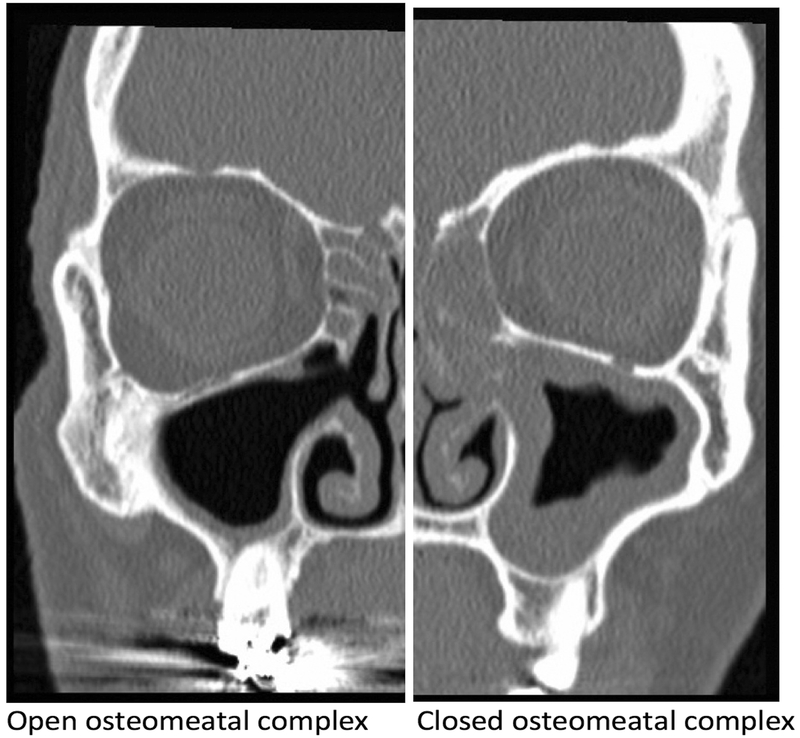
Images were captured in JPEG format and split into left and right hemi-slices using Preview (Apple Inc, Cupertino, CA) to isolate the osteomeatal complex of each side into a separate imaging file. Each image was then flipped along the vertical axis (mirrored) to provide additional training data. This is a common technique in machine learning29 and can be performed by the neural network itself during the training process, although the computational cost is quite high. Given the limits of our system, we elected to perform mirroring separately to allow more rapid training. After collection of our training set of 956 images, each OMC was graded as “open” or “closed” by a fellowship-trained rhinologist. An “open” OMC was defined as one with a fully aerated path from the nasal cavity into the maxillary antrum, while a “closed” OMC was defined as any scan with OMC inflammation completely obstructing the path into the maxillary sinus (Figure 1).
Figure 1:
Open vs. closed osteomeatal complex examples on coronal CT slices
CNN Training and Validation
The base network used for this study was Inception-V3 (Google, Mountain View, CA), a CNN trained using 1.28 million images in 1,000 different categories13. Using a technique known as transfer learning30, the final classification layer of Inception-V3 was retrained using the training data collected previously to classify CT images into either an “open” or “closed” OMC category. A high-level overview of the architecture behind Inception-V3 follows. Each image is first resized into a 299×299 pixel representation and broken down into an array of numbers corresponding to the intensity of each pixel. This creates a mathematical representation of each image that can be processed through the network. Each subsequent node within the neural network learns to recognize progressively complex shapes, which then are aggregated into several categories. For example, the first layer may look for edges, the second for lines, the third for shapes, etc. Eventually, the network is able to recognize groups of features that collectively form a learned structure. For example, the presence of two eye objects, a nose object, and a mouth object would lead the algorithm to detect a face.
The final layer of the neural network is the classification layer. Based on the nodal activations that have occurred previously, each potential output has an associated probability based on how many features the network has detected. For example, when shown an unknown image of a plant, a network designed to distinguish between images of plants and animals would likely have a high number of activations in nodes that detect higher-order structures like leaves and petals, and thereby assign a higher probability to the plant category.
In the present study, the previously trained architecture of Inception-V3 was kept intact except for the final classification layer, which was retrained using these CT images to distinguish between an “open” OMC and a “closed” OMC. This allows the network to leverage lower level information learned from a larger dataset and transfer this to a newly defined task – also known as transfer learning. To understand this, imagine teaching a new student how to read a CT scan. One would already assume that they knew how to recognize shapes, colors, etc., and build on that knowledge to recognize the features of importance on the CT. Similarly, transfer learning uses the prior knowledge the CNN has learned from the original set of 1.28 million images, but teaches the network to apply this to the new task of OMC classification.
This transfer learning method was executed in Python using TensorFlow (Google, Mountain View, CA), an open-source environment created for the purposes of creating and training artificial neural networks. 80% of the training set was used for the learning process, which was set for 100,000 training steps. This determines the number of iterations the algorithm is run prior to finalizing the model. For each training step, the model’s prediction for a set of images is compared to the actual value, and depending on the amount of error, the weights of each node are updated for the next run to continually decrease the error with the goal of obtaining a “perfect” classifier. 10% of the images were set aside and used for validation and adjustment of the learning rate, or the amount each node weight is modified for each step. This rate is arbitrarily set and modified to optimize test accuracy in the validation set, so one can consider the validation set as part of the learning process. The final 10% of the data was kept as a clean test data set on which to assess the performance of the model after training was completed.
Data Management and Analysis
Study data was stripped of all protected health information and each patient was assigned a unique study identification number. Data collection was performed using a HIPAA compliant, closed-environment database (Access, Microsoft Corp., Redmond, WA). Post-hoc analysis of model performance was performed using R 3.2 (R Foundation for Statistical Computing, Vienna, Austria). Accuracy of the model was assessed using the percentage of correct responses given by the algorithm when viewing test set images, with 95% confidence intervals calculated using Wald’s test. Output probabilities from the model on each test image were then used to construct a receiver operating curve to assess the classification ability of the algorithm. An area-under-the-curve (AUC) analysis was then performed to grade the predictive ability of the neural network, and confidence intervals were calculated using DeLong’s method31. Two comparison models were constructed to compare CNN classifier performance: a random-guessing classifier, which randomly picks between open/closed, and a dominant classifier, which always chooses the option with the highest prevalence in the dataset (closed). This is done because any classifier which picks the dominant class every time will have an accuracy equivalent to prevalence of that class in the dataset.
RESULTS
Patient characteristics of the pooled study cohort are shown in Table 1.
Table 1:
Baseline patient characteristics of pooled study cohort (n=239)
| Characteristics: | N (%) |
|---|---|
| Sex | |
| Males | 110 (46%) |
| Females | 129 (54%) |
| Prior surgery | |
| Yes | 83 (35%) |
| No | 156 (65%) |
| OMC Status (per side) | |
| Open | 155 (32%) |
| Closed | 323 (68%) |
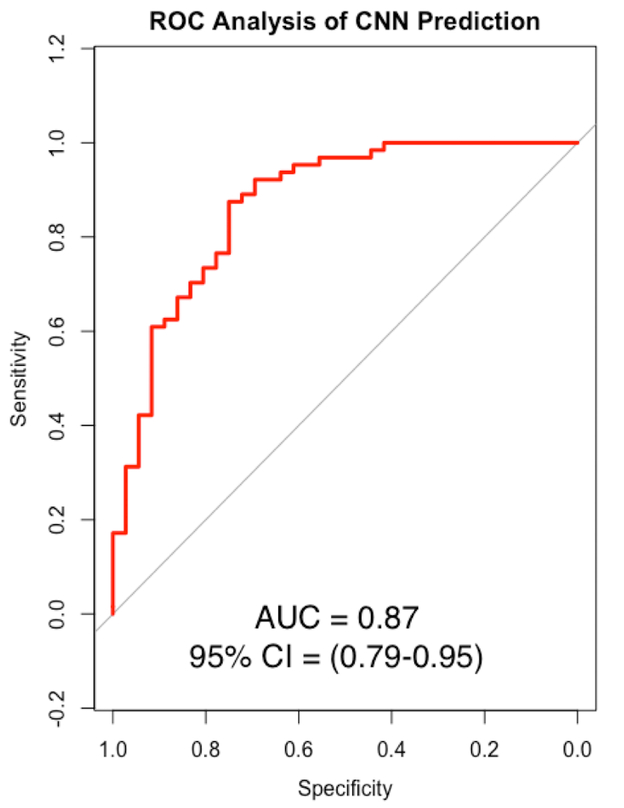
After training the neural network for 100,000 training steps at a learning rate of 0.005, the final accuracy on the test set was 85%. This learning rate was selected due to optimal classification performance after several trial runs on the validation set. Lower rates tended to increase algorithm run time without any added increase in accuracy, and higher rates appeared to decrease accuracy. The 95% confidence interval for algorithm accuracy was 78–92%. Receiver operating curve analysis on the test set showed good classification ability of the CNN with a final area-under-the-curve (AUC) of 0.87 (Figure 3). This was significantly different when compared against the random guessing model (AUC = 0.48, p<0.0001) and the dominant classifier model (AUC = 0.5, p<0.0001).
Figure 3: Receiver operating curve (ROC) analysis of algorithm performance.
ROC, Receiver operating characteristic; AUC, area under the curve; CI, confidence interval (DeLong). The red line represents the algorithm, while the gray line represents the baseline dominant classifier, which predicts the most commonly occurring class in the dataset (p < 0.0001).
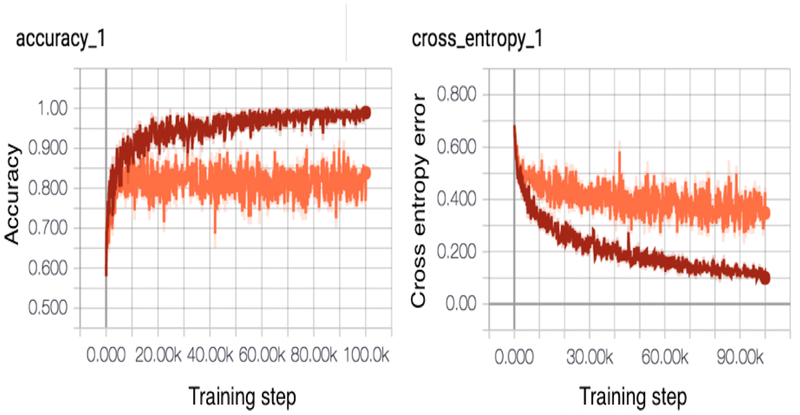
Real time training graphs generated within TensorFlow for the model are shown in Figure 2. The red line indicates performance on the training set, while the orange line indicates performance on the validation set. Note on the left graph that, by the end of training, the algorithm achieves near-perfect accuracy on the training set, but is unable to generalize this to the validation set. The right image shows continually decreasing cross-entropy error, which is a mathematical measure of the difference between the predicted output for an image and the actual value. Towards the end of training, error improvement of the validation set (orange) appears to plateau, suggesting that further training steps at this learning rate are unlikely to improve performance.
Figure 2: Real-time training graphs of training and validation set accuracy/cross-entropy.
Training set curve is visualized in red; validation set in orange. Note that the model achieves perfect performance on the training set, but is unable to fully generalize this success to the validation set. Cross-entropy error is the quantity the network attempts to minimize after each iteration of training. Improvements in cross-entropy on the validation set plateau during the last half of training; thus, the network is “learning less” with each training cycle.
DISCUSSION
This study is the first investigation into the feasibility of using CNNs to automatically identify clinically relevant information from sinonasal CT scans. Surprisingly, even with a limited amount of data, our neural network seems to have the ability to learn and correctly classify the vast majority of cases presented to it without any prior knowledge of the task at hand. While the performance of this AI is no match for a well-trained clinician, the success of the neural network given the circumstance is remarkable. An equivalent human example would involve asking an individual with no prior medical knowledge to learn the features of the OMC from a set CT images labeled “open” and “closed” well enough to consistently identify the same features on an unknown set of images. With further improvements in accuracy from training on larger datasets, an AI similar to the one presented could be useful in extracting clinical information from medical imaging archives in an efficient matter. As these algorithms evolve, it is possible that a properly trained AI could one day learn to evaluate CT scans in a manner similar to a practitioner, and perhaps even detect subtle features or patterns on these tests that are beyond our own current comprehension. As it stands, however, substantial work is needed prior to achieving the goal of human-level performance for this application.
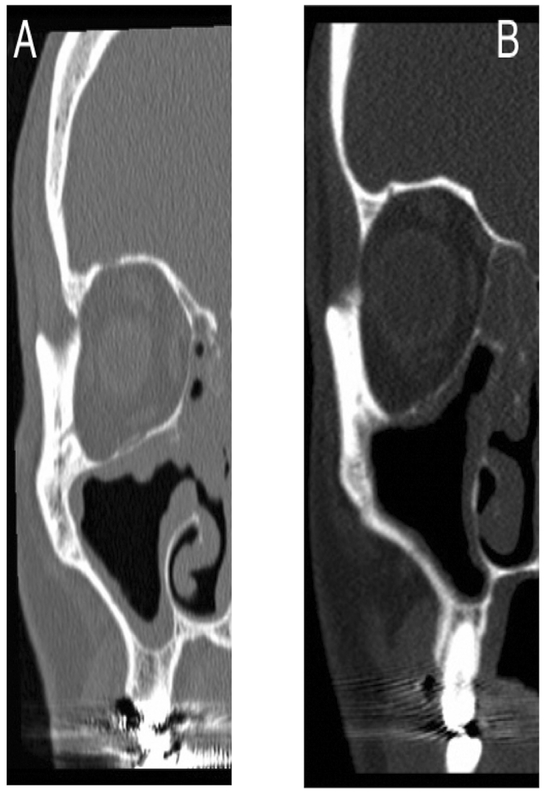
Until then, one barrier to the widespread adoption and acceptance of neural networks in medicine is our inability to understand exactly how it arrives at the correct conclusion32. Due to the complex behavior of CNNs, we are only able to rate the performance of the network using the final answer it generates instead of knowing its “thought process.” Compare this to a well-established statistical technique like logistic regression, where every variable and its precise contribution to the final output is known through a closed-form solution. A CNN, on the other hand, is similar to a medical student who correctly answers every question asked during a rotation. It is thereby assumed that the student has a foundational understanding of the content, but it is impossible to know definitively unless a situation arises where the wrong answer is given to a query, in which case we can correct the error. For our model, some examples of misclassified images are shown below (Figure 4) and give some insight into the network’s logic. The image in A was technically an “open” OMC per our a-priori definition, but a reasonable third-party could argue that it is functionally closed. Similarly, in image B, the rough shape of a patent outflow tract is present although it is visibly not connected to the nasal passages.
Figure 4: Comparison of images misclassified by the CNN.
A: Image of “open” OMC incorrectly classified as “closed”. B: Image of “closed” OMC incorrectly classified as “open.”
Despite these errors, the results from our ROC analysis show that valuable information is learned by the algorithm during training, as seen by the statistically significant differences in AUC (0.87) when compared to the random model (0.48) or the dominant classifier (0.5), which adjusts for class imbalance. An AUC range between 0.8–0.9 is generally considered to be representative of good to excellent classifier performance33.
Several limits of this study should be noted. First, all the scans used were pathological CTs of patients with CRS. Therefore, the performance metrics reported are only applicable to a diseased cohort; the performance of the CNN on CT scans of patients without CRS is unknown. We also only had the ability to analyze a single 2-dimensional CT image due the current state of CNN technology, which does not reflect the way a trained human practitioner would analyze a scan. There also is the potential issue of disagreement regarding the “ground truth” of each CT given that a single 2-dimensional image cannot adequately capture the complexity of the OMC. This was partially mitigated by setting clear, a-priori rules for an “open” and “closed” OMC and having scans evaluated by fellowship-trained attending rhinologists. Additionally, while the dataset is a moderate size compared to many studies in rhinology, it pales in comparison the millions of data points that are routinely used for training computer vision algorithms. This is reflected in our training graphs, which show plateauing of validation accuracy around the 80–90th percentile even as training accuracy approaches 100% – a sign that the network is overfitting the training set and could benefit from additional data. Ideally a dedicated CNN would be developed for reading CT scans of the sinuses using hundreds of thousands of labelled CT images, but in the meantime, the transfer-learning method seems to allow for robust results. Finally, even with a perfect AI, there are probably limits to the utility of CT scans alone as predictive tools for CRS given the weak correlations between CT scoring and symptoms8. We therefore envision AI as a potential way to incorporate information from CT scans into a comprehensive predictive model that includes medical history, patient reported outcomes measures, endoscopy images, and cytokine19/microbiome profiles, although many technological advances are likely needed before this vision can become a clinical reality.
CONCLUSIONS
Current state-of-the-art CNNs may be able to learn clinically relevant information from 2-dimensional sinonasal CT images with minimal supervision. Future work will extend this approach to 3-dimensional CT in order to further refine this technology for possible clinical applications.
Acknowledgments
Financial Disclosures: Timothy L. Smith is supported by a grant for this investigation from the National Institute on Deafness and Other Communication Disorders (NIDCD), one of the National Institutes of Health, Bethesda, MD., USA (R01 DC005805; PI/PD: TL Smith). Public clinical trial registration (www.clinicaltrials.gov) ID# NCT01332136. Justin H. Turner is supported by NIH RO3 DC014809, L30 AI113795 and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. These funding organizations did not contribute to the design or conduct of this study; collection, management, analysis, or interpretation of the data; preparation, review, approval or decision to submit this manuscript for publication. Naweed Chowdhury is a consultant for OptiNose, Inc. There are no relevant financial disclosures for Rakesh K. Chandra.
Footnotes
Potential Conflicts of Interest: None
REFERENCES
- 1.Batra PS, Setzen M, Li Y, Han JK, Setzen G Computed tomography imaging practice patterns in adult chronic rhinosinusitis: survey of the American Academy of Otolaryngology-Head and Neck Surgery and American Rhinologic Society membership. Int Forum Allergy Rhinol. 2015;5(6):506–512. doi: 10.1002/alr.21483. [DOI] [PubMed] [Google Scholar]
- 2.Rudmik L, Soler ZM, Hopkins C, et al. Defining appropriateness criteria for endoscopic sinus surgery during management of uncomplicated adult chronic rhinosinusitis: a RAND/UCLA appropriateness study. Int Forum Allergy Rhinol. 2016;6(6):557–567. doi: 10.1002/alr.21769. [DOI] [PubMed] [Google Scholar]
- 3.Philpott C, Hopkins C, Erskine S, et al. The burden of revision sinonasal surgery in the UK--data from the Chronic Rhinosinusitis Epidemiology Study (CRES): a cross-sectional study. BMJ Open. 2015;5(4):e006680–e006680. doi: 10.1136/bmjopen-2014-006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund VJ, Mackay IS Staging in rhinosinusitus. Rhinology. 1993;31(4):183–184. [PubMed] [Google Scholar]
- 5.Okushi T, Nakayama T, Morimoto S, et al. A modified Lund-Mackay system for radiological evaluation of chronic rhinosinusitis. Auris Nasus Larynx. 2013;40(6):548–553. doi: 10.1016/j.anl.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Zinreich SJ Imaging for staging of rhinosinusitis. Ann Otol Rhinol Laryngol Suppl. 2004;193:19–23. [DOI] [PubMed] [Google Scholar]
- 7.Bhandarkar ND, Sautter NB, Kennedy DW, Smith TL Osteitis in chronic rhinosinusitis: a review of the literature. Int Forum Allergy Rhinol. 2013;3(5):355–363. doi: 10.1002/alr.21118. [DOI] [PubMed] [Google Scholar]
- 8.Hopkins C, Browne JP, Slack R, Lund V, Brown P The Lund-Mackay staging system for chronic rhinosinusitis: how is it used and what does it predict? Otolaryngol Head Neck Surg. 2007;137(4):555–561. doi: 10.1016/j.otohns.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Soler ZM, Pallanch JF, Sansoni ER, et al. Volumetric computed tomography analysis of the olfactory cleft in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol. 2015;5(9):846–854. doi: 10.1002/alr.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim S, Ramirez MV, Garneau JC, et al. Three-dimensional image analysis for staging chronic rhinosinusitis. Int Forum Allergy Rhinol. 2017;7(11):1052–1057. doi: 10.1002/alr.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeCun Y, Bengio Y, Hinton G Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 12.Krizhevsky A, Sutskever I & Hinton G (2012). Imagenet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 25, 1097–1105. 2012. [Google Scholar]
- 13.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z Rethinking the Inception Architecture for Computer Vision. December 2015. [Google Scholar]
- 14.Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 15.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith KP, Kang AD, Kirby JE Automated Interpretation of Blood Culture Gram Stains by Use of a Deep Convolutional Neural Network. Bourbeau P, ed. J Clin Microbiol. 2017;56(3):e01521–17. doi: 10.1128/JCM.01521-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halicek M, Lu G, Little J V, et al. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J Biomed Opt. 2017;22(6):60503. doi: 10.1117/1.JBO.22.6.060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu GS, Zhu MH, Kim J, Raphael P, Applegate BE, Oghalai JS ELHnet: a convolutional neural network for classifying cochlear endolymphatic hydrops imaged with optical coherence tomography. Biomed Opt Express. 2017;8(10):4579–4594. doi: 10.1364/BOE.8.004579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner JH, Chandra RK, Li P, Bonnet K, Schlundt DG Identification of Clinically Relevant Chronic Rhinosinusitis Endotypes using Cluster Analysis of Mucus Cytokines. J Allergy Clin Immunol. February 2018. doi: 10.1016/j.jaci.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Chandra RK, Li P, Hull BP, Turner JH Olfactory and middle meatal cytokine levels correlate with olfactory function in chronic rhinosinusitis. Laryngoscope. February 2018. doi: 10.1002/lary.27112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdhury NI, Mace JC, Bodner TE, et al. Investigating the Minimal Clinically Important Difference for SNOT-22 Symptom Domains in Surgically Managed Chronic Rhinosinusitis. Int Forum Allergy Rhinol. 2017;Early View. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury NI, Mace JC, Smith TL, Rudmik L What drives productivity loss in chronic rhinosinusitis? A SNOT-22 subdomain analysis. Laryngoscope. June 2017. doi: 10.1002/lary.26723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauser LJ, Chandra RK, Li P, Turner JH Role of tissue eosinophils in chronic rhinosinusitis-associated olfactory loss. Int Forum Allergy Rhinol. 2017;7(10):957–962. doi: 10.1002/alr.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marple BF, Stankiewicz JA, Baroody FM, et al. Diagnosis and Management of Chronic Rhinosinusitis in Adults. Postgrad Med. 2009;121(6):121–139. doi: 10.3810/pgm.2009.11.2081. [DOI] [PubMed] [Google Scholar]
- 25.Orlandi RR, Kingdom TT, Hwang PH, et al. International Consensus Statement on Allergy and Rhinology: Rhinosinusitis. Int Forum Allergy Rhinol. 2016;6(S1):S22–S209. doi: 10.1002/alr.21695. [DOI] [PubMed] [Google Scholar]
- 26.International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. ICH Harmonised Tripartite Guideline. Guideline for Good Clinical Practice E6 (R1). Available at: http://www.ich.org/products/guidelines/.
- 27.Kennedy DW Functional endoscopic sinus surgery. Technique. Arch Otolaryngol. 1985;111(10):643–649. [DOI] [PubMed] [Google Scholar]
- 28.Chandra RK, Pearlman A, Conley DB, Kern RC, Chang D Significance of osteomeatal complex obstruction. J Otolaryngol Head Neck Surg. 2010;39(2):171–174. [PubMed] [Google Scholar]
- 29.Simonyan K, Zisserman A Very Deep Convolutional Networks for Large-Scale Image Recognition. September 2014. [Google Scholar]
- 30.Pan SJ, Yang Q A Survey on Transfer Learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 31.DeLong ER, DeLong DM, Clarke-Pearson DL Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 32.Tu JV Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol. 1996;49(11):1225–1231. doi: 10.1016/S0895-4356(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 33.Youngstrom EA A Primer on Receiver Operating Characteristic Analysis and Diagnostic Efficiency Statistics for Pediatric Psychology: We Are Ready to ROC. J Pediatr Psychol. 2014;39(2):204–221. doi: 10.1093/jpepsy/jst062. [DOI] [PMC free article] [PubMed] [Google Scholar]