Abstract
The ever-changing tumor-microenvironment constantly challenges individual cancer cells to balance supply and demand, presenting tumor vulnerabilities and therapeutic opportunities. Everolimus and temsirolimus are inhibitors of mTOR (mTORi) approved for treating metastatic renal cell carcinoma (mRCC). However, treatment outcome varies greatly among patients. Accordingly, administration of mTORi in mRCC is diminishing, which could potentially result in missing timely delivery of effective treatment for select patients. Here we implemented a clinically applicable, integrated platform encompassing a single dose of [1-13C] pyruvate to visualize the in vivo effect of mTORi on the conversion of pyruvate to lactate using hyperpolarized MRI. A striking difference that predicts treatment benefit was demonstrated using two preclinical models derived from clear cell RCC (ccRCC) patients who exhibited primary resistance to VEGFRi and quickly succumbed to their diseases within 6 months after the diagnosis of metastasis without receiving mTORi. Our findings suggest that hyperpolarized MRI could be further developed to personalize kidney cancer treatment.
INTRODUCTION
To meet intrinsically dysregulated high metabolic demands, cancer cells challenge the host’s metabolic homeostasis and competes for fuel sources. The metabolic dominance of cancer cells has long been recognized, including the “Warburg Effect” in which, upon oncogenic signaling, the energy-efficient oxidative phosphorylation is rerouted through the energy-inefficient aerobic glycolysis to produce molecular building blocks for aberrant cellular proliferation1. The heavy reliance of cancer cells on glycolysis renders therapeutic opportunities. The demonstrated efficacy and exemplary approval of everolimus and temsirolimus (allosteric inhibitors of the cellular metabolic epicenter mTORC1 complex) in treating human malignancies including breast and kidney cancers validates targeting cancer metabolism as a feasible anti-cancer therapeutic strategy2. However, treatment outcome varies, and there is no reproducible, predictive biomarker to guide patient selection.
Cancer cells shift glucose utilization and metabolism from oxidative phosphorylation to aerobic glycolysis (Warburg effect). It has been shown that the PI3K/mTOR axis directly controls glycolysis through both transcription (e.g., GLUT1, HK2 and PFK) and post-translational modification of glycolytic enzymes (e.g., LDH, PKM2)3. Activation of this pathway upregulates glycolytic flux, providing the basis for imaging approaches such as [18F]-flurodeoxyglucose (18FDG) PET. In the setting of pathway inhibition, glycolytic flux can be reduced, diverted or unchanged, each of which have broad implications for the future therapeutic effect, whether growth arrest or cell death. Novel approaches, including hyperpolarized magnetic resonance imaging (HP MRI) have the potential to interrogate in vivo metabolic flux non-invasively using hyperpolarized nutrients4,5. Using the substrate HP pyruvate, one is able to image pyruvate’s metabolic products in real time, providing a simultaneous quantitative assessment of the flux of metabolites through multiple pathways, decoupling transport from metabolic fate and rate.
Clear cell renal cell carcinoma (ccRCC) is the most common kidney cancer subtype, accounting for most kidney cancer mortality6 and is known for its conspicuous intratumor genomic heterogeneity which may contribute to the highly varied clinical outcomes observed in kidney cancer patients treated with mTORi. Multi-region genomics of ccRCC tumor samples from long-term responders to single agent mTORi have been performed, which demonstrated the presence of early convergent evolution of mutation events centering on the core mTORC1 pathway in many of these cases7,8. However, multi-regional sequencing of primary and metastatic tumors is not common practice; hence, our ability to predict response to mTORi remains extremely limited. In targeted gene sequencing studies of patient-derived RCC cell lines (MSK IMPACT), we identified cells with differential activation of the PIK3CA pathway via an activating point mutation (JHRCC228) as compared to another patient-derived cell line (JHRCC12)9. In this work, we take advantage of a system that accurately recapitulates RCC response to targeted inhibition and demonstrate that HP derived metabolic flux measurements of lactate generation in vivo indicate on-target inhibition of the PI3K/mTOR pathway. In addition, this clinically translatable method provides a means of detecting early response to targeted inhibition, a major limitation of current efforts to develop novel therapeutics and assess their efficacy in patients.
METHODS
Chemicals
Unless otherwise indicated, all chemicals and solvents were purchase from Sigma Aldrich (St. Louis, MO, USA). 99%-enriched [1,6-13C2]glucose and [1-13C]pyruvic acid were used without further purification.
Cell culture and in vitro treatment
JHRCC12 and JHRCC228 cells were acquired directly from patients and cultured as previously reported9 and their respective clinicopathologic characteristics are included in Supplementary Table 1. Briefly, cells were cultured in advanced DMEM/F12 (Invitrogen) supplemented with 10% FBS, nonessential amino acids, L-glutamine, sodium pyruvate, and antibiotics. For in vitro rapamycin treatment, 4×104 cells were seeded in 6-cm dishes in triplicate. The next day, fresh medium with a 100 nM concentration of rapamycin was added to each dish. The control group was treated with DMSO. Cells were trypsinized and counted on days 2, 4, 6, and 8 by cell counter (Nexcelom, USA). Cell proliferation curves were generated with GraphPad Prism software v.5.02 (La Jolla, CA, USA). Cells were sequenced using MSK IMPACT to confirm they have not significantly changed and additionally mycoplasma testing was routinely conducted to ensure no contamination.
Cell treatment and 1H NMR analysis
For each cell line, 1×106 cells were grown in a 100-mm dish. 24 hours before the day of the experiment, the cells were split in two groups (n=5 replicates per group): vehicle (0.1% DMSO) and treated (100 nM rapamycin). After 21 hours, the media were exchanged either with 5.0 mM non-enriched glucose or 5.0 mM [1,6-13C2] glucose-enriched DMEM (supplemented with 10% dialyzed FBS, 1% Pen/strep). The non-enriched glucose samples were used for cell counting, and the enriched glucose samples were used for tracing measurements. The cells were incubated at 37°C for 3 hours. Then the media were removed and stored at −80°C for further purification and analysis. The cells were quickly washed twice with cold PBS. 2 mL of 80% ice-cold methanol was used to extract the water-soluble component of the cells. The cells were placed at −80oC overnight to ensure optimal metabolite extraction. The following day, the sample was centrifuged at 4000 RPM at 4°C for 30 minutes. The supernatant was isolated from the cell pellet and dried using Genevac™ concentrator EZ-2.3.
Media samples were filtered at 14000 RPM at 4°C for 30 minutes using Amicon Ultra 0.5 mL centrifugal filters (3K) (Merck Millipore Ltd. Darmstadt, Germany) to eliminate protein components that could interfere with the NMR analysis. The filter was pre-washed with 400 μL of water to eliminate glycerol from the cartridge. An internal standard of 140 μL of 5X PBS in D2O, containing 2.5mM of DSS and 50mM imidazole as pH indicator were added to 560 μL of the filtered media. The dried water-soluble intracellular content was dissolved in 700 μL of standard, containing 50 uM DSS as an internal standard and 1mM imidazole as pH indicator. 1H NMR spectra for each cell extract and media samples were acquired with a water presaturation recycle delay of 4 s, acquisition time of 2.67 s, 90° pulse and 256 averages. A residual glycerol peak was identified in our spectra and quantified. Spectral data were processed as previously described using a combination of MestReNova (Spain) and Chenomx (Canada), yielding concentrations and fractional enrichments for the resonances of interest.
Immunoblot analysis
Cells were cultured and treated as described above. Cells were harvested in ice-cold PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4), pelleted, and lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 1% Na deoxycholate, 0.01 M sodium phosphate, pH 7.2, 0.1% SDS, 2 mM EDTA, 50 mM NaF) containing Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Rockford, IL, USA). Protein concentration was determined using a Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL, USA). Equal amounts of proteins (30 μg) were resolved by 10% NuPAGE gel (Life Technologies) and transferred onto PVDF membrane (Immobilon-P, Millipore). Antibody detection was accomplished using the enhanced chemiluminescence method (Western Lightning, PerkinElmer) and the LAS-3000 Imaging System (Fujifilm).
Antibodies used for immunoblot analysis are as follows: anti-pSer-473 AKT (no. 9271, Cell Signaling Technology), anti-pThr-308 AKT1 (no. 9275, Cell Signaling Technology), anti-AKT (no. 9272, Cell Signaling Technology), anti-pThr-389 S6K (no. 9205, Cell Signaling Technology), anti-S6K (no. 9202, Cell Signaling Technology), anti-pSer-65 4EBP1 (no. 9451, Cell Signaling Technology), anti-4EBP1 (no. 9452, Cell Signaling Technology), anti-pSer 235/236 S6 (no. 4858, Cell Signaling Technology), anti-S6 (no. 2217, Cell Signaling Technology), anti-p44/42 MAPK (no. 9101, Cell Signaling Technology), anti-44/42 MAPK (no. 9107, Cell Signaling Technology), anti-pSer9 GSK-3β (no. 9336, Cell Signaling Technology), anti-β-Actin (A-1978, Sigma).
In vivo response to therapy
All murine studies were conducted under an MSK IACUC approved protocol. Patient-derived xenograft models were generated as previously described9. Briefly, tumor tissues were minced to small pieces and digested in dissociation buffer containing 225U/ml Collagenase type III at 37°C for 3 hours. Samples were then washed with advanced DMEM/F12 (Life Technologies), treated with Red Blood Cell Lysis Buffer (Sigma) for 2 minutes, washed again, filtered through 70μm cell strainers, and cultured in F10 medium (advanced DMEM/F12 with 10% FBS, 1X L-glutamine, 1X sodium pyruvate, 1X non-essential amino acids and 1X penicilin/streptomycin). 2 million cells resuspended in PBS: matrigel (1:1) were injected into the flank of the mice subcutaneously. 2 flank tumors were placed per mouse. In vivo treatment was started when tumor volumes reached 100–150 mm3. Mice were then randomly assigned into vehicle and treatment groups (5 mice, 9–10 tumors in each arm) and were treated with either rapamycin (7.5 mg/kg) or vehicle (0.25% PEG-400, 0.25% Tween-80) every other day by intraperitoneal injection as previously reported10. Following guidelines set forth by the IACUC, mice were sacrificed when the tumor length reached 2 cm. Tumor volume was measured using calipers (calculated as 0.5 × L × W2), and growth curves were generated using GraphPad Prism software.
Tumor xenograft 1H NMR data acquisition and processing
A mass of 100 mg of wet weight of xenograft tumor was dissected. Pooled tumors from each mouse were finely ground in a mortar under liquid nitrogen. The samples were combined with perchloric acid (4%; 1:4 w/v) and centrifuged at 14000 RPM for 15 min. The supernatant was transferred to a new tube, where chloroform/tri-n-octylamine (78%/22%; v/v) was added in a 1:2 volumetric ratio. The samples were centrifuged at 5000 RPM for 15 min; the aqueous phase was removed and transferred to a microcentrifuge tube and then lyophilized. Then dried samples were dissolved in 800 μL of standard containing 0.5 mM DSS as a concentration reference standard and 10 mM imidazole as a pH reference and placed into a 5-mm NMR tube for subsequent NMR analysis. Experiments were performed on a 14.1 T NMR spectrometer equipped with an autosampler and 1H cryoprobe (Bruker Biospin, Billerica, MA). 1H NMR spectra for each sample were acquired with a water presaturation recycle delay of 4 s, acquisition time of 2.67 s, 90° pulse, and 256 averages. Resonances of each metabolite were identified and quantified using MestReNova 10.0.2 (Spain)11 and bar plots were generated using GraphPad Prism software.
Hyperpolarized [1-13C]pyruvate magnetic resonance
100 μL of 14.2 M [1-13C]pyruvate (GE Healthcare, Waukesha, WI) was mixed with 15 mM of trityl radical (GE Healthcare, Waukesha, WI) and polarized in SPINlab polarizer (GE Healthcare, Waukesha, WI).11 Following polarization, the frozen sample was rapidly dissolved in 20.0 mL of 40 mM TRIS buffer. The dissolution was neutralized in a pre-chilled receiving vial containing sodium hydroxide such that the final solution was pH 7.4. 200 μL of 100 mM hyperpolarized [1-13C]pyruvate was injected intravenously in the tail vein of a catheterized animal in 10 seconds. After 15 seconds of pyruvate distribution, HP data were acquired on a permanent 1T MRI system (nanoScan PET/MRI, Mediso, Hungary). A dual-tuned 1H/13C coil was used to acquire anatomic reference images and spectroscopic data. T2-weighted fast-spin-echo (FSE) images were acquired for anatomic localization, covering the whole tumor (TE/TR=88.5/200 ms, 50-mm FOV, 256×252 matrix, 2-mm slice thickness). A preliminary dynamic slab study on a small group of mice (n=3) was performed to measure average delivery time of hyperpolarized [1-13C]pyruvate to the tumor region. A two-dimensional chemical shift imaging (2DCSI) sequence was used to acquire the HP spectra (32-mm FOV, 12×12 matrix, 2-cm slab thickness, sweepwidth= 3052 Hz, dwelltime = 0.328 ms, spectral points = 496, echo time = 34 ms, repetition time = 140 ms, flip angle = 20°), with the timing derived from the previous dynamic studies. For each cell line, six mice with two flank tumors were used (3 mice per group of vehicle (DMSO) and rapamycin treatment (15mg/Kg), received via intraperitoneal injection 24 hours before the HP MRS study), resulting in 6 tumor replicates per group. Spectroscopic data were processed using custom software (MATLAB R2015b, Mathworks). The HP [1-13C]pyruvate signal (PYR) and [1-13C]lactate signal (LAC) were calculated from the area under the respective peaks for each voxel. The fraction of HP [1-13C]pyruvate converted to lactate was calculated using the following formula for the lactate ratio: Lac/(Lac+Pyr). Tumor regions were delineated on each slice of the anatomic images and registered with the spectroscopy data. Tumor lactate ratios were normalized to muscle lactate ratios to normalize for perfusion differences (eq.1). Average lactate ratios were calculated for each tumor using spectroscopy voxels containing >80% tumor to limit partial volume effects. Normalized lactate ratio maps were generated for tumor voxels containing <20% muscle. The maps were upsampled to 48×48 (0.67×0.67 mm2) and overlaid on the anatomic images. Bar plots were generated using GraphPad Prism software.
| (eq.1) |
Correlative 18FDG PET imaging
In an additional cohort of mice, JHRCC12 and JHRCC228 cells were xenografted and imaged when tumors were approximately 200–300 mm3. Mice were treated with either DMSO (JHRCC12 n=5 and JHRCC228 n=3) or rapamycin (JHRCC12 n=4, JHRCC228 n=4), analogous to the hyperpolarized magnetic resonance studies. 24 hours post-treatment, mice were injected with approximately 5–8 MBq (150–200 μCi) of 18FDG and imaged 45min post-injection on a nanoScan PET/MRI (Mediso, Hungary). Standard uptake values were calculated by annotating regions of interest (ROIs) around the tumor of interest and bladder. The mean activities were recorded for each region of interest, and the standardized uptake value was calculated by multiplying the region-of-interest activity by the mouse weight divided by the injected dose. Bar plots were generated using GraphPad Prism software.
Histologic analysis
Tumor tissue was removed and fixed in 10% (vol/vol) neutral-buffered formalin overnight. The fixed tissue was then processed, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin (H&E). For immunohistochemistry staining, anti-pThr-389 S6K (no. 9205, Cell Signaling Technology) was used. Briefly, formalin-fixed, paraffin-embedded tissue was cut in 5 μm sections and air-dried overnight. Staining was performed in house by Laboratory of Comparative Pathology at MSKCC.
Statistical analysis
Statistical analysis performed with GraphPad Prism software and data presented as mean ± SD (standard deviation) significance of cell line experiments, tracing and hyperpolarized MRS were determined by Student’s t-test (two-tailed, equal variance). Significance of tumor growth curves was analyzed using two-way ANOVA.
RESULTS
Recently, we reported the generation of patient-derived xenograft (PDX) models and cell lines, JHRCC12 and JHRCC228 from clear cell renal cell carcinoma (ccRCC) patients9,12 (Fig. 1A). We investigated the in vitro and in vivo role of mTORi in these models, characterizing their genomics and clinical characteristics.
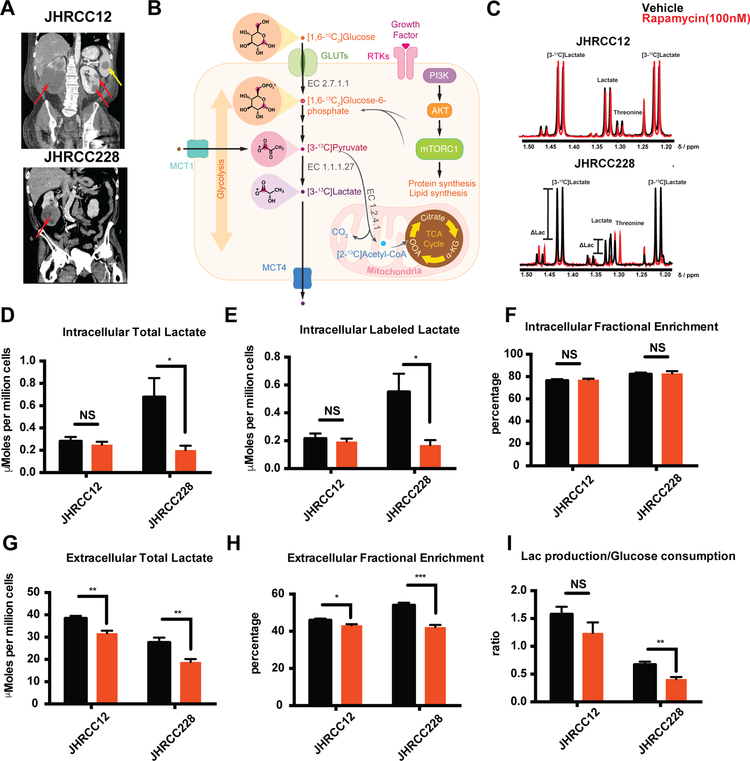
Figure 1.
In vitro characterization of metabolic flux with mTORi. A, CTs of patients from which each line was derived demonstrating widespread metastatic disease (red arrows indicate primary tumors and yellow arrow indicates metastasis). B, [1,6-13C2] glucose metabolism through glycolysis and TCA cycle. EC 2.7.1.1 (hexokinase), EC 1.1.1.27 (L-lactate dehydrogenase), EC 1.2.4.1 (pyruvate dehydrogenase). C, Representative 1HNMR spectra of intracellular JHRCC12 and JHRCC228 after treatment with vehicle (DMSO, black) and rapamycin (100nM, red). D, Total intracellular lactate shows almost no change in JHRCC12 (p<0.54) treatment with rapamycin comparing to DMSO (vehicle) and a significant drop in JHRCC228 treatment (p<0.027) (N=5) (1HNMR studies). E, Labeled intracellular lactate from [1,6-13C2] glucose shows no change in JHRCC12 (p<0.63) treatment with 100 nM rapamycin comparing to DMSO (vehicle) and a decrease in JHRCC228 (p<0.023) (N=5) (1HNMR studies). F, Intracellular fractional enrichment exhibit similar levels of incorporation of 13C carbon in lactate pool. G, Extracellular total lactate has dropped in JHRCC12 (p<0.0008) and JHRCC228 (p<0.0027) (N=5). H, Extracellular fractional enrichment shows a slight change in JHRCC12 (p<0.04) and a greater decrease in JHRCC228 (p<0.0004) (N=5). I, Lactate production/ glucose consumption shows no changes in JHRCC12 (P<0.18) and decreases in JHRCC228 (P<0.006) (N=5). NS (not significant). Data are mean ± SD in D, E, F, G, H and I.
Isotopic tracing studies demonstrate reduced glycolytic metabolism to lactate in with acute rapamycin treatment in vitro
NMR-based metabolomics provides a means of assessing changes in the pool sizes of critical metabolites in vitro13. By combining this approach with 13C-enriched metabolic substrates, the nutrient of origin can readily be identified and this allows for pathway analysis in the setting of evaluating treatment response, where metabolites of interest can be directly quantified (Fig. 1B)14,15. Using a 3-hour incubation of cells with [1,6-13C2] glucose, rapid accumulation [3-13C] lactate is observed, as expected, representing rapid glycolytic flux to lactate (Fig. 1C). The total pool size of lactate was significantly higher in JHRCC228 than in JHRCC12 cells (0.677±0.383, 0.280±0.096 μmoles per million cells, p=0.036), and upon treatment with rapamycin, the level of total lactate significantly decreased only in JHRCC228 cells (Fig. 1D). This was mirrored in isotopic tracing studies, where [3-13C] lactate level was higher in JHRCC228 cells and also decreased nearly 70% after rapamycin treatment (p=0.023) (Fig. 1E). The fractional enrichment (FE) of lactate, measured based on the splitting of the methyl group (1JC-H =128 Hz) (Fig. 1C), was unchanged with treatment in both cell lines reflecting that, while the net flux changes, the observed lactate change is predominantly derived from glucose metabolism.
Excretion of produced lactate to the extracellular space via the monocarboxylate transporters (MCTs) has been shown to be upregulated in cancer and possibly downregulated with rapamycin16,17. The extracellular total lactate significantly decreased in both cell lines, though to a greater degree in JHRCC228 cells (17.4% and 31.6% for JHRCC12 and JHRCC228, Fig. 1G). The FE of lactate was also more greatly reduced in JHRCC228 cells, indicating a substantial decrease in glucose-derived lactate in these cells upon rapamycin treatment (Fig. 1H). Moreover, it is possible that the lactate that is produced is balanced by the glucose consumed, implying that glucose uptake dictates the amount of lactate observed18. Interestingly, the ratio of lactate production to glucose consumption decreases from 0.700 ± 0.095 to 0.410± 0.082 (41.4%) in JHRCC228 cells, while the ratio remains unchanged in JHRCC12 cells (Fig. 1I).
Tyrosine kinase inhibitor resistant ccRCC models demonstrate differential in vitro response to mTORi
To determine whether these cells are sensitive to mTOR inhibition, we treated both cell lines with rapamycin (Fig. 2A, Supp. Table 2). Rapamycin significantly suppressed cell proliferation in both JHRCC12 and JHRCC228 models in vitro. However, JHRCC228 showed complete response to rapamycin, while JHRCC12 showed partial response to rapamycin, with a dose dependent suppression of proliferation.
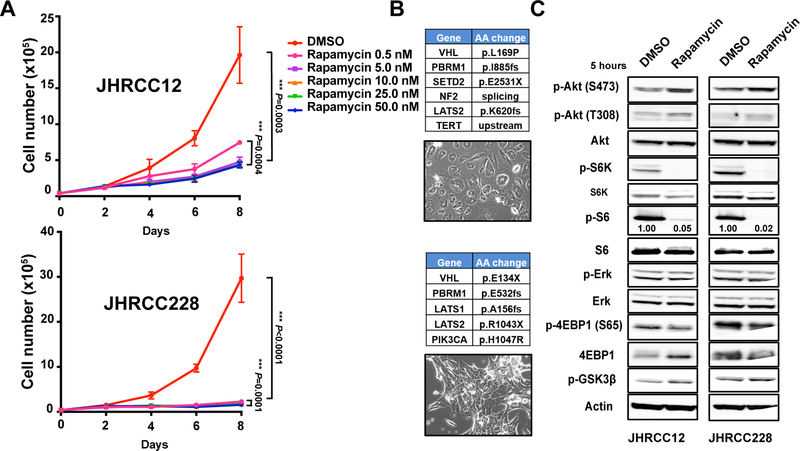
Figure 2.
Comparison of rapamycin response in two primary patient ccRCC cell lines. A, 4×104 JHRCC12 and JHRCC228 cells were seeded, treated with rapamycin at the indicated concentrations, and counted at days 2, 4, 6, and 8 (n=3). Statistical significance was determined by Student’s t-test. B, Tables show genes with mutations in JHRCC12 (top) and JHRCC228 (bottom). Images show the cell morphology of each cell line. C, Immunoblots with the indicated antibodies in JHRCC12 and JHRCC228 cells treated with control and rapamycin (100nM) for 5 hours are shown. Data are mean ± SD in A and B.
We next investigated the underlying mechanisms by which two cell lines showed differential response to rapamycin. One of the major signaling pathways that is affected by rapamycin is PI3K-AKT-mTOR signaling pathway. Rapamycin inhibits the protein kinase target of rapamycin (TOR) by forming a complex with FKBP12, which then binds to mammalian TOR complex 1 (mTORC1). mTORC1 recruits substrates through regulatory-associated protein of mTOR (RAPTOR), which binds a TOR signaling (TOS) motif in substrate proteins such as ribosomal S6 kinase (S6K) and 4E-BP. The binding of rapamycin to mTORC1 suppresses the phosphorylation regulation of mTORC1 to S6K and 4E-BP1, which in turn blocks the downstream signaling that controls protein synthesis and cell proliferation. Western blot analysis revealed that p-S6K was completely blocked in both JHRCC12 and JHRCC228 within 5 hours of treatment (Fig. 2C). However, JHRCC12 maintained a low level of p-S6 expression compared with JHRCC228. These data suggest that the remaining p-S6 might be responsible for the partial response phenotype to rapamycin treatment in JHRCC12. In addition, we found p-AKT (S473), p-AKT (T308) and p-GSK3β levels moderately increase with rapamycin treatment (Fig. 2C). p-GSK3β is a downstream target of AKT signaling and this suggests that there is a negative feedback signaling loop in both cell lines, though this is unlikely to explain the differential sensitivity of the two models to treatment. In summary, both models demonstrate in vitro target inhibition of the mTOR pathway downstream of PI3K.
JHRCC12 and JHRCC228 show differential long-term response to rapamycin in vivo while displaying on-target inhibition
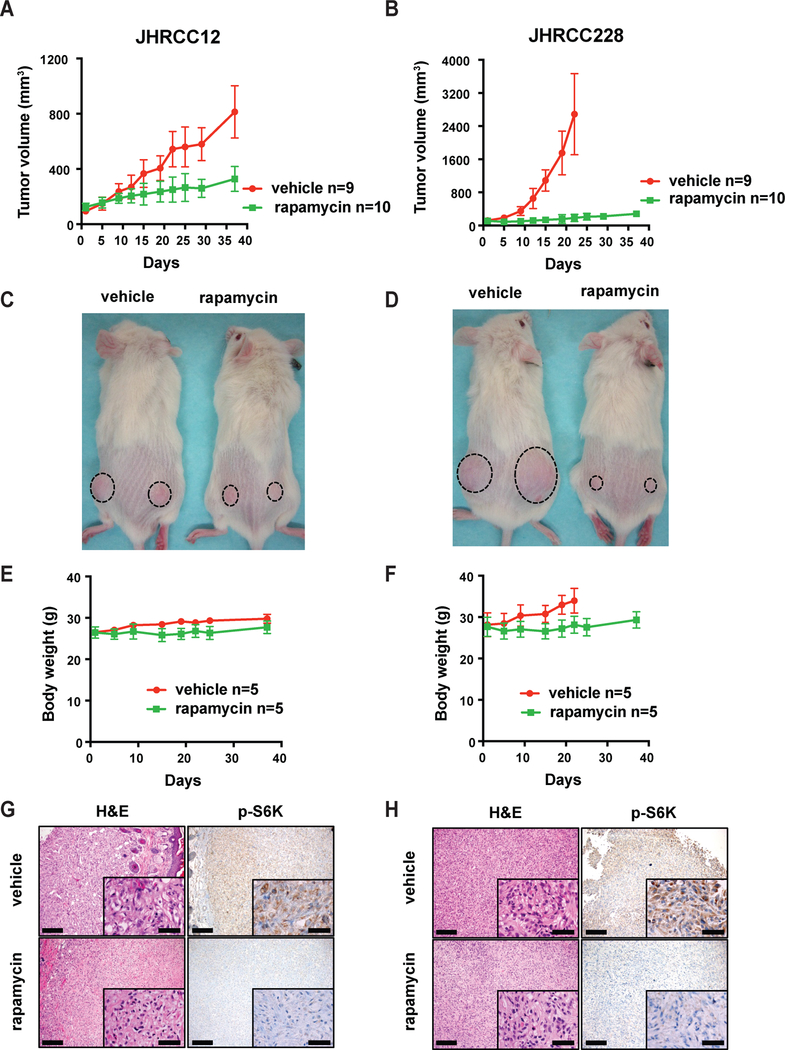
Both clinical data and our previous in vivo data showed JHRCC12 and JHRCC228 are insensitive to anti-angiogenesis treatment9. However, our in vitro data revealed an early differential response to rapamycin in these two models. To further address this difference, we treated tumor-bearing mice for up to 40 days with rapamycin. Consistent with our in vitro results, both models showed significant suppression of tumor growth. However, JHRCC228 tumor growth was fully suppressed by rapamycin and JHRCC12 showed partial inhibition of growth (Fig. 3A, B, C and D). There was no severe body weight loss in both models after rapamycin treatment (Fig. 3E and F). No significant difference was observed in tumor size at 5 days of treatment in either model. p-S6K expression was detected by immunohistochemistry staining in vehicle-treated tumors in both models, but suppressed in rapamycin-treated tumors (Fig. 3G and H).
Figure 3.
Comparison of in vivo rapamycin response. A-D, 2×106 JHRCC12 or JHRCC228 cells were injected subcutaneously into NSG mice. When tumors reached 100–150mm3, mice were randomly divided into two groups. One group was treated with vehicle (n=9) and the other group was treated with rapamycin (n=10). Tumor volume was measured twice per week for 37 days by caliper and tumor growth curve was generated using GraphPad Prism software. Statistical significance of tumor growth curves was analyzed using two-way ANOVA. Animal images were taken 19 days after treatment. E and F, Body weight was measured for 37 days and figures were generated using GraphPad Prism software for both models. G and H, Representative images of vehicle and rapamycin treated tumor grafts stained by H&E and p-S6K for JHRCC12 and JHRCC228. Scale bars indicate 200μm and 50μm, respectively. Data are mean ± SD in A, B, E and F.
Early reduction in hyperpolarized lactate is detected in PIK3CA mutants following treatment
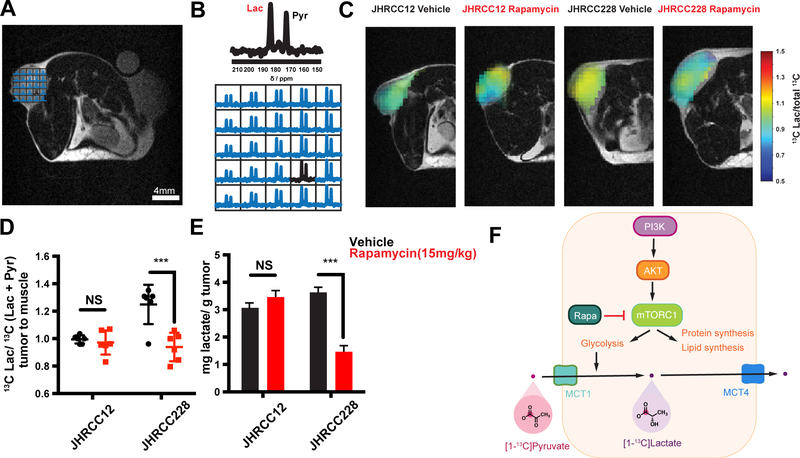
HP MRI using hyperpolarized [1-13C] pyruvate has been used as a method to assess the in vivo metabolic conversion of HP pyruvate to lactate. Recent work has shown that with targeted inhibition of signaling pathways, changes in in vivo HP lactate non-invasively indicate early changes in tumor metabolism, though the relationship between target inhibition and metabolic changes remains unclear19. Figure 4A and B show a representative T2-weighted 1H image with corresponding HP MRI overlay, with the [1-13C] pyruvate peak on the right and [1-13C] lactate on the left. With treatment of a single dose of rapamycin (15 mg/kg) 24 hours before the HP study, JHRCC228 xenografts demonstrated a significant decrease in metabolism (normalized HP lactate: 1.250±0.053 to 0.939±0.042, p=0.0016) compared to non-responding JHRCC12 xenografts (Fig. 4C and D). Following the HP study, tumors were dissected and the total pool size of lactate was measured for comparison to in vivo experiments. The total lactate was significantly decreased more than 50% (p=0.00031) in treated PIK3CA mutant JHRCC228 tumors with rapamycin treatment, whereas there was no detectable difference in JHRCC12 (Fig. 4E). In comparison, when imaged with 18FDG PET, both PDX models demonstrated a reduction in uptake at 24 hr post-rapamycin treatment, though neither was signficant (Supp. Fig. 1). Taken together, this further demonstrates that in vivo HP MRI measurements accurately reflect a metabolic response as early as 24 hours post-treatment, marked by the assessment of a downstream metabolic product, HP lactate.
Figure 4.
Metabolic imaging reveals changes in glycolytic flux within 24 hours of treatment. A and B, Anatomic T2-weighted MRI and representative 2DCSI grid with accompanying HP 13C 2DCSI data demonstrating conversion of HP pyruvate to lactate in a tumor. C, Representative image overlays of normalized HP lactate demonstrating in vivo response to treatment. D, Quantitative comparison of models after 24 hours of treatment with a single dose of rapamycin (15 mg/Kg) in vivo (n=6 replicates per group). The 13C Lac/total 13C carbon represents the lactate signal divided by the sum of pyruvate and lactate as derived by eq. 1. E, Steady-state pool size measurements of lactate in each model derived from tumor extractions and 1H NMR analysis (n=6 replicates per group). All data reported as mean ± SD with significance as a student’s t-test with p<0.05. F, Schematic for in vivo hyperpolarized [1-13C] pyruvate metabolism. NS (not significant)
DISCUSSION
The development and use of targeted therapies have been hindered by the lack of relevant biomarkers for target engagement and, more importantly, on-target efficacy. While the metabolism resulting from inhibition of signaling in the PI3K/mTOR pathway has been interrogated using standard in vitro methods, the ability of metabolic change to predict treatment outcome in a clinically relevant in vivo model has not been explored, and early metabolic biomarkers that would inform on a later response are lacking. In this work, we use hyperpolarized pyruvate MRI to probe in vivo response to a known metabolic target of mTORi and use it to evaluate in vivo response to therapy (Fig. 4F). These metabolic changes, specifically the reduction in glycolytic flux to the product lactate, were confirmed in vitro in our patient-derived model systems and then translated to an in vivo model under conditions analogous to a patient being treated with the same inhibitor. The resulting decrease in HP lactate signal scaled to changes in the pool of lactate prior to tumor size changes20. This change in glycolytic metabolism (a downstream target of the pathway) was not correlated with the status of canonical protein post-translational markers of mTORi (including loss of p-S6K), implying that, while these tumors are dependent on PI3K/mTOR signaling, this metabolism is controlled to some degree outside of these proteins. More importantly, the imaging results demonstrate a biomarker dependent on the pathway and highlights that a PIK3CA mutant, which requires increased glycolytic metabolism to lactate, is sensitive to pathway inhibition and would benefit from such a therapeutic intervention.
Previous work has identified that changes in glycolytic flux can be observed using HP pyruvate MRI, both in vitro and in vivo, though no link has been made to a reliance on the signaling pathway beyond inhibition of post-translational modification of the downstream target protein. Recent work from our group demonstrated that a loss of p-S6K was observed in a range of sarcoma models treated with rapamycin, though a varying sensitivity was observed in metabolic flux to HP lactate11. This along with other work targeting the PI3K/mTOR pathway and using HP MRI to visualize inhibition has not connected overall treatment response to pathway inhibition and has created confusion in the field19,21,22. In this demonstration, a PDX model of ccRCC harboring a PIK3CA activating mutation shows reduced in vivo flux to HP lactate with inhibition, whereas this is not the case for a wild-type PDX. This work highlights that glycolytic flux to lactate likely informs on cancer cells’ requirement for anaerobic energy production to proliferate and that if a targeted treatment directly affects this pathway, reducing the flux, these cancer cells are dependent on that signaling to proliferate.
While this work explores the role of inhibition of the PI3K/mTOR pathway and its effects on glycolytic flux, it is well known that mTOR inhibition can regulate many other metabolic processes in vitro, including amino acid sensing and utilization as well as autophagy23,24. It is likely that these pathways are also rapidly modulated in the setting of mTORi in systems that require this signaling in vivo. With the development of many HP MRI substrates to explore these pathways25, it is possible to take advantage of these approaches to discover other unexpected links between PI3K/mTOR pathway inhibition and metabolic flux as well as translate these metabolic imaging strategies for monitoring response to patient management on therapies that target these aspects of PI3K/mTOR biology in patients.
The use of both in vitro isotopic tracing metabolomics and in vivo HP MRI provides a rapid methodology for characterizing clinically relevant models and their response to treatment. Our future work includes the use of this platform to investigate a range of model genotypes in order to elucidate on-target inhibition of this pathway and link these metabolic changes to a specific signaling cascade in order to inform drug design. Correlative 18FDG PET studies in these same models with mTORi do not demonstrate a marked separation in contrast to the HP pyruvate measurements. This is possibly due to both the potential for dephosphorylation of the trapped 18FDG by high activity of glucose 6-phosphatase (G6Pase)26 which is expressed to a high degree in the kidney27, leading to wash out of the probe, but may also reveal a clear mechanistic requirement for not only control of glucose uptake but downstream glycolytic flux to resulting lactate production. Furthermore, this study is limited in that it only utilizes 2 patient derived models. In further experiments, we aim to develop a deeper understanding of this signaling network using both more models and more highly selective mTOR inhibitors. Moreover, given that significant metabolic inhibition was observed in the PDX model following a single dose of treatment (24 hours post-administration) and that this metabolic change predicted therapeutic sensitivity, this methodology could be readily used in patients with ccRCC by imaging before and after initial treatments to predict response to therapy. HP MRI has recently been translated to the clinic and early studies have demonstrated the feasibility and safety of HP pyruvate for imaging4,28. We are hopeful that this could allow for not only personalized approaches to non-invasively monitor primary ccRCC with treatment, but also assessing the heterogeneity of disease response in vivo.
Supplementary Material
SIGNIFICANCE.
Precision oncology hinges on predictive biomarkers. mTORC1 integrates cancer metabolism, and its inhibition yields varied clinical benefits in ccRCC patients. Here, hyperpolarized [1-13C] pyruvate MRI provides a functional assessment of mTOR treatment.
ACKNOWLEGEMENT
This work was supported by the National Institutes of Health, R01 CA223231 (YD, CR, EHC, and JJH), P30 CA008748 (KRK), R00 EB014328 (KRK), R01 CA195476 (KRK) and R21 CA212958 (KRK), K99 CA226357 (SJ), as well as the Society of Memorial Sloan Kettering (YC), the Center for Molecular Imaging and Nanotechnology (KRK) and the Geoffrey Beene Cancer Research Center (KRK). KRK also serves on the SAB of NVision Imaging Technologies.
Footnotes
Conflict of interest: KRK serves on the SAB of nVision Imaging. The authors have no other potential conflict of interest to disclose.
REFERENCES
- 1.Vander Heiden MG, Cantley LC & Thompson CB Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janku F, Yap TA & Meric-Bernstam F Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15, 273–291 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Taha C, et al. Opposite translational control of GLUT1 and GLUT4 glucose transporter mRNAs in response to insulin - Role of mammalian target of rapamycin, protein kinase B, and phosphatidylinositol 3-kinase in GLUT1 mRNA translation. J Biol Chem 274, 33085–33091 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Nelson SJ, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-C-13]Pyruvate. Sci Transl Med 5(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miloushev VZ, et al. Metabolic Imaging of the Human Brain with Hyperpolarized 13C Pyruvate Demonstrates 13C Lactate Production in Brain Tumor Patients. Cancer Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh JJ, et al. Renal cell carcinoma. Nat Rev Dis Primers 3, 17009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voss MH, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res 20, 1955–1964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei EY & Hsieh JJ A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol 12, 706–712 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Dong Y, et al. Tumor Xenografts of Human Clear Cell Renal Cell Carcinoma But Not Corresponding Cell Lines Recapitulate Clinical Response to Sunitinib: Feasibility of Using Biopsy Samples. European urology focus 3, 590–598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodrum C, Nobil A & Dabora SL Comparison of three rapamycin dosing schedules in A/J Tsc2+/− mice and improved survival with angiogenesis inhibitor or asparaginase treatment in mice with subcutaneous tuberous sclerosis related tumors. J Transl Med 8, 14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Gialleonardo V, et al. Multinuclear NMR and MRI Reveal an Early Metabolic Response to mTOR Inhibition in Sarcoma. Cancer Res 77, 3113–3120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh JJ, et al. Genomic Biomarkers of a Randomized Trial Comparing First-line Everolimus and Sunitinib in Patients with Metastatic Renal Cell Carcinoma. European urology 71, 405–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan TWM & Lane AN Structure-based profiling of metabolites and isotopomers by NMR. Prog Nucl Mag Res Sp 52, 69–117 (2008). [Google Scholar]
- 14.Di Gialleonardo V, et al. High-Throughput Indirect Quantitation of 13C Enriched Metabolites Using 1H NMR. Analytical chemistry 88, 11147–11153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buescher JM, et al. A roadmap for interpreting C-13 metabolite labeling patterns from cells. Curr Opin Biotech 34, 189–201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baek G, et al. MCT4 Defines a Glycolytic Subtype of Pancreatic Cancer with Poor Prognosis and Unique Metabolic Dependencies. Cell Rep 9, 2233–2249 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Keshari KR, et al. Hyperpolarized C-13-Pyruvate Magnetic Resonance Reveals Rapid Lactate Export in Metastatic Renal Cell Carcinomas. Cancer research 73, 529–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edinger AL & Thompson CB Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13, 2276–2288 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaumeil MM, et al. Hyperpolarized C-13 MR spectroscopic imaging can be used to monitor Everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. Neuroimage 59, 193–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuru TH, et al. Initial experience with temsirolimus in 2nd-, 3rd- and 4th-line therapy for metastatic renal cell cancer: good clinical response. Urologia internationalis 86, 256–260 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Ward CS, et al. Noninvasive detection of target modulation following phosphatidylinositol 3-kinase inhibition using hyperpolarized 13C magnetic resonance spectroscopy. Cancer research 70, 1296–1305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radoul M, et al. MR Studies of Glioblastoma Models Treated with Dual PI3K/mTOR Inhibitor and Temozolomide:Metabolic Changes Are Associated with Enhanced Survival. Molecular cancer therapeutics 15, 1113–1122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Kundu M, Viollet B & Guan KL AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshari KR & Wilson DM Chemistry and biochemistry of C-13 hyperpolarized magnetic resonance using dynamic nuclear polarization. Chem Soc Rev 43, 1627–1659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caraco C, Aloj L, Chen LY, Chou JY & Eckelman WC Cellular release of [18F]2-fluoro-2-deoxyglucose as a function of the glucose-6-phosphatase enzyme system. J Biol Chem 275, 18489–18494 (2000). [DOI] [PubMed] [Google Scholar]
- 27.van Schaftingen E & Gerin I The glucose-6-phosphatase system. Biochem J 362, 513–532 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miloushev VZ, et al. Metabolic Imaging of the Human Brain with Hyperpolarized (13)C Pyruvate Demonstrates (13)C Lactate Production in Brain Tumor Patients. Cancer Res 78, 3755–3760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.