Abstract
While the number of proteins effectively targeted for post-translational degradation by PROTAC has grown steadily, the number of E3 ligases successfully exploited to accomplish this has been limited to the few for which small molecule ligands have been discovered. Although the E3 ligase MDM2 is bound by the nutlin class of small molecule ligands, there are few nutlin-based PROTAC. Since a nutlin-based PROTAC should both knockdown its target protein and upregulate the tumor suppressor, p53, we examined the ability of such a PROTAC to decrease cancer cell viability. A nutlin-based, BRD4-degrading PROTAC, A1874, was able to degrade its target protein by 98% with nanomolar potency. Given the complementary ability of A1874 to stabilize p53, we discovered that the nutlin-based PROTAC was more effective in inhibiting proliferation of many cancer cell lines with wild type p53 than was a corresponding VHL-utilizing PROTAC with similar potency and efficacy to degrade BRD4. This is the first report of a PROTAC in which the E3 ligase ligand and targeting warhead combine to exert a synergistic antiproliferative effect. Our study highlights the untapped potential that may be unlocked by expanding the repertoire of E3 ligases that can be recruited by PROTAC.
Keywords: nutlin, BRD4, PROTAC, degradation, p53
Introduction
Recent years have seen post-translational protein degradation harnessed to target and eliminate many cellular proteins of interest to both basic researchers and clinicians. This has been accomplished through development of a variety of heterobifunctional molecules that tether together a ligand that binds the protein of interest with another ligand that engages an E3 ubiquitin ligase (1–3). Upon introduction into living cells, these chimeric molecules facilitate complex formation of the protein of interest with an E3 ligase, resulting in ubiquitination of the former and its subsequent degradation by the 26S proteasome: hence, these heterobifunctional molecules are called “PROteolysis-TArgeting Chimeras”, or “PROTACs” (4,5).
From a basic research standpoint, PROTACs serve as an effective tool enabling small molecule-based target protein knockdown that is more tunable and reversible than some modalities (e.g. RNAi or genetic knockout). Moreover, unlike other small molecule-based strategies for regulating protein levels post-translationally (6,7), PROTACs require no genetic modification of the cells. From a clinical standpoint, PROTACs permit the engagement and elimination of both established drug target proteins as well as the so-called “undruggable” proteome. Because PROTACs can bind to any available surface feature of its protein target to mediate its dimerization with an E3 ligase and the resultant ubiquitination (8), PROTACs are not restricted to engaging their intended target proteins only at their cognate active sites. This is an advantage for PROTAC therapeutics since many disease-related proteins (scaffolding proteins, transcription factors, etc.) lack a tractable active site through which a conventional small molecule inhibitor could work – hence the designation of such disease-related proteins as traditionally “undruggable” (9). Furthermore, because transient interaction with the target protein is sufficient for a PROTAC to commit it for degradation, PROTACs can act catalytically and be effective at lower concentrations that those necessary for the sustained, maximum target occupancy that traditional small molecule inhibitors require to be effective (10,11). Thus, in both contexts – the research lab and the clinic -- PROTACs have received recent attention, leading to the generation of PROTACs that degrade cyclin-dependent kinases (12), lipid kinases (13), peptidases (4), protein isomerases (5), scaffolding proteins (13), transcription factors (3,11,14), protein kinases (1,10,15–18) and epigenetic regulating proteins (2,19).
Beyond expanding the repertoire of proteins successfully degraded, research also led to a better understanding of the finer mechanistic details concerning how PROTACs work and expanding the mechanistic paradigm itself. It is now known that the most effective PROTACs facilitate the formation of protein-protein interactions between target protein and E3 ligase (20). The importance of these cooperative interactions is underscored by the observation that PROTACs with only modest target affinity can nonetheless induce potent target degradation (18). Moreover, design variations led to the creation of PROTACs that bind irreversibly to either of its recruited proteins via an orthogonal ligand (21,22), such that target degradation then depends on only a single binding event to permit an even higher degree of targeting efficiency as well as specificity. Other structural refinements led to the development of PROTACs whose target degradation is conditional on the activation of specific signaling pathways in the cell (13), as well as PROTACs that cause the degradation of the harnessed E3 ligase itself (23).
Paradoxically, while the scope of targeted proteins and refinement of PROTAC functionality has been wide-ranging, the selection of E3 ligases through which PROTACs induce target ubiquitination has remained limited. Although 600 E3 ligases are encoded by the human genome (24), only three have been routinely used for PROTAC-mediated target ubiquitination: von Hippel Lindau, or VHL; cereblon; and the “inhibitor of apoptosis protein”, or IAP. While the first PROTAC (4) worked through recruitment of the E3 ligase, ß-TRCP, it did so by utilization of a phosphopeptide as the E3-binding component and as such was not cell permeable nor projected to be stable in vivo. A recent study proposes that, despite the past emphasis on VHL, cereblon and IAP, many other E3 ligases should be amenable for utilization by PROTACs, including the already mentioned ß-TRCP, as well as parkin and Siah1 (22). The major hurdle for exploiting these E3 ligases is the lack of corresponding small molecule ligands for them that could be incorporated into the PROTAC structure.
In 2008 our group published the first all-small molecule PROTAC, which degrades the androgen receptor through its recruitment to the E3 ligase, MDM2 (25). Previous PROTACs, including the ones that worked through VHL recruitment, had used peptide sequences to deliver their target proteins to an E3 ligase for their ubiquitination. By coupling the small molecule MDM2 inhibitor, nutlin 3, with the androgen receptor antagonist, hydroxyflutamide, an entirely non-peptidic entity was created that induced the degradation of the androgen receptor, albeit at micromolar concentrations. Further inquiry into the use of MDM2 has lagged, in part due to the generation of VHL-binding small molecules (26), the discovery that phthalamide-based ligands (e.g. thalidomide) bind to cereblon (27), and the availability of bestatin methyl ester and other ligands for IAP (28,29). These molecules have been used to create all-small molecule PROTACs that induce target degradation at sub-micromolar and even sub-nanomolar concentrations. However, in light of the potential wide-ranging amenability of E3 ligases for repurposing into resources for PROTAC-mediated protein degradation, we reasoned that renewed effort concerning nutlin-based PROTACs was worthwhile considering that, among the E3 ligases currently used for PROTACs, MDM2 stands out in that its endogenous substrate, the tumor suppressor p53, plays a crucial tumor suppressor role. Indeed, the ability to manipulate p53 levels using nutlins holds therapeutic promise in the field for cancer treatment (30,31).
In this study, we present evidence showing not only that MDM2 is indeed amenable to being used for nanomolar-potency PROTAC mediated target degradation, but moreover a nutlin-based PROTAC retains the p53-stabilizing activity of the parent molecule allowing a MDM2-recruiting PROTAC to be more active against certain cancers than a counterpart VHL-utilizing PROTAC directed against the same target protein.
Methods and Materials
Chemical syntheses
See Supplementary Data for synthetic schemes and validations of PROTACs A1874, A1875 and A743.
Reagents
Antibodies against BRD4 (cat. no. 13440), GAPDH (cat. no. 2118) and p21CIP1/WAF1 (cat. no. 2947) were purchased from Cell Signaling Technology (Danvers, MA). Antibody recognizing c-Myc (cat. no. ab32072) was from Abcam (Cambridge, MA) and antibody for p53 (cat. no. OP43) was obtained from EMD Millipore (Burlington, MA). HRP-linked anti-mouse (cat. no. NA931V) and anti-rabbit (cat. no. NA934V) secondary antibodies were from GE Life Sciences (Marlborough, MA). JQ1 (cat. no. HY-13030) and idasanutlin (cat. no. HY-15676) were obtained from MedChem Express (Monmouth Junction, NJ) and all other reagents were procured from Sigma (St. Louis, MO) unless otherwise specified.
Cell lines
HT29 colon cancer cells, NCI-H2030 lung cancer cells, Daudi cells and SJSA1 osteosarcoma cells were purchased from ATCC (Manassas, VA). A375 cells were a gift from Neal Rosen (Memorial Sloan Kettering Cancer Center, New York, NY) and HCT116 cell lines were donated by Gary Kupfer (Yale University, New Haven, CT). MOLM-13 cells were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). NCI-H2030, SJSA1, MOLM-13 and Daudi cells were cultured in RPMI Medium 1640 from ATCC (Manassas, VA), HT29 cells were grown in McCoy’s 5A Medium and A375 and HCT116 cells were grown in DMEM from GIBCO (Gaithersburg, MD). All media was supplemented with 10% heat-inactivated fetal bovine serum (Biological Industries, USA) and 100 units/ml penicillin and 100 μg/ml streptomycin (GIBCO). Multiple early passage frozen stocks of each cell line in this study were stored under liquid nitrogen: all cell lines were used in experimentation for no longer than 10 passages from thaw. Mycoplasma testing of all cultured cell lines was performed every three weeks using the MycoAlert detection kit from Lonza (Walkersville, MD). Cultures that yielded a test ratio score of < 1 were considered negative for mycoplasma exposure; a test ratio score of 1 to 2 indicated an early stage mycoplasma exposure and use of the culture was discontinued; cultures that yielded a test ratio score of > 2 were designated as testing positive for significant mycoplasma contamination, and affected experiments were repeated using mycoplasma-negative cultures.
Immunoblotting
Cultured cells were incubated in the presence of the PROTACs or component ligands for 24 hours, after which they were rinsed once with ice cold PBS and harvested in buffer containing 25 mM Tris HCl pH 7.4, 1% Nonidet P-40, 0.25% deoxycholic acid and supplemented with 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 30 μg/ml bestatin and 0.3 TIU/ml aprotinin. Lysis proceeded on ice for 15 minutes with occasional vortexing, followed by centrifugation at 16,000 x g for 15 minutes. Supernatant was removed and subjected to protein SDS polyacrylamide gel electrophoresis (Tris-glycine buffer), followed by electrophoretic transfer to nitrocellulose membrane. Membranes were probed for the proteins indicated and visualized using a ChemiDoc MP Imaging System. Protein band intensity on immunoblots was quantitated using Image Lab software v5.2.1 (Bio Rad Labs) and graphed using Prism v7.0 (GraphPad Software).
Cell viability assay
Cells were seeded into 96-well plates and incubated with the indicated concentrations of the PROTACs or component ligands for 48 hrs., at which time MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] (Promega Corp., Madison, WI) and PMS [phenazine methosulfate] (Sigma, MO) were added to a final concentration of 330 μg/ml and 25 μM, respectively. Viable cells converted the MTS to its colored formazan derivative, which was quantitated by measuring absorbance at 490 nm using a Victor2 plate reader (PerkinElmer – Shelton, CT). Data was graphed and analyzed using Prism v7.0 software (GraphPad Software). The Combination Index (CI) value to determine whether the observed activity of A1874 (EA) at each treatment concentration was synergistic compared to the calculated additive effect of JQ1 (EJ) and idasanutlin (EN) was determined using the Bliss Independence model equation: CI = (EJ + EN – EJEN)/ EA as previously described in (32).
Results
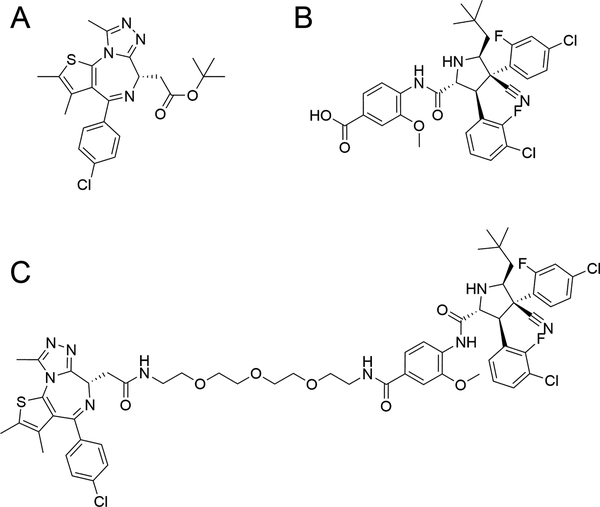
We chose the bromodomain-containing protein, BRD4 for our degradation target in this study for practical reasons: due to its role in driving hematological and solid tumors (33,34), BRD4 has already been successfully targeted for degradation by PROTAC recruitment of either cereblon (2,19) or VHL (20,35), providing a solid foundation from which to begin our investigation. In order to create a PROTAC capable of degrading BRD4 by recruiting it to MDM2, we elected to tether the BET (“bromodomain and extra-terminal”) inhibitor JQ1 (Figure 1A) (36) from its diazepine ring to the methoxyphenyl group of the MDM2 inhibitor, idasanutlin (Figure 1B) (37), by way of a 13-atom long PEG-based linker. The full synthesis protocol and scheme for the resultant PROTAC, A1874 (Figure 1C) are included in the Supplemental Information.
Figure 1: Schematic of nutlin-based PROTAC and its component ligands.
(A) structure of the BRD4/BET inhibitor, JQ1; (B) structure of idasanutlin (RG7388), an MDM2 antagonist; (C) structure of A1874, an MDM2-recruiting, BRD4-degrading PROTAC.
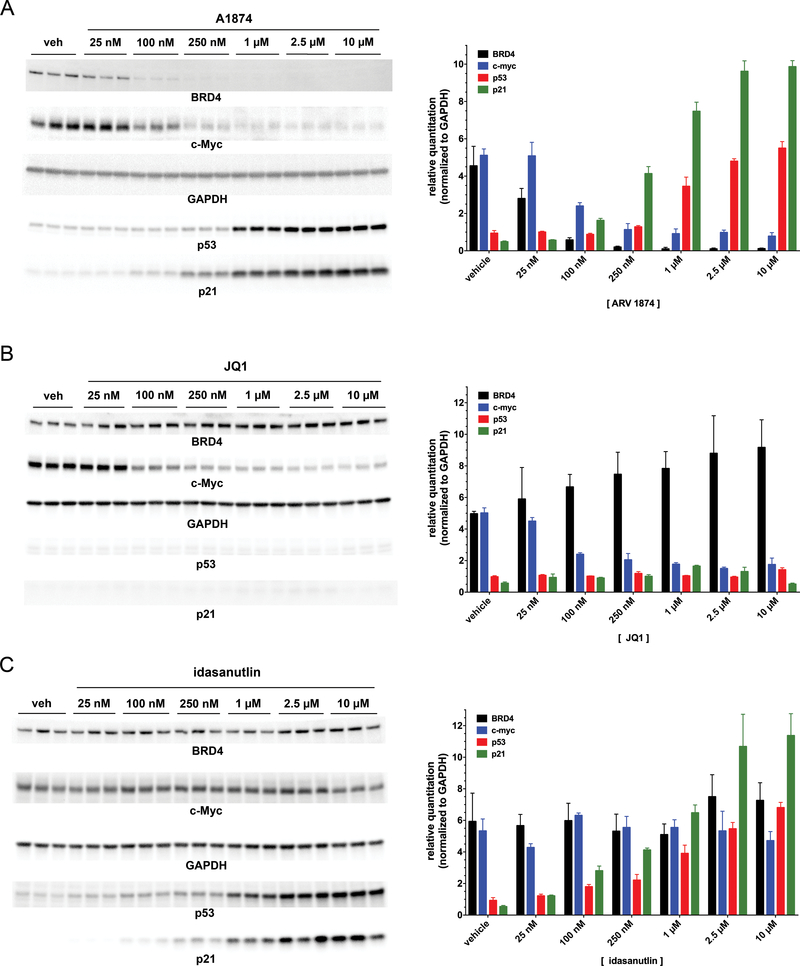
Once synthesized, the ability of the new PROTAC to induce BRD4 degradation in cells was evaluated by immunoblotting (Figure 2A). The colon cancer cell line, HCT116, was selected for initial evaluation of A1874 given the reported relationship of BRD4/c-Myc signaling in colon cancer (33,38) and the wild type status of p53 of this cell line. Treatment of HCT116 cells for 24 hours with increasing concentrations of A1874 induced a dose-dependent knockdown of BRD4 levels, with near-maximum knockdown by 100 nM and a maximum degradation (Dmax) of BRD4 of 98% of the levels in control (0.1% DMSO-treated) cells. By contrast, treatment with equivalent concentrations of unmodified inhibitor JQ1 did not cause BRD4 loss – indeed, at higher concentrations of JQ1 there appears to be compensatory BRD4 upregulation (Figure 2B). There was no detectable rebounding of BRD4 expression levels at higher PROTAC concentrations (up to 10 μM), which contrasts with other PROTACs in which we and others have reported such a “hook effect” (1,21,39). It is worth highlighting that the extent of target protein knockdown and potency (DC50 = 32 nM) are substantially improved compared to our previous report on nutlin-based PROTACs (25), in which 10 μM was needed to produce ~ 50% target knockdown. Thus, A1874 is a much improved nutlin-based protein degrader, approaching the activity of our other reported PROTACs that work through recruitment of the more frequently-used E3 ligases, cereblon and VHL.
Figure 2: A1874 combines the biochemical activities of both JQ1 and idasanutlin into a single molecule.
(A) Representative immunoblots from HCT116 cells treated with increasing concentrations of A1874 (left panel) and quantified results (right panel). (B) Representative immunoblots from HCT116 cells treated with increasing concentrations of JQ1 (left panel) and quantified results (right panel). (C) Representative immunoblots from HCT116 cells treated with increasing concentrations of idasanutlin (left panel) and quantified results (right panel).
To determine whether BRD4 knockdown by A1874 has a functional impact on downstream signaling, c-Myc expression was measured by immunoblotting (Figure 2A). c-Myc, a transcription factor that drives cell proliferation, is itself dependent on functional BRD4 for its own expression (40,41); thus, BRD4 knockdown is predicted to reduce c-Myc expression. In response to treatment with A1874, c-Myc expression is reduced by 85% relative to control HCT116 cells, which is greater than the 70% suppression observed in response to JQ1 itself (Figure 2B) as well as consistent with observations with other BRD4-targeting PROTACs (2,19,35). Thus, despite the limited degradation demonstrated by the first all small molecule PROTAC (25), we show here that using MDM2 to ubiquitinate target proteins for proteasomal degradation can be an effective PROTAC strategy.
In addition to enabling engagement of MDM2 for the purpose of target ubiquitination, we hypothesized that idasanutlin incorporation into the PROTAC could also have an additional, potentially complementary biological activity: the stabilization of the tumor suppressor, p53. The natural protein target of MDM2 is p53, a transcription factor that causes cell cycle arrest and/or activates apoptotic signaling in cells that have damaged DNA. While p53 is constitutively synthesized, it is normally present only at low levels in normal cells because it is also constitutively degraded via the action of MDM2 and the ubiquitin-proteasome system. However, DNA damage, hypoxia and activation of oncogenes, which could result in transformation into a cancerous phenotype, cause disruption of the interaction between p53 and MDM2. This disruption prevents degradation of the former and permits its level to rise, triggering the aforementioned tumor suppressing actions. Immunoblotting for p53 levels in the HCT116 cells as treatment concentration with A1874 increased showed dose-dependent p53 stabilization (Figure 2A), up to 5.9-fold over steady-state levels. The p53 increase was slightly less than the 7.2-fold increase seen upon treated with unmodified idasanutlin (Figure 2C). On the other hand, this p53 upregulation is not detected in HCT116 cells treated with JQ1 (Figure 2B), just as BRD4 knockdown and c-Myc suppression are not observed in cells treated with idasanutlin (Figure 2C). Only the PROTAC itself combines these activities into a single agent. Stabilization of p53 by A1874 was sufficient to upregulate the levels of its well-characterized effector protein, p21CIP1/WAF1 (Figure 2A). It is noteworthy that significant amounts of p21CIP1/WAF1 are induced at 250 nM of A1874, a concentration causing only small increases in p53 levels, reflecting the amplification often associated as a signal is transduced downstream. Thus, from the perspective of downstream effectors, the effective dose ranges of the two anti-mitogenic mechanisms of A1874 (c-Myc suppression and p21 induction) begin to overlap, allowing for the possibility of a combined biological effect even in the nanomolar concentration range.
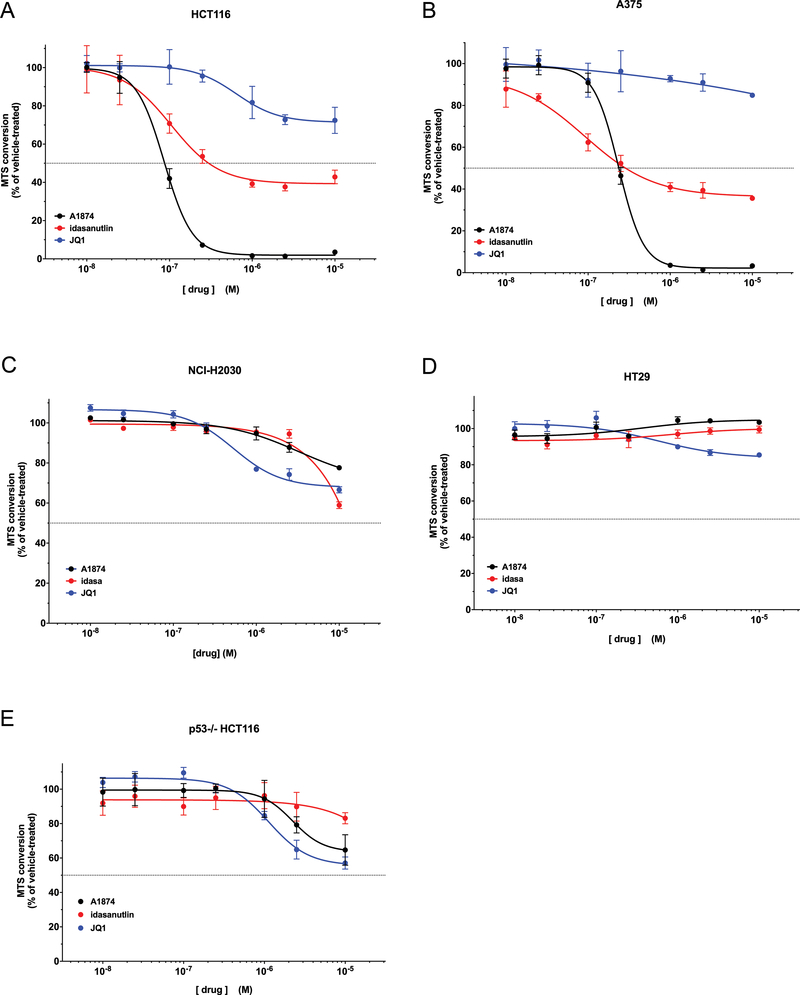
Suppression of c-Myc or induction of p21CIP1/WAF1 -- either one alone -- will inhibit cell proliferation. HCT116 cells incubated with JQ1 or idasanutlin for 48h showed clear dose-dependent loss of viability (Figure 3A). Treatment with JQ1 resulted in 25% loss of MTS signal compared to control cells; and idasanutlin treatment caused a 62% loss. However, treatment with A1874, which combines the activities of the two inhibitors, ultimately resulted in a 97% loss in HCT116 cell viability. That the effect of A1874 was greater than either idasanutlin or JQ1 alone, and even slightly more effective than a combined treatment (Supplementary Figure S1), demonstrates that combining both activities into a single PROTAC does not result in one activity diminishing the other. In fact, the evidence demonstrates the opposite – that their combined effect in A1874 can be “synergistic”: were the two activities in A1874 merely additive, their predicted combined effect would be an 87% reduction (the sum of the individual effects of JQ1 and idasanutlin). That the actual loss of HCT116 viability is greater than that suggests a synergistic anti-proliferative effect of the PROTAC; and indeed, subjecting the viability losses to Bliss Independence analysis of drug effects interactions (32) determined that at concentrations of 100 nM or greater, the antiproliferative effects of the two activities of A1874 are synergistic (Bliss Independence Combination Index values are < 1) (Table 1). This reflects the observation that 100 nM is the lowest concentration at which A1874 begins to affect both c-Myc and p21 CIP1/WAF1. To confirm this finding, A1874 was applied to a different cell line: A375 melanoma cells (Figure 3B). Similar to the HCT116 cells, the 98% loss of cell viability when A375 cells are treated with A1874 is greater than the sum of the effects of JQ1 alone and idasanutlin alone (15% loss and 64% loss, respectively). Analysis showed that in A375 cells, the activities of A1874 synergize at concentrations of 250 nM or greater (Supplementary Table S1). Thus, the superior anti-cancer activity of A1874 is not unique to a single cell line.
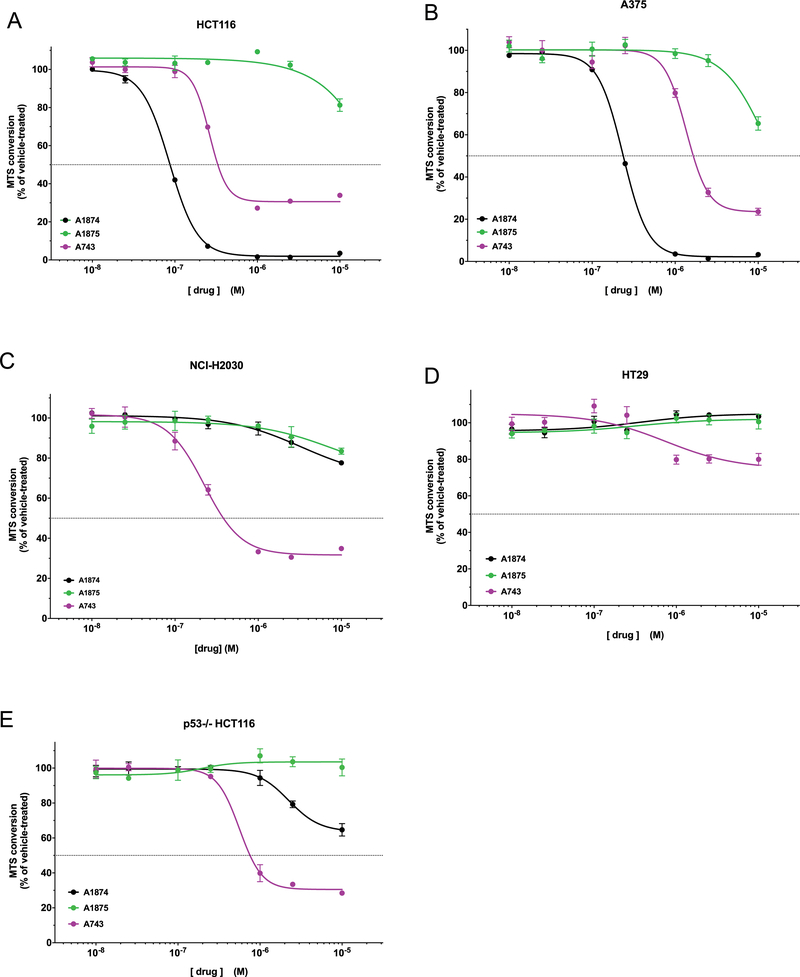
Figure 3: Cell viability assay of cell lines treated with A1874 compared to its component ligands.
(A) HCT116 colorectal cancer cells; (B) A375 melanoma cells; (C) NCI-H2030 lung cancer cells; (D) HT29 colorectal cancer cells; (E) p53−/− HCT116 colorectal cancer cells (isogenic with panel (A)). A1874 data points that are asterisked exhibit synergistic activity compared to the sum of the component warhead activities at the same concentration according to Bliss Independence model (32).
Table 1: Bliss Independence analysis of A1874 activities in HCT116 cells.
The percentage cell inhibition (compared to vehicle-treated control cells) for each compound at each concentration from Figure 3A was converted to their decimal equivalents (EJ for JQ1; EN for idasanutlin; EA for A1874) and the Combination Index (CI) value was derived as explained in the Experimental Methods and (32). CI values <1 indicate synergy.
| compound concentration(μM) | % inhibitionby JQ1 | EJ | % inhibition by idasanutlin | EN | % inhibition by A1874 | EA | CI |
|---|---|---|---|---|---|---|---|
| 0.01 | 0.0 % | 0.000 | 0.9 % | 0.009 | 0.0 % | 0.000 | ∅ |
| 0.025 | 0.1 % | 0.001 | 6.5 % | 0.065 | 5.1 % | 0.051 | 1.29 |
| 0.10 | 0.0 % | 0.000 | 29.2 % | 0.292 | 58.0 % | 0.580 | 0.50 |
| 0.25 | 4.4 % | 0.044 | 46.4 % | 0.464 | 92.8 % | 0.928 | 0.53 |
| 1.00 | 18.2 % | 0.182 | 60.8 % | 0.608 | 98.4 % | 0.984 | 0.69 |
| 2.50 | 27.2 % | 0.272 | 62.3 % | 0.623 | 98.6 % | 0.986 | 0.74 |
| 10.0 | 27.6 % | 0.276 | 57.2 % | 0.572 | 96.5 % | 0.965 | 0.71 |
While A375 and HCT116 cells have different oncogenic driving mutations, they both express wild type p53 -- given the targeted protein level changes caused by A1874 treatment, this would appear to be a key element for its maximum effectiveness. To investigate whether the maximum effect of A1874 requires wild type p53, the treatments were performed in two other cell lines: NCI-H2030 lung cancer cells and HT-29 colon cancer cells. These cell lines were chosen because (1) they both possess p53 mutations that render it inactive, and (2) their driving oncogenic mutations match those of the previously tested A1874-sensitive cell lines. More specifically, HT-29 cells possess the same B-Raf-activating V600E mutation as A375 cells; while NCI-H2030 cells and HCT116 cells both possess constitutively active K-Ras. However, when treated with up to 1 μM A1874, the NCI-H2030 and HT-29 cells exhibited nominal loss of viability (Figures 3C and3D, respectively). Only at 10 μM A1874, a concentration that is far above that necessary to maximally decrease viability in the wild type p53 cells, was there limited reduction (22.4%) in viability, and then in only the NCI-H2030 cells. In fact, the mutant B-Raf expressing cells seemed to be affected less potently than the mutated K-Ras expressing cells regardless of p53 status. To confirm that the p53-null status that underlies the loss of effectiveness of A1874, we tested the PROTAC’s activity against an isogenic, p53-deficient version of the HCT116 cells and observed diminished loss of viability (Figure 3E). The effectiveness of A1874 in this cell line was similar to that of unmodified JQ1, consistent with the predicted absence of p53 signaling in these cells (corroborated by their insensitivity to idasanutlin). In this cellular context, the PROTAC has been reduced to having a single biological activity – targeting BRD4 to suppress c-Myc levels. These data clearly indicate that the maximum activity of A1874 depends on the cells ability to effectively mobilize p53 in response to the idasanutlin moiety.
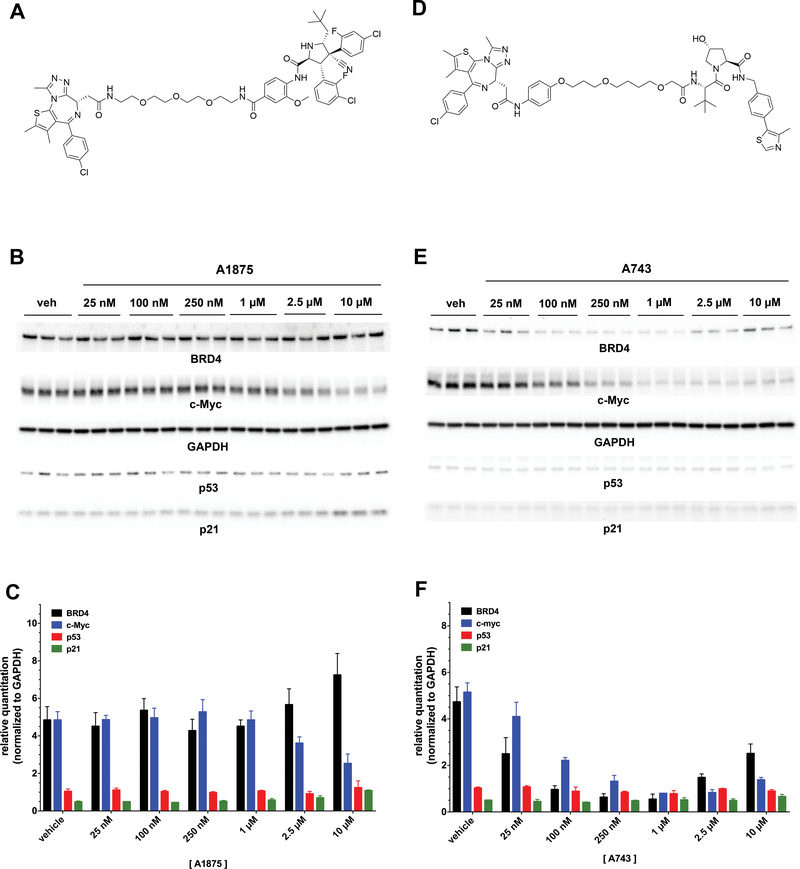
Further support that MDM2 recruitment is crucial for the PROTAC’s pronounced biological activity was provided by testing A1875, an inactive version of the PROTAC with identical physicochemical properties but discrete stereochemical alterations in the idasanutlin moiety that diminish its MDM2 binding (Figure 4A and Supplemental Information). This reduces its activity both to degrade BRD4 and to stabilize p53 (Figure 4B and4C). Another approach to the evaluation of nutlins as E3 ligase-targeting ligands would be comparing the activity of A1874 against a PROTAC that degrades BRD4 with similar DC50 and suppresses c-Myc with a similar efficacy, but that works through a different E3 ligase. A743 is an analogous BRD4 degrader but it incorporates a VHL-binding ligand (Figure 4D and Supplemental Information) rather than idasanutlin. As constructed, A743 degrades BRD4 with similar potency (DC50 = 23.1 nM) and efficacy (Dmax = 89%) as A1874 (Figure 4E and4F). Like some PROTACs, A743 demonstrates a “hook effect” at concentrations ≥ 2.5 μM; however, the hook effect on BRD4 degradation at higher concentrations does not influence the downstream effect of A743 since it suppresses c-Myc levels by 84% -- nearly equal to A1874. Importantly, A743 does not affect p53 levels since it induces BRD4 ubiquitination by VHL recruitment and not MDM2 recruitment.
Figure 4: Structures and activities of PROTAC variants of A1874.
(A) Structure of A1875, a diastereomeric analog of A1874 with greatly reduced affinity for MDM2. (B) Representative immunoblots from HCT116 cells treated with increasing concentrations of A1875. (C) Quantified results of immunoblots in panel (B). (D) Structure of A743, a VHL-recruiting BRD4-degrading PROTAC. (E) Representative immunoblots from HCT116 cells treated with increasing concentrations of A743. (F) Quantified results of immunoblots in panel (E).
In each of the cell lines tested (Figure 5A-E), the epimeric A1875 had substantially diminished effectiveness in MTS assay relative to A1874, reinforcing that the activity of the PROTAC is dependent on efficient engagement of MDM2. The results from comparisons of A743 against A1874 across the five cell lines were even more showed a dose-dependent loss of viability (Figure 5). However, in each case, the more effective PROTAC was dependent on the p53 status of the cell line. In the wild type p53 cell lines (Figure 5A&B), A1874 was more effective than A743: in HCT116 cells, A1874 caused a 97% decrease in viability compared to the 69% loss resulting from A743 treatment; and in A375 cells, A1874 caused a 98 % loss of viability, which is greater than that 76% loss caused by A743. However, in the mutant or null p53 cell lines (Figure 5C-E), A743 was the more effective PROTAC at decreasing viability: in NCI-H2030 cells, HT29 cells and the p53−/− HCT116 line, A743 decreased viability by 68%, 25% and 69%, respectively. Conversely, A1874 decreased viability by only 22%, 0%, and 36%, respectively, making it the less effective PROTAC in these cell lines.
Figure 5: Engagement of MDM2 by PROTAC enhances anticancer activity beyond that of BRD4 degradation in cells with wild type p53.
(A) HCT116 colorectal cancer cells; (B) A375 melanoma cells; (C) NCI-H2030 lung cancer cells; (D) HT29 colorectal cancer cells; (E) p53−/− HCT116 colorectal cancer cells (isogenic with panel (A)).
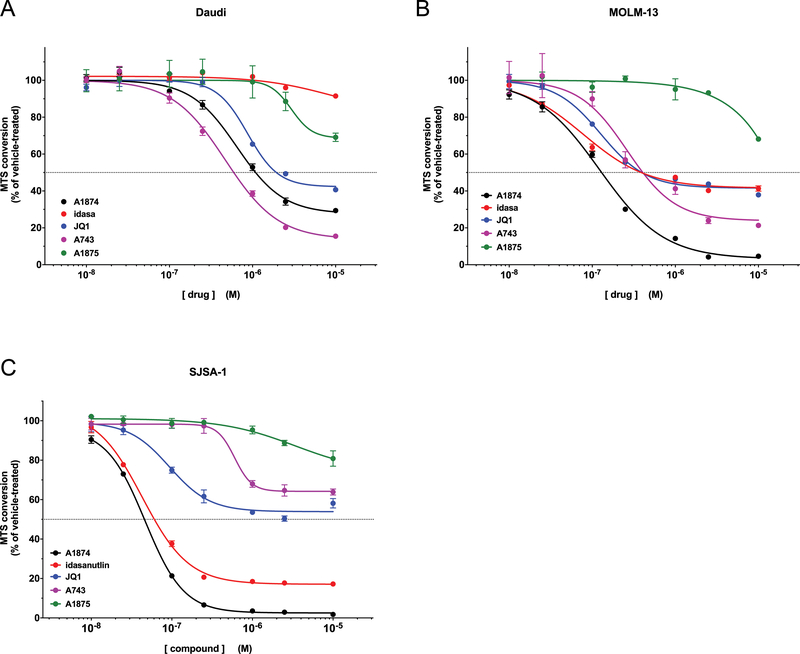
To determine whether different cellular contexts beyond p53 status impact the relative effectiveness of A1874, we tested it against another, smaller panel of cells that are known to have a more pronounced sensitivity to either of its component ligands (Figure 6). Daudi cells and MOLM-13 are hematological cancer cells that are reported to possess an accentuated sensitivity to BRD4 inhibition (34,42). On the other hand, SJSA1 osteosarcoma cells (wild type p53) express unusually high levels of MDM2, which renders them especially susceptible to MDM2 inhibition. Against both hematological cancer lines (Figure 6A and6B), JQ1 reduced viability similarly by 58%. It is, perhaps, a product of this enhanced dependency on BRD4 that even A1875, whose ability to bind to MDM2 is greatly reduced such that it can only inhibit BRD4, was able to cause some loss of viability at the highest concentration tested -- 31% and 32% in Daudi and MOLM-13 cells, respectively. While these two lines reacted similarly to the JQ1 inhibitors, their sensitivities to idasanutlin were quite distinct from each other: MOLM-13 cells, which are wild type p53, were inhibited by idasanutlin to an equal extent as they were by JQ1 (58% reduction), whereas the mutant p53-expressing Daudi cells were minimally affected (9% reduction) by idasanutlin up to 10 μM. Mirroring this pattern, MOLM-13 cells were also more susceptible to inhibition by A1874 than were the Daudi cells, although in both cases the PROTAC was more active than either of the component ligands: A1874 reduced viability of Daudi cells by 70% and of MOLM-13 cells by 95%. While there was no synergism observed, A1874 was clearly the most active against these cancer lines. Interestingly, in Daudi cells the VHL-recruiting A743 outperformed the MDM2-recruiting A1874, although this appears to be more a result of a relatively diminished activity of the latter than a substantial increase in activity of A743 in this cell line.
Figure 6: Comparative effectiveness of MDM2-recruiting PROTAC versus component ligands and VHL-recruiting PROTAC in cells with accentuated JQ1 sensitivity and/or elevated MDM2 levels.
(A) Daudi lymphoma cells; (B) MOLM-13 myeloid leukemia cells; (C) SJSA-1 osteosarcoma cells.
Predictably, idasanutlin was effective at reducing viability (down by 82%) of the MDM2-overexpressing SJSA-1 cells (Figure 6C); unexpectedly, JQ1 was also more effective against SJSA1 cells (46% reduction in viability) than it had been against the cells in Figures 3 and 5, and approached the sensitivity exhibited by the Daudi and MOLM-13 cells. Nevertheless, the MDM2-recruiting A1874 demonstrated the highest effectiveness to decrease SJSA-1 cell viability (97.5% viability loss), also outperforming the VHL-recruiting A743 (36% viability loss). The increased levels of MDM2 clearly narrowed the effectiveness differential between A1874 and idasanutlin, such that the latter was nearly as effective as the former, limiting how far the PROTAC could outperform the MDM2 inhibitor. The increased MDM2 levels in the SJSA1 cells only modestly increased the potency of A1874 itself: down to an IC50 of 46.5 nM in SJSA1 cells vs. 86.3 nM and 236 nM in HCT116 cells and A375 cells, respectively. In the end, A1874 was the most effective PROTAC at reducing viability of p53 wild type cells here, consistent with the earlier results in Figure 5.
Discussion
PROTAC-mediated protein degradation combines the lasting effects of nucleic acid-based methods of protein knockdown with the more readily adjustable attributes of application of small molecules – i.e. delivery, dosing, timing, etc. Given their modular design, PROTACs are also adjustable. Studies have shown that changes in all three PROTAC components– target protein ligand, linker and E3 ligand – can have a profound impact on their target specificity as well as their potency and efficacy to cause degradation. Changes in linker length and hydrophilicity can mean the difference between highly effective versus nominal degradation (21,39,43). Linkers that are too short will prevent formation of the trimer complex (target protein:PROTAC:E3 ligase) to favor only dimers (target protein:PROTAC or PROTAC:E3 ligase); while linkers that are too long provide excess steric freedom and fail to facilitate a sufficiently tight interaction of its binding partners to establish stabilizing protein:protein interactions. Likewise, we have documented that structurally dissimilar ligands with high affinity for a common protein will not necessarily each give rise to an effective target degrader when incorporated into PROTACs (1). Nevertheless, effective PROTACs have been synthesized and reported to degrade an ever-increasing number of target proteins, including but not limited to FKBP12 (5), the androgen receptor (11,44), HER1/HER2 (16), BRD4 and other BET proteins (2,19,39), RIPK2 (10), the estrogen receptor (3), p38MAPK (18), the retinoic acid binding protein (45), MetAP2 (4), c-Met (18), CDK9 (12), BCR-Abl (1), FRS2α (13) and BTK (15,17).
Compared to the variety of proteins successfully degraded, the paucity of options for recruiting an E3 ligase is notable. With few exceptions, PROTACs to date have been constructed to recruit one of the following E3 ligases: VHL, cereblon or IAP. This is striking since the human genome encodes for hundreds of different E3 ligases (46); while some may not be amenable for use with PROTACs, a recent study suggests that many of them can mediate target protein degradation if successfully harnessed (22). However, the limiting factor has been the availability of small molecule ligands to harness them. In this report, we have revisited the strategy of constructing PROTACs using the small molecule, idasanutlin -- a ligand with high affinity for the E3 ligase, MDM2 (37). Nutlins were initially developed to act as a pharmacological means to elevate expression of the tumor suppressor p53 and there have been numerous reports (47–49) concerning the potential of nutlins as anti-cancer drugs/adjuvants. Our original study (25) indicated that nutlin-based PROTACs were limited in their ability to facilitate degradation: a PROTAC that recruited MDM2 to ubiquitinate the androgen receptor caused only limited target degradation at micromolar concentrations. However, that study was limited in its scope, and given what has been learned since about the structural and mechanistic considerations of PROTAC functioning (50,51) we have revisited nutlin-based PROTACs here. This study has shown is that not only can nutlin-based PROTACs mediate degradation far more potently and effectively than previously realized, but that by virtue of the p53-stabilizing activity particular to nutlins, the PROTACs derived from them can have biological activity surpassing that of equipotent degraders that harness other E3 ligases.
We synthesized a PROTAC by joining the BET protein inhibitor, JQ1, to the MDM2 antagonist, idasanutlin. JQ1 inhibits BRD4 resulting in c-Myc suppression and subsequent reduced proliferation of a variety of cancers (52–54). Idasanutlin disrupts the interaction between MDM2 and the tumor suppressor protein, p53, leading to accumulation of the latter and activation of tumor suppressing mechanisms that also inhibit cell proliferation and reduce viability. We reasoned that if these complementary activities would be incorporated in a PROTAC synthesized from both molecules, a degrader capable of dual-mode anti-cancer activity would be created. Indeed, the resultant PROTAC, A1874, mediates both BRD4 degradation/c-Myc suppression and p53 stabilization when incubated on HCT116 cells. The degree of c-Myc suppression mediated by A1874 was greater than that by JQ1 itself in the same cells, perhaps owing to degradation having a more profound effect than inhibition of the same target protein (16,19). Moreover, the level of p53 upregulation in response to A1874 approaches that of unmodified idasanutlin. Unsurprisingly, when HCT116 cells were measured for viability following A1874 incubation, the reduction in viability was striking compared to that due to either of the individual ligand components; and even synergistic compared to the sum of their effects.. Enhanced effects has been demonstrated between nutlins combined with other anti-cancer drugs (30,31,55), however such studies to date have relied on protein inhibition rather than PROTAC-mediated protein degradation.
A similar result as with the HCT-116 cells was obtained in testing the PROTAC on A375 melanoma cells. Since both of these cell lines have wild type p53, the greater effect of the combined reagent seems intuitive. Even in instances where the effect of either component ligand alone was substantial in terms of reducing cancer cell viability, the PROTAC A1874 was nonetheless the most effective treatment in cells with wild type p53 (e.g. MOLM-13 and SJSA-1 cells), indicating a combined effect. In order to confirm the involvement of MDM2 and the p53 response in mediating the profound anti-cancer response of A1874, the PROTAC was both (1) tested in comparable cell line contexts that were p53 mutant or null and (2) compared to a similarly potent BRD4-degrading PROTAC, A743, that works through recruitment of VHL as its E3 ligase. In cell lines that lacked functional p53, A1874 was much less capable to reduce cell viability than it had been in wild type p53 cell lines; similarly, in cell lines with wild type p53, A1874 was more active at reducing cell viability than the cognate VHL-recruiting PROTAC, while the reverse was true in cell lines with mutant p53. Hence, in utilizing idasanutlin, there has developed a selectivity of cell susceptibility to the PROTAC that stands apart from that typically determined from the expression pattern of the target protein itself; in this instance, selectivity is also reliant on signaling events downstream from the E3 ligase. This recently has been shown for some IAP-recruiting degraders (29) although for those molecules, the contribution from the E3 ligase side of the molecule towards the overall observed activity is minor.
To summarize, this study shows that the E3 ligase MDM2 can serve as a valuable addition to the small number of E3 ligases that when harnessed can produce nanomolar PROTAC-mediated target protein degradation. Moreover, with further refinement, nutlin-based PROTACs could become extremely effective candidate anticancer therapeutics due to their dual-mode mechanism of action – elimination of a proto-oncogene/oncogene and activation of a tumor suppressor – where one anticipates that development of resistance would be more difficult to because a single mutation in either partner binding protein may not abrogate all activity of the PROTAC.
Supplementary Material
Significance.
Findings present the first BRD4-targeting MDM2-based PROTAC that possesses potent, distinct and synergistic biological activities associated with both ends of this heterobifunctional molecule.
Acknowledgements
The authors would like to thank Ashton Lai, M.D./Ph.D. and Irene Ojini, Ph.D. for their assistance in the early stages of this study, and George Burslem, Ph.D. for assistance in preparing this manuscript..J. Hines is supported by a grant from the NCI (R50 CA211252), and C. M. Crews is supported by an award from the NIH (R35 CA197589).
Footnotes
Conflict of Interest Statement:
C.M.C. is founder, shareholder and consultant to Arvinas, Inc. In addition, his lab receives sponsored research support from Arvinas. H.D. and Y.Q. are Arvinas employees.
References
- 1.Lai A, Toure M, Hellerschmied D, Salami J, Jaime-Figueroa S, Ko E, et al. Modular PROTAC design for the degradation of oncogenic BCR-Abl. Angew Chem Int Ed Eng 2016;55(2):807–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winter G, Buckley D, Paulk J, Roberts J, Souza A, Dhe-Paganon S, et al. Pthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015;348:1376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okuhira K, Demizu Y, Hattori T, Ohoka N, Shibata N, Nishimaki-Mogami T, et al. Development of hybrid small molecules that induce degradation of estrogen receptor-alphia and necrotic cell death in breast cancer cells. Cancer Sci 2013;104(11):1492–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakamoto K, Kim K, Kumagai A, Mercurio F, Crews C, Deshaies R. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA 2001;98(15):8554–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneekloth J, Fonseca F, Koldobskiy M, Mandal A, Deshaies R, Sakamoto K, et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc 2004;126(12):3748–54. [DOI] [PubMed] [Google Scholar]
- 6.Bonger K, Chen L, Liu C, Wandless T. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat Chem Biol 2011;7(8):531–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banaszynski L, Chen L, Maynard-Smith L, Ooi A, Wandless T. A rapid, reversible and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 2006;126(5):995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raina K, Crews C. Targeted protein knockdown using small molecule degraders. Curr Opin Chem Biol 2017;39:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crews C Targeting the undruggable proteome: the small molecules of my dreams. Chem Biol 2010;16(6):551–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bondeson D, Mares A, Smith I, Ko E, Campos S, Miah A, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 2015;11(8):611–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salami J, Alabi S, Willard R, Vitale N, Wang J, Dong H, et al. Androgen receptor degratation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol 2018;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson C, Jiang B, Erb M, Liang Y, Doctor Z, Zhang Z, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 2018;14(2):1630170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hines J, Gough J, Corson T, Crews C. Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc Natl Acad Sci, USA 2013;110(22):8942–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puppala D, Lee H, Kim K, Swanson H. Development of an aryl hydrocarbon receptor antagonist using the proteolysis-targeting chimeric molecules approach: a potential tool for chemoprevention. Mol Pharmacol 2008;73(4):1064–71. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Dobrovolsky D, Paulk J, Yang G, Weisberg E, Doctor Z, et al. A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem Biol 2018;25(1):88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burslem G, Smith B, Lai A, Jaime-Figueroa S, McQuaid D, Bondeson D, et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem Biol 2018;25(1):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buhimschi A, Armstrong H, Toure M, Jaime-Figueroa S, Chen T, Lehman A, et al. Targeting the C481S ibrutinib-resistance mutation in Bruton’s tyrosine kinase using PROTAC-mediated degradation. Biochemistry 2018;57(26):3564–75. [DOI] [PubMed] [Google Scholar]
- 18.Bondeson D, Smith B, Burslem G, Buhimschi A, Hines J, Jaime-Figueroa S, et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem Biol 2018;25(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 2015;22(6):755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadd M, Testa A, Lucas X, Chan K, Chen W, Lamont D, et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 2017;13(5):514–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckley D, Raina K, Darricarrere N, Hines J, Gustafson J, Smith I, et al. HaloPROTACs: use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem Biol 2015;10(8):1831–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ottis P, Toure M, Cromm P, Ko E, Gustafson J, Crews C. Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem Biol 2017;12(10):2570–78. [DOI] [PubMed] [Google Scholar]
- 23.Maniaci C, Hughes S, Testa A, Chen W, Lamont D, Rocha S, et al. Homo-PROTACs: bivalent small molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation. Nat Commun 2017;8(1):830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burslem G, Crews C. Small-molecule modulation of protein homeostasis. Chem Rev 2017;117(17):11269–301. [DOI] [PubMed] [Google Scholar]
- 25.Schneekloth A, Pucheault M, Tae H, Crews C. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg Med Chem Lett 2008;18(22):5904–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckley D, Gustafson J, Van Molle I, Roth A, Tae H, Gareiss P, et al. Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew Chem Int Ed Engl 2012;51(46):11463–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, et al. Identification of a primary target of thalidomide teratogenicity. Science 2010;327:1345–50. [DOI] [PubMed] [Google Scholar]
- 28.Sekine K, Takubo K, Kikuchi R, Nishimoto M, Kitagawa M, Abe F, et al. Small molecules destabilize cIAP1 by activating auto-ubiquitylation. J Biol Chem 2008;283(14):8961–68. [DOI] [PubMed] [Google Scholar]
- 29.Ohoka N, Morita Y, Nagai K, Shimokawa K, Ujikawa O, Fujimori I, et al. Derivatization of inhibitor of apoptosis protein (IAP) ligands yields improved inducers of estrogen receptor alpha degradation. J Biol Chem 2018;293(18):6776–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X-H, Zhang S-F, Bao J-T, Liu F-Y. Oridonin synergizes with Nutlin-3 in osteosarcoma cells by modulating the levels of multiple Bcl-2 family proteins. Tumor Biol 2017;39(6):1–9. [DOI] [PubMed] [Google Scholar]
- 31.Laroche A, Chaire V, Algeo M-P, Karanian M, Fourneaux B, Italiano A. Mdm2 antagonists synergize with PI3K/mTOR inhibition in well-differentiated/dedifferentiated liposarcomas. Oncotarget 2017;33:53968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect 2015;3(3):e00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCleland M, Mesh K, Lorenzana E, Chopra V, Segal E, Watanbe C, et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J Clin Invest 2016;126(2):639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Ther Adv Hematol 2015;6(3):128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi A, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA 2016;113(26):7124–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith W, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468(7327):1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Q, Zhang Z, Liu J, Jiang N, Zhang J, Ross T, et al. Discovery of RG7388, a potent and selective p53-mdm2 inhibitor in clinical development. J Med Chem 2013;56(14):5979–83. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Zhou J, Ye F, Xiong H, Peng L, Zheng Z, et al. BRD4 inhibitor inhibits colorectal cancer growth and metastasis. Int J Mol Sci 2015;16(1):1928–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan K, Zengerle M, Testa A, Ciulli A. Impact of target warhead and linkage vector on inducing protein degradation: comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J Med Chem 2018;61:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyoshima M, Howie H, Imakura M, Walsh R, Annis J, Chang A, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci USA 2012;109(24):9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuber J, Shi J, Wang E, Rappaport A, Hermann H, Sison E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478(7370):524–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abedin S, Boddy C, Munshi H. BET inhibitors in the treatment of hematologic malignancies: current insights and future prospects. Onco Targets Ther 2016;9:5943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cyrus K, Wehenkel M, Choi E, Han H, Lee H, Swanson H, et al. Impact of linker length on the activity of PROTACs. Mol Biosyst 2011;7(2):359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cyrus K, Wehenkel M, Choi E, Swanson H, Kim K. Two-headed PROTAC: an effective new tool for targeted protein degradation. Chembiochem 2010;11(11):1531–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh Y, Ishikawa M, Naito M, Hashimoto Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J Am Chem Soc 2010;132(16):5820–26. [DOI] [PubMed] [Google Scholar]
- 46.Zheng N, Shabek N. Ubiquitin ligases: structure, function and regulation. Ann Rev Biochem 2017;86:129–57. [DOI] [PubMed] [Google Scholar]
- 47.Hui W, Liu S, Zheng J, Fang Z, Ding Q, Feng C. Nutlin-3a as a novel anticancer agent for adrenocortical carcinoma with CTNNB1 mutation. Cancer Med 2018;7(4):1440–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Esfandiari A, Ho Y, Wang N, Mahdi A, Aputullahoglu E, et al. Targeting negative regulation of p53 by mdm2 and WIP1 as a therapeutic strategy in cutaneous melanoma. Br J Cancer 2018;118(4):495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalan S, Amat R, Schachter M, Kwiatkowski N, Abraham B, Liang Y, et al. Activation of the p53 transcriptional program sensitizes cancer cells to cdk7 inhibitors. Cell Rep 2017;21(2):467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neklesa T, Winkler J, Crews C. Targeted protein degradation by PROTACs. Pharmacol Ther 2017;174:138–44. [DOI] [PubMed] [Google Scholar]
- 51.Churcher I Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J Med Chem 2018;61(2):444–52. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Liu Z, Wang Z, Wang S, Chen Z, Li Z, et al. Targeting c-myc: JQ1 as a promising option for c-myc-amplified esophageal squamous cell carcinoma. Cancer Lett 2018;419:64–74. [DOI] [PubMed] [Google Scholar]
- 53.Karakashev S, Zhu H, Yokoyama Y, Zhao B, Fatkhutdinov N, Kossenkov A, et al. BET bromodomain inhibition synergizes with PARP inhibitor in epithelial ovarian cancer. Cell Rep 2017;21(12):3398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhadury J, Nilsson L, Muralidharan S, LC G, Li Z, Gesner E, et al. BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc Natl Acad Sci USA 2014;111(26):E2721–E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He S, Dong G, Wu S, Fang K, Miao Z, Wang W, et al. Small molecules simultaneously inhibiting p53-Murine Double Minute 2 (MDM2) Interaction and Histone Deacetylases (HDACs): Discovery of Novel Multitargeting Antitumor Agents. J Med Chem 2018;61:7245–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.