Summary
Stromalin, a cohesin complex protein, was recently identified as a novel memory suppressor gene but its mechanism remained unknown. Here, we show that Stromalin functions as a negative regulator of synaptic vesicle (SV) pool size in Drosophila neurons. Stromalin knockdown in dopamine neurons during a critical developmental period enhances learning and increases SV pool size without altering the number of dopamine neurons, their axons, or synapses. The developmental effect of Stromalin knockdown persists into adulthood, leading to strengthened synaptic connections and enhanced olfactory memory acquisition in adult flies. Correcting the SV content in dopamine neuron axon terminals by impairing anterograde SV trafficking motor protein Unc104/KIF1A rescues the enhanced learning phenotype in Stromalin knockdown flies. Our results identify a new mechanism for memory suppression and also reveal that the size of the SV pool is controlled genetically and independently from other aspects of neuron structure and function through Stromalin.
Keywords: cohesin, dopamine, olfactory learning, synaptic vesicles, KIF1A
Graphical Abstract
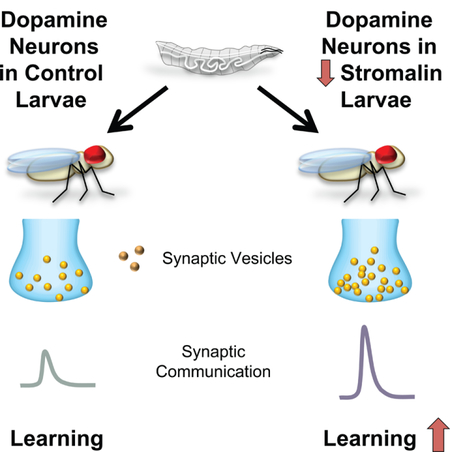
eTOC Blurb
Phan et. al., report cohesin complex gene stromalin developmentally and negatively regulates synaptic vesicle number, independent of axonal morphology, synapse number or volume. Stromalin and Unc104/KIF1A offer tools to bidirectionally alter synaptic vesicle numbers and learning in Drosophila.
Learning and memory are tightly regulated processes that require the activity of hundreds of genes to orchestrate the proper development of neural circuits and the underlying physiological changes necessary for cellular and synaptic plasticity. While many genes are known that define mechanisms required for the formation and consolidation of memory (Alberini and Kandel, 2014; Davis, 2005; Kandel, 2001), far less is known about the genetic factors that constrain memory formation and their molecular and cellular mechanisms (Abel et al., 1998). Important conceptual insights about memory formation might come from elucidating the cellular mechanisms underlying this class of genetic element. Memory suppressor genes, named so by analogy to tumor suppressor genes (Abel et al., 1998), could in principal function by limiting memory acquisition, consolidation or retrieval; or by participating in active forgetting processes (Davis and Zhong, 2017).
Several dozen and novel memory suppressor genes were recently identified in a large RNAi screen for effects on 3h aversive olfactory memory expression in Drosophila (Walkinshaw et al., 2015). They were classed as such because knockdown of these genes led to increased memory expression. Stromalin, a member of the cohesin complex, was one such gene identified in the screen. The highly conserved cohesin complex is comprised of Stromalin (STAG1/2 in mammals) and three other subunits named structural maintenance of chromosomes 1 (SMC1), SMC3, and Rad21 (Dorsett, 2011; Peters et al., 2008). Although the complex was first identified for its role in the proper segregation of chromosomes during cell division (Peters et al., 2008), evidence has emerged showing that the complex has other important biological functions. Cohesion complex mRNAs and proteins are present at moderate to high levels in both the Drosophila and mouse nervous system (Fukuda and Hoog, 2010; Lee et al., 2002; Valdeolmillos et al., 1998), revealing potential roles beyond chromosome segregation. Elegant studies from two research groups have clearly shown that members of the complex have a post-mitotic role in the proper pruning of axons in Drosophila mushroom body neurons (Pauli et al., 2008; Schuldiner et al., 2008). Other studies have provided evidence for roles in gene expression, DNA repair, and cancer susceptibility (Dorsett, 2011; Dorsett and Strom, 2012; Losada, 2014). It is notable that absent from this list are clear and specific roles for the complex in learning and memory processes, other than the general cognitive disturbances observed in humans with cohesinopathies (see Discussion).
We report here that RNAi knockdown of Stromalin in mushroom body and dopamine neurons leads to enhanced aversive olfactory memory in adult flies. We show that stromalin functions during development as a negative regulator of both synaptic and dense-core vesicle number in the nervous system, limiting the strength of synaptic connections to suppress memory acquisition. Reducing Stromalin levels specifically increases the number of vesicles in neurons, without detectably altering other features of the targeted neurons in adult flies, including synapse number, synapse volume, or neurite branching. These observations offer evidence that the size of the synaptic vesicle pool is regulated independently of other structural features of the neuron.
Results
Aversive olfactory memory is limited by Stromalin
Western blot analyses on fly head lysates confirmed that pan-neuronal expression of an RNAi against stromalin (SA) from a targeted insertion site in the genome resulted in a ~50% decrease in Stromalin protein levels in the adult head compared to control flies, which contain an empty landing site (Figure 1A, Figure S1A). Interestingly, we find that pan-neuronal expression of stromalinRNAi resulted in an increase in 3hr aversive olfactory memory scores compared to control flies (Figure 1B), which was also replicated by a second independently derived stromalinRNAi fly line (stromalinRNAi2; Figure S1B). Moreover, we found no difference in odor and shock avoidance behavior of the knockdown (KD) flies that could account for the increased memory performance (Figure S1C).
Figure 1. Stromalin knockdown enhances aversive olfactory memory.
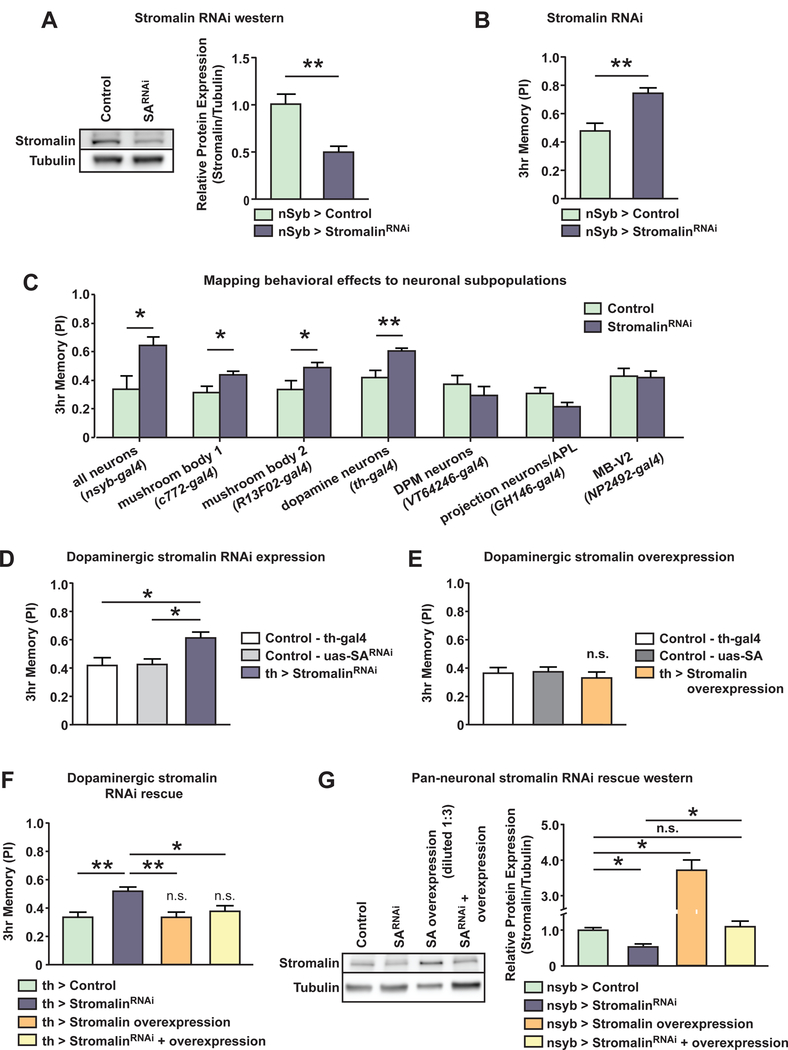
(A) Western blot of fly head lysates demonstrated pan-neuronal stromalinRNAi expression significantly reduced Stromalin protein levels. A full western blot is shown in Supplementary Figure 1B. Control: nSyb-GAL4>+, UAS-Dcr2. StromalinRNAi: nSyb-GAL4>UAS-SARNAi, UAS-Dcr2. Mann-Whitney U, **p<0.01. n=5. Mean±SEM.
(B) Pan-neuronal expression of stromalinRNAi increased 3hr aversive olfactory memory scores. Control: nSyb-GAL4>+, UAS-Dcr2. StromalinRNAi: nSybGAL4>UAS-SARNAi, UAS-Dcr2. Student’s t-test, **p<0.01. n=8. Mean±SEM.
(C) Knockdown of Stromalin in MBn (c772-GAL4, R13F02-GAL4) or DAn (THGAL4) enhanced 3hr memory scores. Control: GAL4>+, UAS-Dcr2. StromalinRNAi: GAL4>UAS-SARNAi, UAS-Dcr2. Student’s t-test, *p<0.05, **p<0.01. n=8–14. Mean±SEM.
(D) Stromalin knockdown in DAn enhanced 3hr memory scores compared to GAL4 and UAS genetic controls. Control-GAL4: TH-GAL4>+, UAS-Dcr2. ControlUAS: + >UAS-SARNAi. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2. One-way ANOVA, Bonferroni post hoc, *p<0.05. n=10. Mean±SEM.
(E) Overexpression of Stromalin in DAn did not alter 3hr memory scores compared to GAL4 and UAS genetic controls. Control-GAL4: TH-GAL4>+. Control-UAS: + >UAS-SA. Stromalin overexpression: TH-GAL4>UAS-SA. n=12. Mean±SEM.
(F) Stromalin knockdown in DAn increased 3hr memory scores compared to controls, while stromalin overexpression in DAn had no effect (not significant compared to controls). The enhanced memory phenotype in Stromalin KD animals was rescued when stromalinRNAi expression was combined with Stromalin overexpression in DAn (yellow bar; not significantly different from the control). Control: TH-GAL4>+, UAS-Dcr2. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2. Stromalin Overexpression: TH-GAL4>UAS-SA, UAS-Dcr2. StromalinRNAi + Overexpression: TH-GAL4>UAS-SARNAi, UAS-SA, UAS-Dcr2. One-way ANOVA, Bonferroni post hoc, *p<0.05, **p<0.01. n=10. Mean±SEM.
(G) Western blotting experiments using fly head lysates revealed that Panneuronal stromalinRNAi expression and Stromalin overexpression resulted in significantly lower and higher levels of Stromalin protein compared to the control, respectively. A full western blot is shown in Supplementary Figure 1D. Most importantly, stromalinRNAi + Stromalin overexpression in neurons produced Stromalin protein levels similar to the control, both are are significantly higher than Stromalin knockdown samples. Control: nSyb-GAL4>+, UAS-Dcr2. StromalinRNAi: nSyb-GAL4>UAS-SARNAi, UAS-Dcr2. Stromalin overexpression: nSyb-GAL4>UAS-SA, UAS-Dcr2. StromalinRNAi + overexpression: nSybGAL4>UAS-SARNAi, UAS-SA, UAS-Dcr2. Kruskal-Wallis, Dunn’s post hoc, *p<0.05. n=8–9. Mean±SEM.
See also Figure S1.
Using a panel of GAL4 drivers, flies expressing stromalinRNAi in specific subpopulations of neurons previously shown to be important for aversive olfactory conditioning were tested for 3hr memory performance. We found that Stromalin KD in both mushroom body neurons (MBn) and dopaminergic neurons (DAn) led to enhanced 3hr memory scores, while expression elsewhere in the olfactory circuit had no effect (Figure 1C). Because DAn are critical mediators of learning and memory in Drosophila (Davis, 1993, 2011; Heisenberg, 2003), we chose to focus on Stromalin effects in DAn in subsequent experiments. The THGAL4 driver expresses in a subset of Drosophila DAn, including the PPL1 cluster that innervates the vertical lobe, heel and junction neuropil of the MB, a structure known to be important for the formation of aversive olfactory memory (Davis, 2011). Behavioral testing of flies carrying a TH-GAL4/+ driver with UAS-Dicer2/+, or flies carrying UAS-SARNAi/+ alone revealed that the behavioral effect was not due to leaky expression of stromalinRNAi, as flies only displayed an increased memory score when the GAL4 driver and UAS-SARNAi (along with UAS-Dicer2) were present in the same fly (Figure 1D). Interestingly, overexpression of Stromalin in DAn using a UAS-SA construct did not alter memory expression (Figure 1E), despite experimental results showing that expression of the UAS-SA construct significantly increases Stromalin abundance (Figure 1G, Figure S1D). Thus, wild-type levels of Stromalin are not limiting for memory performance. We also tested the KD of SMC1, another subunit of the cohesin complex, using THGAL4 and discovered that this mimicked the enhanced memory phenotype of Stromalin KD flies (Figure S1E). This observation suggests that the cohesion complex itself, or major components of it, is responsible for the increased memory when expressed at less than normal abundance levels.
We then attempted to rescue the enhanced memory phenotype in Stromalin KD flies using the overexpression construct. Replicating our previous results, Stromalin KD in DAn increased, while overexpression of Stromalin in DAn did not alter, 3hr memory scores (Figure 1F). However, flies containing both UAS-SARNAi and UAS-SA transgenes had memory scores indistinguishable from control flies, effectively rescuing the enhanced memory phenotype of Stromalin KD flies. These behavioral results are consistent with Stromalin protein levels in these transgenic fly lines measured by Western blotting. Pan-neuronal Stromalin KD reduced and overexpression increased Stromalin protein levels (Figure 1G, Figure S1D). Flies expressing both UAS-SARNAi and UAS-SA transgenes had protein levels similar to control flies. Thus, normalizing Stromalin protein levels in KD flies rescued aversive olfactory memory expression.
Memory acquisition, but not memory retention, is suppressed by Stromalin
Several distinct operations underlie the formation of behavioral memory (Davis, 1993, 2011; Heisenberg, 2003), which include acquisition, consolidation, retrieval and forgetting. To dissect the operations of learning and memory that are altered by Stromalin KD in DAn, we performed memory acquisition and retention experiments. Memory retention was examined by varying the time between training and testing (3min-24hr). Flies with Stromalin KD in DAn displayed significantly higher memory scores 1–6hr after training compared to control flies (Figure 2A), although there remained a trend towards elevated performance at 12hr. Memory scores at 24hr after conditioning are very low and it is difficult to extract true differences. Similarly, it is often difficult to extract true differences between groups at 3min after conditioning, since ceiling levels of performance are reached at this time point. Overall, these results suggest a general increase in memory performance after conditioning.
Figure 2. Stromalin knockdown in dopamine neurons during development enhances memory acquisition and retention.
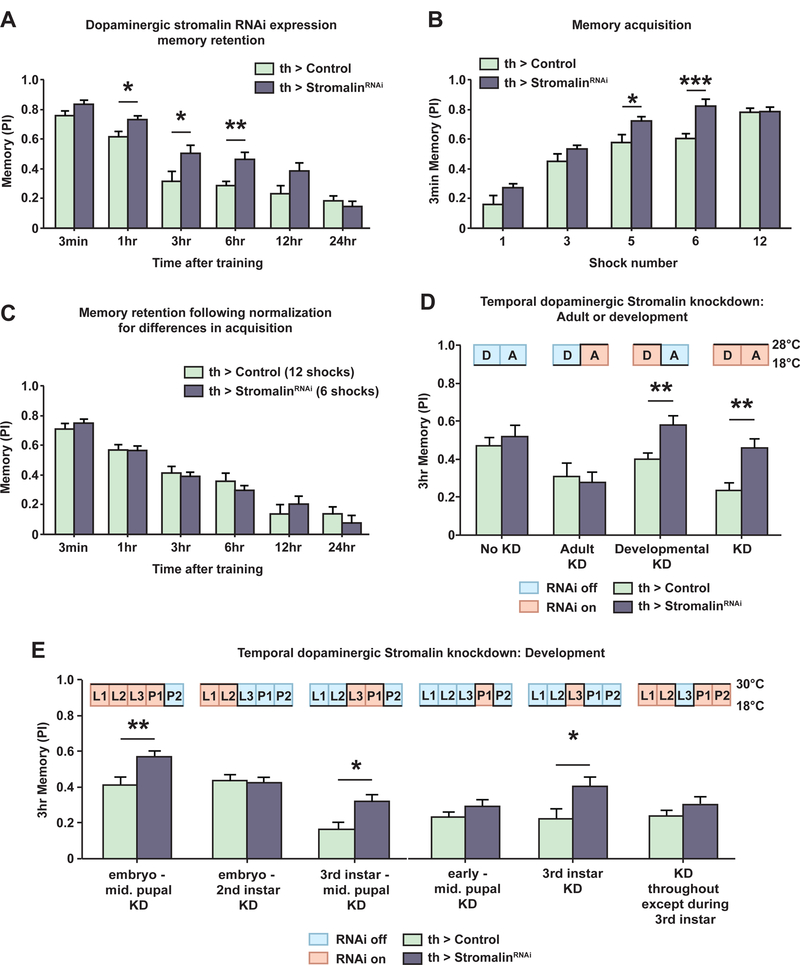
(A) Knockdown of Stromalin in DAn increased memory retention 1–6hr after training. Control: TH-GAL4>+, UAS-Dcr2. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2. Student’s t-test, *p<0.05, **p<0.01. n=8–10. Mean±SEM.
(B) Expression of stromalinRNAi in DAn increased memory acquisition when flies were trained with 5 or 6 electric shock pulses. Memory acquisition was measured by testing 3min memory after varying the number of electric shock pulses received during training. Control: TH-GAL4>+, UAS-Dcr2. StromalinRNAi: THGAL4>UAS-SARNAi, UAS-Dcr2. Student’s t-test, *p<0.05, ***p<0.001. n=8–10. Mean±SEM.
(C) After normalizing for differences in memory acquisition by varying the number of electric shock pulses received by each genotype during training, Stromalin KD in DAn did not alter memory retention. Control: TH-GAL4>+, UAS-Dcr2. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2. n=9–11. Mean±SEM.
(D) Stromalin KD in DAn increased 3hr memory scores in adult flies only when stromalinRNAi was expressed during development. The temporal control of stromalinRNAi expression using incubation temperature and tub-gal80ts (TARGET) is schematized above the bar graphs. Control: tub>gal80ts, TH-GAL4>+, UASDcr2. StromalinRNAi tub>gal80ts, TH-GAL4>UAS-SARNAi, UAS-Dcr2. Student’s ttest, **p<0.01. n=8–9. Mean±SEM.
(E) Stromalin KD in DAn during the 3rd instar larval stage was necessary and sufficient for the enhanced memory phenotype in adult flies. The temporal control of stromalinRNAi expression for windows of Drosophila development is schematized above bar graphs. Control: tub>gal80ts, TH-GAL4>+, UAS-Dcr2. StromalinRNAi tub>gal80ts, TH-GAL4>UAS-SARNAi, UAS-Dcr2. Student’s t-test, *p<0.05, **p<0.01. n=8–14. Mean±SEM.
See also Figure S2.
We then probed acquisition in these flies by varying the number of electric shock pulses they received and testing their performance immediately after training. We found that flies expressing stromalinRNAi in DAn had significantly higher rates of memory acquisition compared to controls when they received an intermediate number of electric shock pulses during training (5–6 shock pulses; Figure 2B). Ceiling levels of performance were reached with 12 shock pulses. These results indicate that Stromalin KD flies appeared to learn just as well as control flies when given only half the number of training associations. Because reduction of Stromalin in DAn enhanced both acquisition and retention, it raised the question of whether the increased memory retention scores are a consequence of enhanced acquisition, or whether both acquisition and retention of memory are enhanced independently. We distinguished between these two possibilities by normalizing for the genotype differences in memory acquisition. Control flies were presented with 12 electric shock pulses while stromalinRNAi flies were presented 6 electric shock pulses during training before testing for memory retention. When trained in this manner, the genotype differences in memory retention disappeared (Figure 2C). Thus, Stromalin KD in DAn appears to specifically enhance memory acquisition, with increased memory retention occurring as a consequence of increased acquisition.
Stromalin alters memory during a narrow developmental window
To unravel the time course for Stromalin’s limiting effects on olfactory memory, we employed the TARGET system, a temperature sensitive repressor of GAL4 (tub-gal80ts; McGuire et al., 2003), to temporally control stromalinRNAi expression in DAn neurons by manipulating incubation temperatures. Using this system, we surprisingly found that Stromalin KD in DAn during development, but not during adulthood, led to an enhanced memory phenotype in adult flies (Figure 2D). Drosophila development consists of several well-defined stages: embryo, 1st, 2nd, and 3rd instar larval stages (L1, L2, L3) followed by the pupal stage (divided into two halves for this experiment; P1, P2). We limited Stromalin expression in DAn during increasingly narrow developmental stages and tested adult flies for aversive olfactory memory performance. This allowed us to determine that Stromalin KD in DAn specifically during the 3rd instar larval stage was both necessary and sufficient for the enhanced memory scores displayed by these flies as adults (Figure 2E). Remarkably, KD of Stromalin only during the 3rd instar produced enhanced memory during adulthood, where KD at all developmental stages except for the 3rd instar was without effect (Figure 2B).
To further delimit Stromalin’s spatial role in memory beyond the PPL1 DAn in the TH-GAL4 expression pattern (Figure 1C), we employed a series of GAL4 drivers exhibiting more limited expression across the PPL1 DAn cluster (Figure S2). However, our initial results based on the known adult expression patterns of these GAL4 drivers were inconsistent. For example, two GAL4 drivers (c061GAL4 and c150-GAL4) both expressed in adult PPL1 DAn innervating the MB heel gave inconsistent results. Since Stromalin KD during the 3rd instar larval stage is critical for its behavioral effects, we examined the GAL4 expression patterns during the 3rd instar larval stage using a membrane-GFP label. In doing so, we found that most GAL4 drivers for DAn PPL1 subpopulations have different expression patterns during development compared to adult flies (Figures S2). This developmental analysis of GAL4 expression patterns, and the enhanced memory performance with the ∆TH-D’-GAL4 and c061-GAL4 drivers, led us to conclude that stromalinRNAi expression in the DAn innervating the heel of the MB was sufficient to produce enhanced memory scores in adult flies.
Stromalin suppresses the synaptic communication between DAn and MBn
We wondered whether the enhanced memory due to Stromalin KD in the THGAL4 subset of DAn might occur from increased functional connections between the PPL1 DAn and the axons of MBn in the lobes, since this connection is known to be important for aversive olfactory memory (Davis, 2011). To test for this possibility, we performed ex vivo experiments using adult brains in which the ATP-gated ion channel, P2X2, was expressed in DAn neurons, allowing us to artificially activate the neurons with increasing concentrations of ATP by bath application. We simultaneously expressed the cAMP sensor, Tepacvv, in the MBn (Boto et al., 2014; Klarenbeek et al., 2011), since DA is known to increase cAMP in the MB lobes (Tomchik and Davis, 2008). Increasing the ATP concentrations produced the predicted increase in Tepacvv fluorescence. When stromalinRNAi was expressed in DAn, ATP-induced DAn activation produced a ~2-fold increase in cAMP response over the control (at 1mM ATP) in the MB heel, junction and upper stalk regions (Figure 3A, Figure S3). These experiments confirmed that the PPL1 DAn to MB synaptic communication was strengthened in Stromalin KD flies.
Figure 3. Stromalin knockdown in dopamine neurons increases the cAMP signal generated in the MB neuropil.
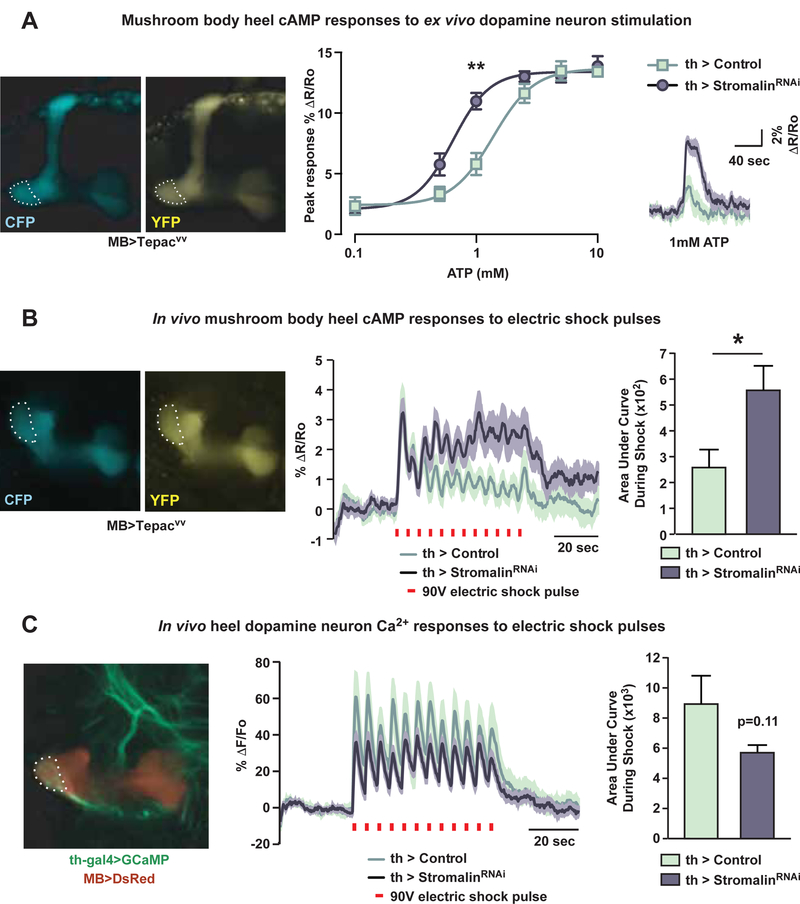
(A) Stromalin KD in DAn increased cAMP responses in the MB heel (dotted outline) upon DAn activation using the ATP-gated P2X2 ion channel in an ex vivo preparation. The mean responses in the right panel are depicted as dark lines with the SEM as the lighter shading. R=ratio of CFP/YFP fluorescence intensity of cAMP reporter Tepacvv. Control: MB>Tepacvv, TH-GAL4>+, UAS-Dcr2, UASP2X2. StromalinRNAi: MB>Tepacvv, TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-P2X2. Mann-Whitney U with Bonferroni correction for multiple comparisons, **p<0.01. n=8–9. Mean±SEM.
(B) Stromalin KD in DAn increased the electric shock-induced cAMP responses in the heel of the MB (dotted outline). The mean responses in the middle panel are shown with dark lines and the SEM as lighter shading. R=ratio of CFP/YFP fluorescence intensity of cAMP reporter Tepacvv. Red bars indicate electric shock delivery (90V, 1.25sec). The bar graph shows the quantitation of the response as the area under the curve between the two genotypes. Control: MB>Tepacvv, TH-GAL4>+, UAS-Dcr2. StromalinRNAi: MB>Tepacvv, TH-GAL4>UAS-SARNAi, UAS-Dcr2. Mann-Whitney U, *p<0.05. n=18. Mean±SEM.
(C) Stromalin KD did not significantly alter electric shock-induced calcium responses in DAn axon terminals innervating the MB heel (dotted outline). F=fluorescence intensity of calcium reporter GCaMP. Red bars indicate electric shock delivery (90V, 1.25sec). The mean response in the middle panel is shown as the dark lines with the SEM as the lighter shading. Bar graph shows the quantitation of the response as the area under the curves between the two genotypes. Control: MB>DsRed, TH-GAL4>+, UAS-Dcr2, UAS-GCaMP6m. StromalinRNAi: MB>DsRed, TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-GCaMP6m. n=18. Mean±SEM.
See also Figure S3.
This observation provided a sound explanation for how reducing Stromalin in the DAn increases acquisition and memory, since current evidence indicates that DAn activation represents the unconditioned stimulus (US) of electric shock for aversive olfactory conditioning (Davis, 2011). The increased synaptic communication between DAn and MBn would lead to an increase in potency of the US to produce an enhancement in acquisition and memory. Nevertheless, the ex vivo conditions and artificial stimulation methods might not accurately reflect the neuronal responses in flies during behavioral training. Therefore, we developed an in vivo, under the microscope, electric foot shock apparatus that mimics behavioral training conditions. Through a window in the fly head, cAMP responses in the MB heel region were imaged during the delivery of electric shock pulses to the fly in the absence of any odor presentation. We were able to see clear peaks of cAMP increases for each electric shock pulse delivered to the fly (12 × 90V at 5sec intervals). Interestingly, control flies showed the largest cAMP response to the first shock, while subsequent shock pulses produced steadily decreasing cAMP responses thereafter (Figure 3B). In contrast, while DAn Stromalin KD flies showed a similar amplitude peak at the first electric shock pulse, cAMP responses to subsequent shock pulses remained elevated (Figure 3B). Calculations of the area under the response profile during shock pulse delivery (from 30–90sec) revealed that Stromalin KD in DAn significantly increased the cAMP signal generated in the MB heel in response to shock. These data argue that Stromalin KD in DAn during a critical developmental period enhances memory acquisition by potentiating the shock-induced DAn to MB communication.
Stromalin does not alter DAn numbers, excitability, or morphology
How does Stromalin alter synaptic communication between the DAn and MBn? One possibility is that it may affect the number of PPL1 DAn in the Drosophila brain during development, changing the potency of the US. This possibility was attractive because of the role for the cohesin complex in proper cell division (Peters et al., 2008). To visualize and count PPL1 DAn, we expressed membrane or nuclear localized GFP in these neurons using TH-GAL4. Stromalin KD in DAn did not significantly alter the numbers of PPL1 neurons in adult flies (Figure 4A–B). Therefore, differences in DAn numbers do not account for Stromalin’s effects on learning.
Figure 4. Stromalin knockdown in dopamine neurons increases levels of presynaptic synaptotagmin:GFP without altering other neuroanatomical markers.
(A) Schematic diagram of the DAn (PPL1) innervation pattern of the MB lobe neuropil. The PPL1 DAn send axonal projections to the vertical but not the horizontal lobes of the MB neuropil.
(B) The number of PPL1 DAn soma, visualized in the dorsal posterior brain using membrane-localized or nuclear-localized GFP, was not altered by Stromalin KD. Control: TH-GAL4>+, UAS-Dcr2, UAS-mCD8:GFP or TH-GAL4>+, UAS-Dcr2, UAS-GFPnls. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-mCD8:GFP or TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-GFPnls.
(C) Stromalin KD in DAn did not affect dopaminergic innervation of the of the MB neuropil, visualized using membrane-localized GFP. Quantitation of fluorescence intensity across MB lobe regions is shown in the right bar plot. MB lobes are outlined with a dotted line. Control: TH-GAL4>+, UAS-Dcr2, UAS-mCD8:GFP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-mCD8:GFP. n=19–20. Mean±SEM. Scale bar 20µm.
(D) Stromalin KD in DAn resulted in elevated levels of the presynaptic marker, syt:GFP across all sections of the vertical lobes and heel. Quantitation of fluorescence intensity across regions of interest are shown in the right bar plot. MB lobes are outlined with a dotted line. Control: TH-GAL4>+, UAS-Dcr2, UASsyt:GFP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-syt:GFP. MannWhitney U, ***p<0.001. n=20. Mean±SEM. Scale bar 20µm.
(E) Stromalin KD increased dopaminergic syt:GFP levels during the middle to late 3rd instar larval stage in the MB heel (arrowhead). MB vertical lobe, junction and heel are outlined with a white dotted line. Quantitation of fluorescence intensity in the heel region is shown in the right bar plot. Quantitation of fluorescence intensity in the vertical lobes is presented in Figure S4. Control: THGAL4>+, UAS-Dcr2, UAS-syt:GFP, UAS-myr-tdTom. StromalinRNAi: THGAL4>UAS-SARNAi, UAS-Dcr2, UAS-syt:GFP, UAS-myr-tdTom. Mann-Whitney U, *p<0.05. n=11–15. Mean±SEM. Scale bar 20µm.
(F) Pan-neuronal stromalinRNAi expression increased syt:GFP across the whole adult Drosophila brain. Control: nSyb-GAL4>+, UAS-Dcr2, and UAS-syt:GFP. StromalinRNAi: nSyb-GAL4>UAS-SARNAi, UAS-Dcr2, and UAS-syt:GFP. MannWhitney U, ***p<0.001. n=13–14. Mean±SEM. Scale bar 100µm.
(G) Reducing Stromalin in MBn increased syt:GFP levels in the adult Drosophila MBn. Control: nSyb-GAL4>+, UAS-Dcr2, and UAS-syt:GFP. StromalinRNAi: nSyb-GAL4>UAS-SARNAi, UAS-Dcr2, and UAS-syt:GFP. Mann-Whitney U, ****p<0.0001. n=13. Mean±SEM. Scale bar 50µm.
See also Figure S4.
A second possibility is that Stromalin KD increases the excitability of the DAn and/or their responsiveness to shock stimuli. To test this, we expressed the cytoplasmic Ca2+ reporter, GCaMP6m (Chen et al., 2013), in the DAn and quantified the Ca2+ responses to electric shock pulses. Interestingly, we found that the Ca2+ responses in the axons of DAn innervating the MB heel region were slightly reduced in stromalinRNAi flies, although this difference was not statistically significant (Figure 3C). Even if this difference were real, it does not account for the increased synaptic communication between DAn and MBn (Figure 3A–B).
Knockout (KO) of the cohesin complex proteins SMC1 and stromalin have previously been reported to cause axon-pruning deficits during development in a subset of Drosophila MBn (Schuldiner et al., 2008). Therefore, we examined whether stromalinRNAi expression in DAn resulted in neuroanatomical defects using membrane-localized GFP. We were unable to detect any obvious morphological changes of DAn projections to the MB in adult fly brains upon visual inspection of confocal images. We then measured membrane-localized GFP fluorescence intensity of DA projections innervating the MB lobes as a surrogate for axon ramification. We again failed to find any differences between the control and Stromalin KD brains, suggesting that Stromalin reduction does not alter the innervation of the MB lobes by DAn axons (Figure 4C). Although we cannot rule out the possibility that subtle morphological differences exist using this methodology, large changes in the morphology of DAn do not appear to underlie enhanced memory acquisition.
Stromalin suppresses presynaptic marker levels in neurons
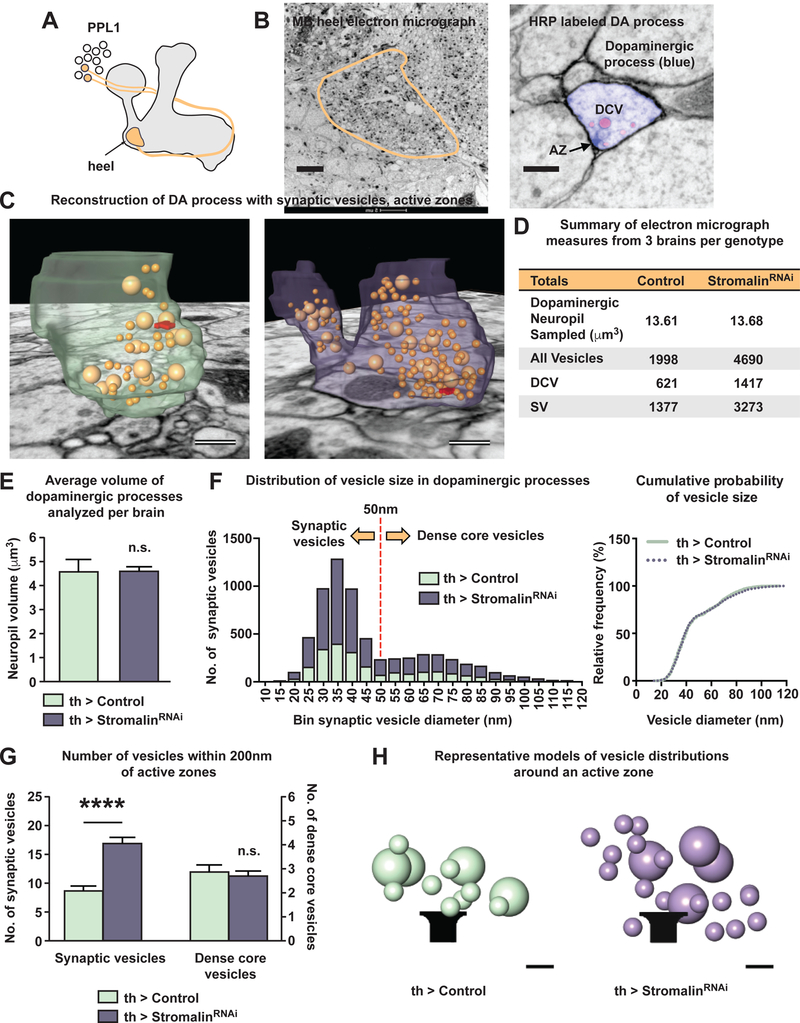
Our inability to detect Stromalin KD-induced neuroanatomical changes in the DAn and the observation of increased communication between DAn and MBn led us to wonder whether Stromalin might instead affect presynaptic proteins or structures. We initially tested this idea by expressing a presynaptic marker localized to synaptic vesicles, Synaptotagmin:GFP (Syt:GFP), in DAn and examining the fluorescence intensity in the regions where PPL1 DAn innervate the MB neuropil (Figure 4A). Remarkably, we discovered that Stromalin KD in DAn resulted in significantly elevated levels of this presynaptic marker in all MB neuropil regions measured (Figure 4D). These results suggested that increased presynaptic function might be at play in Stromalin KD neurons, a result consistent with our increased memory acquisition and increased MB responsiveness detected by functional imaging. If correct, this hypothesis predicts that the difference in presynaptic marker staining should emerge in the KD animals during the 3rd instar larval stage, co-mapping to the critical developmental period for Stromalin KD effects on behavior (Figure 2E).
We therefore examined DAn Syt:GFP expression during the 2nd instar, as well as the early, middle, and late stages of the 3rd instar larvae. As we predicted, larval DAn expressing stromalinRNAi had Syt:GFP levels similar to control larvae during the 2nd to early 3rd instar stages (Figure 4E, Figure S4A–B). However, significantly higher levels of Syt:GFP were detected during the middle to late 3rd instar stages in DAn axons innervating the heel and upper stalk regions of the MB (Figure 4E, Figure S4). Furthermore, repression of stromalinRNAi expression during development using the TARGET system, or during the critical 3rd instar stage, resulted in no significant increases in Syt:GFP expression in the MB heel (Figure S4C). These Syt:GFP developmental results nicely mirror our behavioral data, strongly suggesting that these processes are related, and assigning a critical developmental window in DAn during which Stromalin acts to set baseline levels of adult synaptic strength.
Our aforementioned behavioral data showed that both pan-neuronal and MBn KD of Stromalin produces enhanced olfactory memory (Figure 1C), suggesting that the effects of Stromalin on presynaptic function are not specific to DAn. Therefore, we probed the effect of Stromalin KD on Syt:GFP in the adult brain using the nSyb-GAL4 and R13-GAL4 drivers. We observed elevated levels of the presynaptic marker across the entire brain and in MBn (Figure 4F–G), indicating that Stromalin regulates these markers across multiple types of neurons, and therefore is likely to affect a wide range of behaviors.
Stromalin does not alter the number and volume of dopaminergic axon presynaptic sites
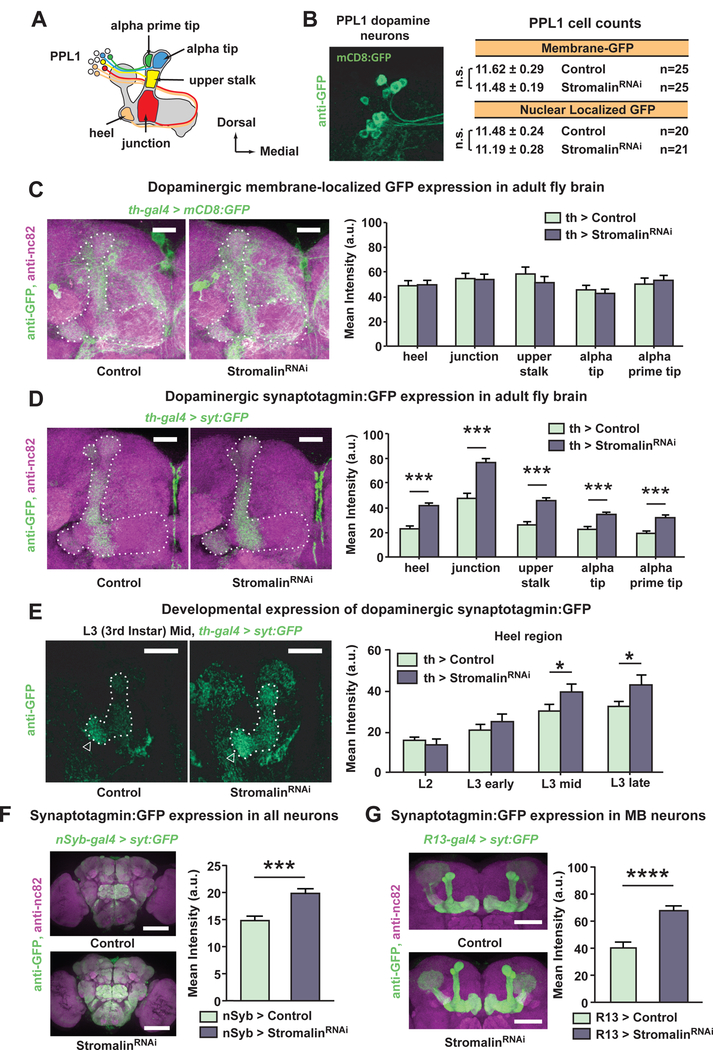
We considered three possible explanations for the increased levels of the Syt:GFP presynaptic marker: 1) Stromalin KD increases the number of dopaminergic synapses on their axonal processes, 2) Stromalin KD increases the volume of dopaminergic pre-synaptic sites, and 3) Stromalin KD increases the number of synaptic vesicles at dopaminergic synapses. To probe the first two possibilities, we used the synapse-labeling tool, GFP reconstituted across synaptic partners (GRASP; Feinberg et al., 2008; Gordon and Scott, 2009). Using GRASP, we found no differences in GFP fluorescence intensity between control and DAn Stromalin KD fly brains (Figure 5A), indicating that Stromalin does not dramatically alter the number or size of PPL1 DAn to MBn connections in the MB neuropil.
Figure 5. Stromalin knockdown in dopamine neurons does not alter the number or volume of dopaminergic presynaptic sites.
(A) The GFP Reconstitution Across Synaptic Partners (GRASP) signal, which indicates the presence of structural connections between DAn and MBn, was not altered by Stromalin KD in DAn. Quantitation of fluorescence intensity across regions of interest is shown in the right bar plot. Control: MB>spGFP11, THGAL4>+, UAS-Dcr2, UAS-spGFP1–10. StromalinRNAi: MB>spGFP11, THGAL4>UAS-SARNAi/, UAS-Dcr2, UAS-spGFP1–10. n=12. Mean±SEM. Scale bar 20µm.
(B) Diagram of the heel region imaged in this experiment (left). Maximum projection of structured illumination microscopy (SIM) images of syt:GFP expression in DAn that innervate the heel region of the MB (middle). Maximum projection images of syt:GFP particle centroids detected in 3D (right) color coded through the z-plane of a representative control brain. Scale bar 5µm.
(C) Stromalin KD in DAn did not alter the number of presynaptic sites, visualized using syt:GFP. Control: TH-GAL4>+, UAS-Dcr2, UAS-syt:GFP, UAS-myr-tdTom. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-syt:GFP, UAS-myrtdTom. n=11. Mean±SEM.
(D) Stromalin knockdown in DAn did not alter the volume of presynaptic sites, measured using the volume of the syt:GFP puncta. Control: TH-GAL4>+, UAS-Dcr2, UAS-syt:GFP, UAS-myr-tdTom. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-syt:GFP, UAS-myr-tdTom. n=11. Bar graph: mean±SEM.
See also Figure S5.
While the GRASP fluorescence intensity signal provides a valuable initial indication of structural connectivity, it offers only an indirect measure of number, and to some extent volume, of the structural connections. To obtain direct measures of DAn presynaptic features, we turned to super resolution structured illumination microscopy (SIM) of Syt:GFP in PPL1 DAn. We focused on the DAn innervation of the MB heel region for these analyses (Figure 5B), since Stromalin KD in this region was sufficient for its behavioral effects (Figure S2). SIM imaged Syt:GFP particles were reconstructed in 3D using image analysis algorithms to estimate the number and volume of presynaptic sites. Expression of stromalinRNAi in the DAn innervating the MB heel did not alter the number or volume of presynaptic sites (Figure 5C–D). We confirmed these results using a presynaptic active zone marker, Bruchpilot:GFP (Brp:GFP), which labels sites on the presynaptic cell membrane where synaptic vesicles are released. This is distinct from Syt:GFP, which labels the synaptic vesicles themselves. Consistent with our GRASP and SIM data, Brp:GFP levels in the MB heel were not affected by stromalinRNAi expression in DAn (Figure S5). Therefore, Stromalin does not appear to alter the number or size of DAn presynaptic boutons.
Stromalin is a master negative regulator of synaptic vesicle number
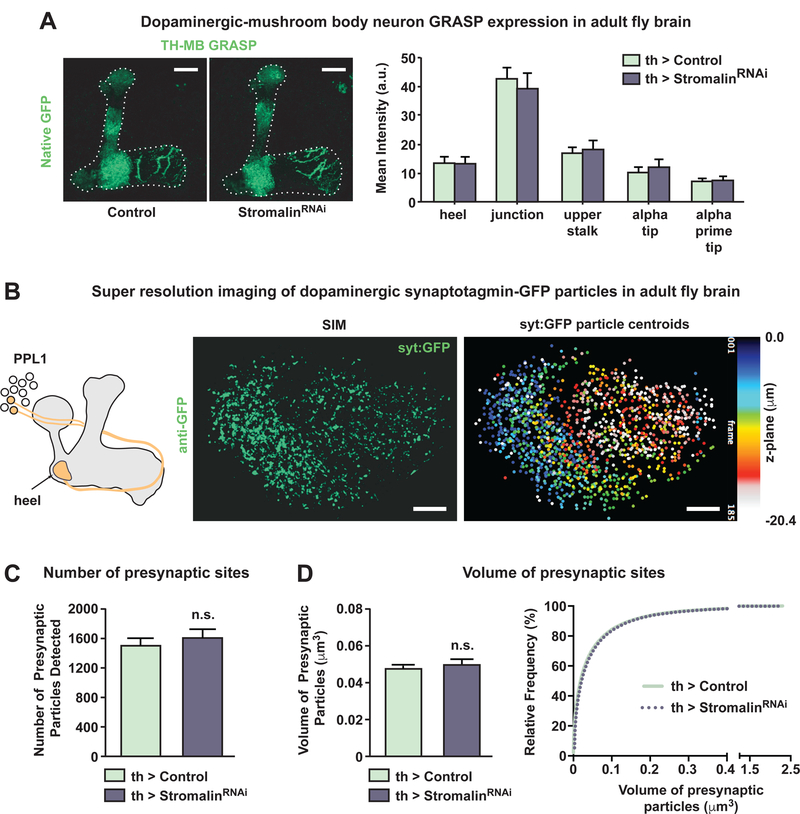
To test directly whether Stromalin KD in DAn alters the number of synaptic vesicles (SVs) present in the DAn axons, we turned to electron microscopy. We expressed membrane-localized HRP using TH-GAL4, which was used to label DAn membranes with an electron dense stain, and examined the DAn innervating the MB heel region (Figure S2). The Drosophila brain was sectioned and serial transmission electron micrographs were collected of the darkly labeled dopaminergic axonal processes (Figure 6A–B). Labeled processes, SVs, and dense core vesicles (DCVs) were traced to obtain measures of DAn axonal process volume, and the number, size and 3D location of dopaminergic vesicles (Figure 6C, Figure S6B).
Figure 6. Stromalin knockdown increases the number of dopaminergic synaptic vesicles.
(A) Diagram of the heel region imaged in this experiment.
(B) Electron micrograph of the MB heel (left; scale bar 5µm). DAn axons expressing membrane-localized HRP were labeled with an electron dense stain (right; scale bar 250nm). Serial EM images were used to trace DAn processes (blue), synaptic vesicles (SV; red), larger sized dense core vesicle (DCV; red), and active zones as identified by T-bars (arrow).
(C) 3D reconstructions of serial EM images of a section of labeled DAn processes in the MB heel region from a control and a Stromalin KD brain. Scale bar 150nm.
(D) Sum of measures obtained from 3 control and 3 stromalinRNAi brains. Control: TH-GAL4>+, UAS-Dcr2, UAS-CD4:HRP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-CD4:HRP.
(E) Volume of DAn processes in the MB heel region sampled from control and stromalinRNAi brains. Control: TH-GAL4>+, UAS-Dcr2, UAS-CD4:HRP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-CD4:HRP. n=3. Mean±SEM.
(F) Despite sampling the same volume of DAn processes from both genotypes, Stromalin KD DAn processes contained 2.3-fold more vesicles than the controls (left). Frequency of DA vesicle sizes indicates the presence of 2 populations of vesicles: synaptic vesicles (≤50nm) and dense core vesicles (>50nm). Cumulative probability graph of vesicle sizes reveals Stromalin KD does not alter the proportion or size of vesicles (right). Control: TH-GAL4>+, UAS-Dcr2, UASCD4:HRP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-CD4:HRP.
(G) Stromalin KD DAn contain 2-fold more synaptic vesicles within 200nm of an active zone compared to control DAn. The numbers of DCVs within this proximity to an active zone was not different between genotypes. Control: TH-GAL4>+, UAS-Dcr2, UAS-CD4:HRP. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2, UAS-CD4:HRP. Mann-Whitney U, ****p<0.0001. n=52 control, n=74 StromalinRNAi. Mean±SEM.
(H) Graphical cartoon of representative control and Stromalin KD vesicle distributions within 200nm of an active zone. Scale bar 50nm.
See also Figure S6.
DAn axonal processes occupied the same volume in both control and Stromalin KD brains (Figure 6D–E), consistent with the light microscopic measures of ramification presented above (Figure 4C). However, DAn expressing stromalinRNAi contained 2.3-fold greater numbers of small diameter SVs (≤50nm) and large diameter DCV (>50nm; Figure 6F). The KD did not alter the proportion of SVs (70%) to DCVs (30%), but increased their overall numbers equally (Figure 6F right panel). In addition, stromalinRNAi expression in MBn increased the levels of two different synaptic vesicle markers, Syt:GFP (Figure 4G) and syp:pHTom (Figure S6A) in the MBn, again indicating that Stromalin KD increases SV numbers in other types of neurons. Thus, stromalin appears to be a master negative regulator of vesicle number.
SVs are released at active zone sites, which can be morphologically identified as electron dense ‘T-bars’ in Drosophila neurons (Figure 6B). We measured the distance from the center of each SV to the nearest active zone in 3D, and calculated the numbers of SVs and DCVs within 100 or 200nm of an active zone (Figure 6G–H, Figure S6C–D). Surprisingly, we discovered that on average, only 8.6±0.1 SVs reside within 200nm of DAn active zones in control flies (Figure 6G). This is a dramatic reduction compared to the number of SVs found near active sites at the neuromuscular junction (~200/active site; Rizzoli and Betz, 2005) and presumably simply reflects differences in the physiology of DAn and motor neurons. Stromalin KD in DAn, however, resulted in a nearly 2-fold increase in the number of SVs within 200nm of an active zone (16.7±0.1; Figure 6G). The doubling of the SV pool in DAn explains the increased amount of DA available for release across multiple stimulations (Figure 3B). The number of DCVs with proximity to active zones was not significantly different between control and Stromalin KD DAn (Figure 6G, Figure S6C–D). However, because DCVs can be released at extrasynaptic sites (Harris and Littleton, 2015), their distribution within the neuropil is likely independent of active zone proximity.
Knockdown of the Drosophila KIF1A homologue prevents proper synaptic vesicle localization to the axon terminals and impairs learning
To directly test whether the SV increases detected in DAn were responsible for the learning enhancements in Stromalin KD flies, we searched for independent methods to manipulate SV numbers in DAn axons. The Drosophila homologue of the mammalian KIF1A neural motor protein, unc104, was shown to be a kinesin3 family protein responsible for trafficking SV precursors and DCVs in the anterograde direction from the cell body to the axon terminals of motor neurons (Kern et al., 2013; Pack-Chung et al., 2007). We thus tested whether the protein plays the same role in PPL1 DAn.
We measured the Syt:GFP fluorescence intensity in the axon terminals of DAn innervating the MB heel in flies expressing UAS-unc104RNAi TRiP. The Unc104 KD neurons had significantly lower Syt:GFP levels in DAn axons innervating the MB heel (Figure 7A left bar graph). In addition, the Syt:GFP signal was increased in the DAn cell bodies, consistent with a block of Syt:GFP axonal transport. These results parallel the effect of unc104 mutants on axonal transport of Syt1 in motor neurons (Pack-Chung et al., 2007). The effects of the UAS-unc104RNAi TRiP transgene were modest compared to a second UAS-unc104RNAi GD transgene, which produced very little Syt:GFP staining in the axons of DAn (Figure S7A), and may have failed to develop synaptic boutons given the severity of the phenotype (Kern et al., 2013; Pack-Chung et al., 2007). We then examined whether mislocalization of SVs in DAn impairs aversive memory acquisition. Indeed, unc104RNAi TRiP expression in DAn resulted in impaired memory acquisition scores when flies were trained with 6 or 12 shocks (Figure 7B). The reduced memory scores of the two unc104RNAi fly strains tested were consistent with the severity of the Syt:GFP effects. Flies expressing unc104RNAi GD produced memory acquisition scores very close to zero (Figure S7B).
Figure 7. Stromalin knockdown learning enhancements are rescued by inhibiting synaptic vesicle transport to dopamine neuron axon terminals.
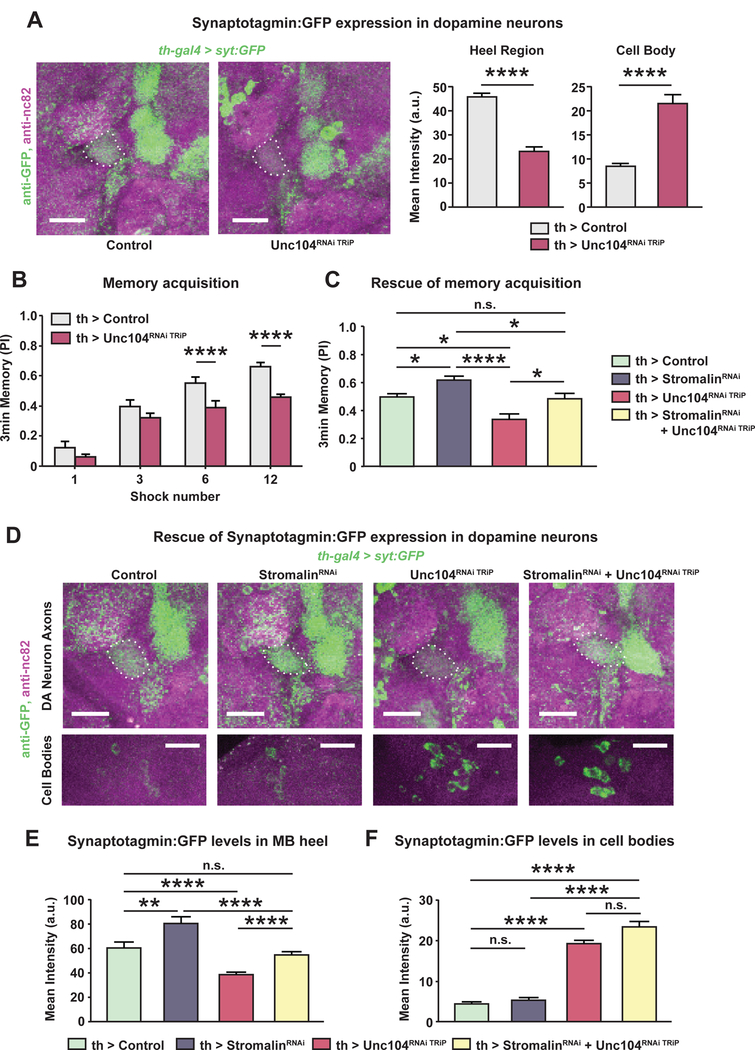
(A) unc104RNAi TRiP (TRiP RNAi library) expression in DAn reduced axonal syt:GFP levels in the MB heel. Concurrently, the level of syt:GFP in the cell bodies (white triangles) increased due to the impaired anterograde transport of synaptic vesicles in Unc104 KD neurons. Dotted white line outlines the MB. The bar graphs quantitate the relative GFP fluorescence. Control: TH-GAL4>+, UASDcr2, UAS-syt:GFP. Unc104RNAi TRiP: TH-GAL4>UAS-Unc104RNAi TRiP, UAS-Dcr2, UAS-syt:GFP. Mann-Whitney U, ****p<0.0001. n=14–15. Mean±SEM. Scale bar 20µm.
(B) Knockdown of Unc104 in DAn impaired aversive memory acquisition with 6 or 12 shock training. Control: TH-GAL4>+, UAS-Dcr2. Unc104RNAi TRiP: THGAL4>UAS-Unc104RNAi TRiP, UAS-Dcr2. Student’s t-test, ****p<0.0001. n=8. Mean±SEM.
(C) Reduced synaptic vesicle transport in DAn rescued the enhanced memory of Stromalin KD flies. DAn stromalinRNAi expression increased, while unc104RNAi TRiP expression impaired, memory acquisition when flies were trained with 6 shocks. Co-expression of both RNAi’s normalized memory acquisition (yellow bar), indicating mutual suppression of the individual phenotypes. The memory scores from this genotype were not different from control flies. Control: TH-GAL4>+, UAS-Dcr2. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-Dcr2. Unc104RNAi TRiP: TH-GAL4>UAS-Unc104RNAi TRiP, UAS-Dcr2. StromalinRNAi + Unc104RNAi TRiP: THGAL4>UAS-SARNAi, UAS-Unc104RNAi TRiP, UAS-Dcr2. One-way ANOVA, Bonferroni post hoc, *p<0.05, ****p<0.0001. n=11–12. Mean±SEM.
(D) Syt:GFP levels in DAn from control, StromalinRNAi, Unc104RNAi TRiP, and StromalinRNAi + Unc104RNAi TRiP MB. The top row of images show the outlined MB lobes (white dotted line) and the syt:GFP expression in the DAn axon terminals. The bottom row of images show the PPL1 DAn cell bodies located in the posterior, dorsal lateral region of the Drosophila brain. The green channel brightness and intensity of the control and Stromalin KD cell body images (lower 2 left images) were enhanced for better visualization. Scale bar 20µm.
(E) Reduced anterograde transport of synaptic vesicles in DAn rescued the increased syt:GFP signal in Stromalin KD flies, mirroring the memory acquisition results in panel C. Reducing Stromalin in DAn increased, while unc104RNAi TRiP expression decreased, syt:GFP levels in the MB heel. Simultaneous expression of stromalinRNAi and unc104RNAi TRiP rescued the syt:GFP increase from stromalinRNAi expression, and the syt:GFP decrease from unc104RNAi TRiP expression (yellow bar). Kruskal-Wallis with a priori planned comparisons using Mann-Whitney U with Bonferroni correction for multiple comparisons was performed to reduce type I errors. **p<0.01, ****p<0.0001. n=16–19. Mean±SEM.
(F) The syt:GFP signal remained significantly elevated in the cell bodies of Unc104RNAi TRiP and the StromalinRNAi + Unc104RNAi TRiP groups. Kruskal-Wallis with a priori planned comparisons using unequal variances t-test with Bonferroni correction for multiple comparisons was performed to reduce type I errors. ****p<0.0001. n=11–15. Mean±SEM The genotype of flies in panels D-F were as follows: Control: TH-GAL84>+, UASsyt:GFP, UAS-Dcr2. StromalinRNAi: TH-GAL4>UAS-SARNAi, UAS-syt:GFP, UASDcr2. Unc104RNAi TRiP: TH-GAL4>UAS-Unc104RNAi TRiP, UAS-syt:GFP, UAS-Dcr2. StromalinRNAi + Unc104RNAi TRiP: TH-GAL4>UAS-SARNAi, UAS-Unc104RNAi TRiP, UAS-syt:GFP, UAS-Dcr2.
See also Figure S7.
Manipulating synaptic vesicle numbers at the dopamine neuron axon terminals alters aversive olfactory learning
In our final set of experiments, we used the unc104RNAi and stromalinRNAi transgenes to manipulate SV numbers in the PPL1 DAn axons and measured the effects of this manipulation on aversive memory acquisition. Control flies contained the corresponding empty landing site for both the stromalinRNAi (KK RNAi library) and unc104RNAi TRiP (TRiP RNAi library). Each respective RNAi line was also combined with the empty landing site of its counterpart (i.e., stromalinRNAi flies containing the TRiP landing site and unc104RNAi TRiP flies containing the KK docking site were generated), while a 4th group of flies contained both the stromalinRNAi and the unc104RNAi TRiP transgenes. The memory acquisition of these flies was tested after training with 6 electric shocks. We replicated our previous findings demonstrating DAn stromalinRNAi expression increased, while unc104RNAi TRiP expression impaired, memory acquisition (Figure 7C). However, the memory scores of flies expressing both stromalinRNAi and unc104RNAi TRiP simultaneously in DAn were not different from controls, demonstrating mutual suppression of the individual phenotypes (Figure 7C). Flies co-expressing stromalinRNAi and the more potent unc104RNAi GD had reduced memory scores not different from flies expressing unc104RNAi GD alone (Figure S7C).
We then measured the Syt:GFP fluorescence in flies expressing one or both RNAi’s. We confirmed that stromalinRNAi expression in DAn significantly increased, while unc104RNAi TRiP expression significantly decreased, Syt:GFP levels in the MB heel (Figure 7D–E), replicating our results above (Figure 4D and 7A). Importantly, simultaneous expression of stromalinRNAi and unc104RNAi TRiP in DAn resulted in Syt:GFP fluorescence in the DAn axon terminals indistinguishable from the control group (Figure 7E), following the memory acquisition results of this genotype (Figure 7C yellow bar). Co-expression of stromalinRNAi and the more potent unc104RNAi GD significantly reduced Syt:GFP levels in the MB heel compared to controls (Figure S7D yellow bar). Therefore, reducing the SV levels in DAn axon terminals by impairing the Unc104 motor suppresses the enhanced learning phenotype observed with Stromalin KD flies, offering strong evidence supporting the claim that Stromalin KD enhances memory acquisition by increasing SV number in DAn axon terminals.
Finally, we quantified the Syt:GFP intensity of the cell bodies in the posterior dorsal lateral part of the brain, where the PPL1 neurons are localized. While control and stromalinRNAi DAn did not differ in the Syt:GFP levels in their cell bodies, flies expressing either unc104RNAi TRiP alone or with stromalinRNAi had significantly higher levels of Syt:GFP in the cell bodies (Figure 7D,F). These observations suggest that Stromalin alters SV content in ways independent of axonal transport.
In sum, our data identify Stromalin as a memory suppressor gene and provide a mechanistic explanation for its actions: suppressing the available synaptic vesicle pool and consequently the synaptic strength between DAn and MBn.
Discussion
Memory suppressor genes offer a unique window for understanding the molecular and cellular mechanisms that constrain memory formation. In contrast to the many genes and gene products known to be required for acquisition and memory consolidation (Davis, 1996; Kandel, 2001), there are but a handful of memory suppressor genes studied to the point of providing new conceptual insights into the processes of memory formation. Some function at the transcriptional level to control the formation of protein synthesis-dependent longterm memory (LTM) (Abel et al., 1998; Lee and Silva, 2009). For instance, isoforms of the Creb transcription factor (Creb repressors) exist that inhibit the normal function of Creb activators to limit LTM (Bartsch et al., 1995; Yin et al., 1994). These are thought to function after initial memory acquisition through biochemical cascades that mobilize new protein synthesis required for LTM. Other memory suppressor genes actively repress communication between neurons (Lee and Silva, 2009). For instance, Drosophila SLC22A encodes a plasma membrane transporter that removes neurotransmitter from the synaptic cleft to terminate synaptic communication (Gai et al., 2016). Cyclin-dependent kinase 5 (Cdk5) promotes proteolysis of the NMDA receptor subunit NR2B, attenuating NMDA receptor signaling in mammalian neurons (Hawasli et al., 2007). It is notable that these and other previously described memory suppressor genes limit the memory capacity of adult organisms, while developmental negative regulation of adult memory is rare (Abel et al., 1998; Cervantes-Sandoval et al., 2016; Guven-Ozkan et al., 2016; Iobbi et al., 2017; Lee, 2014; Lee and Silva, 2009; Lei et al., 2017; Lin et al., 2017). Stromalin is unique, acting during a critical developmental window to constrain the strength of synaptic communication between neurons by limiting the size of the synaptic vesicle pool. This phenotype then persists into adult life.
Our data argue that Stromalin regulates the SV in DAn independent of other structural features of the neuron, such as cell number, the apparent ramification of DAn neuropil in the MB, synapse number or size. These observations lead to the novel and important conclusion that the SV pool is under its own genetic regulation through Stromalin function. Prior to these results, SV pool size was thought to be a function of synapse or active zone size, or some other aspect of neuronal morphology (Garner et al., 2006; Welzel et al., 2011).
Surprisingly, Stromalin alters the number of both SVs and DCVs, suggesting it has a shared role in the biosynthetic or degradative pathways for both types of vesicles that are distinct from the piccolo-bassoon transport vesicles that contain active zone proteins. Components of SVs and DCVs are generated in the endoplasmic reticulum (ER) and processed through the Golgi apparatus, but are sorted separately into SV transport precursor vesicles containing SV proteins and into DCVs for anterograde transport towards the axon terminals (Barkus et al., 2008; Pinto and Almeida, 2016; Rizzoli, 2014). Our study provides the first evidence for a developmental genetic program that specifically controls the strength of synaptic connections by constraining the SV pool in neurons. We hypothesize that Stromalin regulates SV and DCV number through its role in regulating gene expression (Dorsett, 2011; Losada, 2014).
Interestingly, the critical window for Stromalin’s effects on the SV pool size occurs during the 3rd instar larval period. This developmental time point is well after the integration of DAn into the larval olfactory memory circuit and after the initial onset of DAn synaptogenesis onto MBn, since these synapses are already present at the 1st instar larval stage (Eichler et al., 2017). The γ MBn that are present in the larval brain prior to the mid-3rd instar developmental stage undergo extensive axonal and dendritic restructuring during the pupal stage, such that the structural organization and connectivity of the larval γ MBn is distinct from that in the adult (Lee et al., 1999). It is during the mid-3rd instar larval stage that the α’β’ MBn develop, and these appear to persist into the adult fly relatively unchanged in structure. This developmental transition maps directly onto the critical window for Stromalin’s effects on limiting synaptic vesicle pool size in DAn. Stromalin does not affect SV number at the earliest stages of neural circuit development and synaptogenesis, but rather, only upon emergence of the first set of MBn that persist and integrate into the adult neural circuitry. Stromalin may thus be specifically involved in a developmental program that adjusts the strength of DAn synaptic connectivity for adult-relevant neural circuitry and functions.
Insults that produce a loss-of-function of cohesin complex genes have been previously shown to cause developmental axonal and dendritic pruning defects in the γ subset of MBn (Pauli et al., 2008; Schuldiner et al., 2008). Our membraneGFP data (Figure 4C), as well as our EM analysis on DAn neuropil volumes (Figure 6D–E), failed to find similar differences in the DAn axonal ramifications of adult fly brains, arguing that DAn axons either do not undergo the same Stromalin-dependent axonal pruning that occurs with γ MBn, or that such pruning is transient and fails to persist into adulthood. We did express stromalinRNAi in the γ MBn and examined adult γ MBn morphology using membrane-bound GFP staining but did not detect a pronounced morphological difference in these neurons (data not shown). Presumably, a complete loss-of-function is required to detect the pronounced pruning defects observed previously (Pauli et al., 2008; Schuldiner et al., 2008). Moreover, impairing synaptic vesicle transport to axon terminals reversed the enhanced memory phenotype. Taken together, our data fail to support the hypothesis that developmental axonal pruning deficits of DAn lead to the enhancement in learning and memory scores in flies with Stromalin KD in these same neurons.
While our study focused on DAn, our data indicate that Stromalin’s role in constraining synaptic vesicle pool size extends to other neurons of the Drosophila brain, since we also detected Syt:GFP increases with pan-neuronal KD and with KD in the cholinergic MB Kenyon cell neurons (Figure 4F–G). The alteration of neurotransmitter release in a variety of neurons with Stromalin KD is likely to have a profound effect on a range of different behaviors, since mutations in genes affecting synaptic communication have been associated with many behavioral/cognitive disorders, neurodevelopmental, neurodegenerative, and neuropsychiatric disorders (Lepeta et al., 2016). Similarly, we predict that expression of the unc104RNAi transgene would alter other behaviors and generally in ways opposite of stromalinRNAi with an appropriate level of expression. Thus, these transgenes offer valuable new tools for modulating SV content across neurons to probe effects on synaptic communication and behavioral processes. Interestingly, our stromalinRNAi effects were able to rescue the modest learning impairments caused by unc104RNAi expression in DAn, which suggests that increasing synaptic vesicle content may provide a potential symptomatic treatment for patients with KIF1A mutations, who display intellectual disability, sensory and autonomic neuropathy, and spastic paraplegias (Ohba et al., 2015).
Mutations in the highly conserved cohesin complex genes SMC1, SMC3, Rad21 and stromalin (STAG1/2 in mammals) are known to cause cohesinopathies such as Cornelia de Lange Syndrome (Dorsett, 2011; Lehalle et al., 2017; Liu and Krantz, 2009; Mullegama et al., 2017). Our observations prompt the important question of whether alterations in the synaptic vesicle pool and synaptic communication underlie some of the phenotypes associated with the cohesinopathies. The increased memory performance that we observe with Stromalin and SMC1 KD seems at odds with some phenotypes like intellectual disability found in patients. However, an increase in the SV pool across many different types of cells in the human brain resulting from a genomic mutation may produce a more complex and opposite phenotype for learning. Other behavioral phenotypes associated with cohesinopathies, including attention deficit disorder, hyperactivity, repetitive behaviors, and autistic behaviors (Lehalle et al., 2017; Liu and Krantz, 2009; Mullegama et al., 2017) might also be explainable by altered synaptic vesicle pools, and can interfere with learning and memory processes. Furthermore, the increased SV phenotype may also explain the susceptibility of individuals with cohesinopathies to seizures, since SV depletion following repeated neural stimulation is a common mechanism for synaptic depression, important for limiting synaptic hyperactivity that can otherwise lead to runaway network activity (Alabi and Tsien, 2012). Thus, cohesin complex gene mutations may attenuate SV depletion, thereby impairing normal synaptic depression and contributing to the development of seizures and behavioral dysfunction in humans.
STAR Methods
Contact for reagent and resources sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ronald Davis (rdavis@scripps.edu).
Experimental model and subjects details
Fly Conditions and Strains
Flies stocks were cultured on standard Drosophila medium at room temperature. Experimental fly crosses were kept at 25°C, unless otherwise stated, and 70% relative humidity on a 12hr light-dark cycle (lights on at 07:00). Unless otherwise stated, flies of both sexes were used in experiments.
The following GAL4 flies were used in this study: nSyb-GAL4 (gift from Julie Simpson), TH-GAL4 (8848, Bloomington Drosophila Stock Center; BDSC), c772-GAL4 (6494, BDSC), R13F02-GAL4 (48571, BDSC), VT64246-GAL4 (204311, Vienna Drosophila Resource Center; VDRC), GH146-GAL4 (30026, BDSC ), NP2492-GAL4, NP7198-GAL4, NP7135-GAL4 (104219, 114140, 105377, respectively, Kyoto Stock Center), ∆TH-D’ (Liu et al., 2012), c061-GAL4 (30845, BDSC), c150-GAL4 (Dubnau et al., 2003).
The majority of UAS-RNAi fly lines used in this study were obtained from the KK RNAi library (VDRC), including the control line (60100, VDRC), which contains the empty docking sites. The RNAi lines used were UAS-SARNAi (106046, VDRC) and UAS-SMC1RNAi (108922, VDRC). The KK RNAi control line from the VDRC was recently described to have two landing sites, the originally annotated site in the 5’-untranslated region of the tiptop gene, which could lead to false positive phenotypes, as well as a non-annotated site into which the majority of RNAi constructs integrated (Green et al., 2014). Both the UAS-SARNAi and UASSMC1RNAi lines integrated into the non-annotated site. A few RNAi lines were also obtained from the National Institute of Genetics – Fly stocks library (NIG), which include a UAS-GFPRNAi (GFP-IR-1, NIG) control line, and UAS-SARNAi2 (3423R-2, NIG). We also utilized the Unc104 RNAi line from the Harvard TRiP RNAi library (available from the BDSC) UAS-unc104RNAi TRiP (43264, BDSC), as well as its empty landing site control (36303, BDSC), and an Unc104 RNAi line obtained from the GD library, UAS-unc104RNAi GD (23465, VDRC). Additional UAS transgene stocks used in this study are: UAS-Dcr2 (60008 and 60009; VDRC), UAS-P2X2 (Yao et al., 2012), UAS-mCD8:GFP (Lee and Luo, 1999), UAS-myrtdTomato (32221, BDSC), UAS-syt:GFP (6926, BDSC), UAS-brp:GFP (36292, BDSC), UAS-GCaMP6m (42750, BDSC), UAS-spGFP1–10 (Gordon and Scott, 2009), UAS-GFPnls (4776, BDSC), UAS-CD2:HRP (Larsen et al., 2003), UAS-ts pHTom (Pech et al., 2015). Additional stocks utilized include tub-gal80ts (McGuire et al., 2003), MB-gal80 (64306, BDSC), MB-Tepacvv (Boto et al., 2014), MBspGFP11 (Pech et al., 2013).
A UAS-SA fly line was created using stromalin cDNA obtained from the Drosophila Genomics Resource Center (Cat. #5315). The cDNA was amplified and EcoRI and Xhol restriction sites were added using the following primers: Forward 5’- ACTGCACTGAATTCATGATGGCGCGTCGCGGTGGA-3’ and reverse 5’- AGTCGATCCTCGAGCTAGCCCCTGTAATCAGGATC-3’. The PCR product was digested with EcoRI and Xhol, then ligated into the pUAST vector (Brand and Perrimon, 1993) downstream of the Hsp70 promoter and upstream of SV40 terminator sequence. These were used to create transgenic fly lines in a w1118 background (Rainbow Transgenic Flies, Inc.). After genetically mapping the insertions to a chromosome, we tested Stromalin overexpression in 4 independent transformant lines with UAS-SA inserted in the 3rd chromosome using western blots. All 4 lines produced similar levels of Stromalin protein and one was chosen at random for use in behavioral studies.
Method details
Western Blots
Whole adult fly heads were frozen in liquid nitrogen, then homogenized in RIPA buffer (Cell Signaling Technology, cat. #9806) and a volume of homogenate containing ~3 fly heads was loaded into each well of an SDS-PAGE gel, with the exception of Stromalin overexpression lines which were diluted 1:3 due to high levels of Stromalin protein expression (nSyb-GAL4 > UAS-Dcr2, UAS-SA; Figure 1G, Figure S1D). Western blot samples, run in duplicate (technical replicate), were transferred onto PVDF membrane. The membrane was cut horizontally at the 75kDa ladder, so that the top and bottom halves (containing Stromalin and our Tubulin loading control, respectively) could be incubated separately in their respective primary and secondary antibodies. Membranes were blocked for 2hr with 5% skim milk, followed by primary antibody incubation in 5% skim milk overnight at 4°C. Rabbit polyclonal anti-peptide an tibodies were created against Stromalin by immunizing rabbits with four synthetic peptides of Stromalin (PDYEELHSDALNE, DEKAKLTEHFIVT, IEHIRGLEYKSRMD, and SKRAYTRKRRDHD; New England Peptide, Inc.). The rabbit anti-sera were purified by affinity chromatography to GST-SA. Mouse anti-αTubulin (1:5000, Sigma-Aldrich, cat. #T9026) was used as a loading control for western blots. The membrane was incubated with HRP-conjugated secondary antibodies (1:5000 HRP-conjugated goat anti-rabbit IgG, Jackson ImmunoResearch cat. #111–035144, or 1:5000 HRP-conjugated goat anti-mouse IgG, Abcam cat. #AB97250) for 1hr and then visualized using WesternBright Quantum (Advansta, cat. #K-12042D10). The resulting bands were quantified using ImageJ gel analysis tool, and protein expression was calculated relative to tubulin loading controls.
Behavioral Experiments
Flies 1–6day old were used for standard olfactory aversive conditioning experiments. Behavioral experiments were conducted under red light, and in ~65–75% humidity at 23–25°C. Flies were acclimated for >15min in a new food vial under behavioral testing conditions before training. Groups of ~60 flies were tapped into tubes where they received 30sec of air, 1min of odor1 paired with electric shock (12 shocks 1.25sec each, at 5sec intervals at 90V; CS+), 30sec of air, 1min of odor2 with no electric shock (CS-), and 30sec of air. After the specified time (3min-24hr), flies were loaded into a T-maze and given 1min to acclimate, then given 2min to choose between the arms containing odor1 and odor2. The number of flies in each arm were counted and used to calculate a preference index ((CS-) – (CS+))/(CS- + CS+). Odors used were counterbalanced and included 3-octanol (OCT; 0.10–0.20%) and benzaldehyde (BENZ; 0.035–0.09%) diluted in mineral oil. Odor concentrations were adjusted slightly per genotype and study, such that flies displayed similar aversion scores to both odors when tested in the T-maze. Stromalin KD animals did not differ from controls in their naïve odor or shock avoidance (Figure S1C). Memory acquisition was tested as described above with the following differences. One ‘training unit’ was defined as 5s of odor + 1 electric shock pulse (90V; CS+) or 5s of odor with no electric shock (CS-). Flies received 30s of air, n (CS+ training units), 30s of air, n (CS- training units), 30s of air, where n=number of training units (specified in Figure 2B). For TARGET experiments used to determine the developmental stage at which stromalin KD is necessary for behavioral effects, fly crosses were restricted to a 6hr egg laying time window in order to synchronize the development of experimental flies. The development of larvae was monitored daily and their incubation temperatures were shifted at the desired developmental phases, specified above graph bars in Figure 2D–E.
Immunohistochemistry and Imaging
Adult brains from 1–6day old flies or larval brains of the indicated ages were used for these experiments. Whole brains were dissected and processed as previously described (Jenett et al., 2012). Brains were dissected in S2 medium (Life Technologies, cat. #21720–024), then placed in 1% paraformaldehyde in S2 medium overnight at 4°C. Brains were washed in PAT3 (0.5% TritonX-100, 0.5% bovine serum albumin, in phosphate buffered saline), and then incubated with primary antibodies and 3% normal goat serum in PAT3 for 3hr at room temperature then overnight at 4°C. Brains were then washed in PAT3, incubated with secondary antibodies and 3% normal goat serum in PAT3 for 3hr at room temperature then at 4°C for 3–5 days. Brains were w ashed with PAT3 and then with phosphate buffered saline before mounting between two coverslips held apart by spacers (102µm for adult brains, 38µm for larval brains). Primary antibodies used include: rabbit anti-GFP (1:1000, Invitrogen, cat. #A11122), mouse monoclonal anti-GFP (1:1000, Invitrogen, cat. #A11120), rabbit antiDsRed (1:1000, Clonetech Labs, cat. #632496), or mouse monoclonal anti-NC82 (1:20, Developmental Studies Hybridoma Bank, AB_2314866). Secondary antibodies used included: Alexa 488 goat anti-rabbit IgG (1:800, Invitrogen, cat. #A11008), Alexa 488 goat anti-mouse IgG (1:800, Invitrogen, cat. #A11029), Alexa 633 goat anti-rabbit IgG (1:400, Invitrogen, cat. #A21070), or Alexa 633 goat anti-mouse IgG (1:400, Invitrogen, cat. #A21052). Samples were imaged on Leica TCS SP5 II or SP8 confocal microscope using a 25× water emersion objective and 488nm and/or 633nm laser excitation. ImageJ was used to analyze images. Mean fluorescence intensity was obtained by drawing a circular ROI of the same size per experiment, per area examined across all brains within the boundaries of the structure of interest. The intensity values were averaged between the anterior and posterior limits of the structure. Data was collected from both hemispheres of the brain and averaged to obtain one value per region per animal. For structured illumination microscopy (SIM) images, immunostained brains underwent further processing. Brains were washed in PBS then quickly rinsed in dH2O and mounted on poly-lysine coated high-precision coverslips. Samples were then dehydrated using an ethanol series (20%, 30%, 50%, 70%, 95%, 100%x2, 5min each) and cleared in methyl salicylate overnight and then mounted as above. Samples were imaged using a Zeiss ELYRA PS1, 63× oil emersion objective, with 488nm and 642nm laser excitation. To count synapse numbers and measure the volume of synaptic sites of the DAn innervating the heel region using ImageJ, ROIs were drawn around the heel and outside areas were digitally cleared. ImageJ’s 3D object counter (Bolte and Cordelieres, 2006) was used to obtain the number and volume of the Synaptotagmin:GFP particles. The values obtained from both hemispheres of a brain were averaged to obtain one value per animal. A brain was excluded from analysis if the region of interest was obscured (air bubble, debris, etc.).
Functional Imaging
Ex vivo experiments were performed as previously described (CervantesSandoval et al., 2017). Brains from 4–6day old female flies expressing the ATPgated P2X2 ion channel in TH-GAL4 DAN were dissected and placed in physiological saline solution (103mM NaCl, 3mM KCl, 5mM HEPES, 1.5mM CaCl2, MgCl2, 26mM NaHCO3, 1mM NaH2PO4, 10mM trehalose, 7mM sucrose, and 10mM glucose [pH 7.2]). Brains were continuously perfused (2mL/min) with saline, and increasing concentrations of adenosine triphosphate (ATP; 0.1mM, 0.5mM, 1mM, 2.5mM, 5mM, 10mM) were bath applied for 30sec under the confocal microscope, and then washed out for 10–15min with saline. ATP responses were captured in 10min recordings collected at 2Hz. Data was collected from both hemispheres of the brain whenever possible (when regions in both hemispheres were within the same plane of focus) and averaged to obtain one value per region per animal. We previously found that there is leaky expression of UAS-P2X2 in some cells (Cervantes-Sandoval et al., 2017). However, this would not alter our data interpretation for several reasons. First, we observed that the 3rd chromosome insertion of UAS-P2X2 used in this study is less leaky than the 2nd chromosome insertion used in Cervantes et al, 2017. Second, any leaky expression of the P2X2 channel should be the same in both control and Stromalin KD brains, simply adding to noise and not contributing to the genotype differences in cAMP responses that we observed. Finally, we performed in vivo functional imaging experiments with electric shock that did not employ the P2X2 transgene and replicated the differences between genotypes. In vivo functional imaging experiments were performed as previously described (Berry et al., 2012; Cervantes-Sandoval et al., 2017). Female flies 5–6day old were used for the in vivo imaging experiments. The fly was fixed in place using myristic acid in a custom-designed recording chamber without the use of anesthesia. A window was dissected in the dorsal cuticle of the fly head using a syringe needle and physiological saline added on top of this window. Sharp forceps were used to remove fat from the surface of the fly brain. The saline volume was reduced and a drop of low melting point agarose was placed on the exposed brain to reduce movement. Physiological saline was added on top of the agarose. A strip of plastic with a copper grid surface modified from the same material used to deliver electric shocks during behavioral training sessions touched the legs of the immobilized fly. The copper grid was connected to a Grass stimulator and 12 electric shock pulses at 90V and at 5sec intervals were delivered, mirroring the standard behavioral training protocol. The heel region of one hemisphere of the MB was imaged using a Leica TCS SP5 II confocal microscope in 2 × 2min recordings 5min apart at 4Hz and averaged for each animal. One fly was excluded from analysis due to gradual and continuous movement in the z-plane during recording and one fly was excluded because it was unresponsive to touch stimuli after recording (in vivo epac experiment). Fluorescence intensity was quantified using ImageJ.
Electron Microscopy
Female flies 5–6day old carrying TH-GAL4>UAS-CD2:HRP were dissected in 0.1M phosphate buffer (PB; pH 7.4) and placed into 1% paraformaldehyde + 0.1% gluteraldehyde in 0.1M PB overnight at 4°C. Th e HRP-labeled membrane was biotinylated using the TSA Biotin System (Perkin Elmer, cat. #NEL700A001KT) for 10min. The HRP label was amplified by incubating the brains with an avidin-biotin complex HRP kit (Vector Labs, cat. #PK-4000) for 2hr and stained using diaminobenzedine (Vector Labs, cat. # SK-4105). Brains were washed between each step using 0.1M PB. After washing in water, brains were treated with 1% aqueous OsO4 on ice for 60min and en bloc stained with 1% aqueous uranyl acetate for 30min. Brains were dehydrated with an ascending ethanol series, followed by acetone, propylene oxide, and overnight infiltration of Durcupan resin (Sigma). Samples were then flat embedded and polymerized at 60˚C for 2 days. Serial ultrathin coronal sections (50nm) were prepared (UC7; Leica) from 3 brains/genotype and counter stained with 3% uranyl acetate and 0.5% lead citrate for 1min each. Sections were examined in a Tecnai G2 Spirit BioTwin transmission electron microscope (FEI) at 80 kV acceleration voltage. Images were taken at 43,000X magnification with a Veleta CCD camera (Olympus) operated by TIA software (FEI). The MB heel was confirmed morphologically as a crescent shaped neuropil lateral to the horizontal lobe and anterior to the peduncle, surrounded by a thin sheath of glia (Figure 6B). Serial images of all labeled processes within the MB heel were taken for a tissue depth of 700–750nm. Every effort was made to obtain images from the same region of the heel across brains. DAn processes, SVs and DCVs were traced, and active zones, presenting as an electron dense T-bar with or without a platform, were identified using ImageJ by an observer blind to the experimental conditions. ImageJ was used to obtain measures of vesicle numbers and sizes. Cartesian coordinates of vesicle centroids were used to mathematically calculate their distances to the nearest active zone (base of T-bar), using the standard formula: , where d is the distance between point 1 (P1) and point 2 (P2), and xn, yn, zn are the Cartesian coordinates of point n. Measures of the numbers of vesicles within a 100 or 200nm spherical radius of an active zone were obtained. 3D reconstructions and models were created using Amira (FEI).
Quantification and Statistical Analyses
Data were compiled using Excel and/or Matlab and analyzed using Prism 6. The sample sizes per group, p values, and statistical tests used for each experiment are stated in the figures or figure legends. The significance was set at α=0.05, and all tests were two-tailed. The statistical tests employed provide measures for the reliability of the data. The sample sizes chosen were determined from similar experiments that were previously published. For electron microscopy imaging experiments, we determined sample sizes based on prior experience of the Electron Microscopy Core Facility Director (Max Planck Florida Institute for Neuroscience). In general, behavioral experiments were analyzed using parametric statistics, since aversive memory scores are normally distributed (Walkinshaw et al., 2015), while functional imaging and anatomical imaging data were analyzed using non-parametric statistics. Multiple comparisons were completed using Bonferroni corrections. All data points were included in our analyses (no exclusion of animals or data points) unless otherwise stated. The animals selected for experiments were randomly chosen from the population of flies with the required genotype.
Supplementary Material
Highlights.
Stromalin genetically constrains adult memory acquisition and synaptic strength.
Stromalin negatively regulates synaptic vesicle number during development.
Synaptic vesicle number is controlled independently of synapse number and volume.
Stromalin and Unc104 bidirectionally alter synaptic vesicle numbers at synapses.
Acknowledgements
We thank our colleagues that have supplied Drosophila stocks. Supported by grants 5R37NS019904, 2R01NS052351 and 1R35NS097224 from the NINDS to R.L.D. A.P. is the recipient of a CIHR postdoctoral fellowship 358761.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing financial interests.
References
- Abel T, Martin KC, Bartsch D, and Kandel ER (1998). Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science 279, 338–341. [DOI] [PubMed] [Google Scholar]
- Alabi AA, and Tsien RW (2012). Synaptic vesicle pools and dynamics. Cold Spring Harbor perspectives in biology 4, a013680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM, and Kandel ER (2014). The regulation of transcription in memory consolidation. Cold Spring Harbor perspectives in biology 7, a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkus RV, Klyachko O, Horiuchi D, Dickson BJ, and Saxton WM (2008). Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Molecular biology of the cell 19, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, and Kandel ER (1995). Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83, 979–992. [DOI] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, and Davis RL (2012). Dopamine is required for learning and forgetting in Drosophila. Neuron 74, 530542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, and Cordelieres FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc-Oxford 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Boto T, Louis T, Jindachomthong K, Jalink K, and Tomchik SM (2014). Dopaminergic modulation of cAMP drives nonlinear plasticity across the Drosophila mushroom body lobes. Current biology 24, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401415. [DOI] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Chakraborty M, MacMullen C, and Davis RL (2016). Scribble Scaffolds a Signalosome for Active Forgetting. Neuron 90, 1230–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Phan A, Chakraborty M, and Davis RL (2017). Reciprocal synapses between mushroom body and dopamine neurons form a positive feedback loop required for learning. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (1993). Mushroom bodies and Drosophila learning. Neuron 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Davis RL (1996). Physiology and biochemistry of Drosophila learning mutants. Physiological reviews 76, 299–317. [DOI] [PubMed] [Google Scholar]
- Davis RL (2005). Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annual review of neuroscience 28, 275–302. [DOI] [PubMed] [Google Scholar]
- Davis RL (2011). Traces of Drosophila memory. Neuron 70, 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, and Zhong Y (2017). The Biology of Forgetting-A Perspective. Neuron 95, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D (2011). Cohesin: genomic insights into controlling gene transcription and development. Current opinion in genetics & development 21, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, and Strom L (2012). The ancient and evolving roles of cohesin in gene expression and DNA repair. Current biology : CB 22, R240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. (2003). The staufen/pumilio pathway is involved in Drosophila long-term memory. Current biology : CB 13, 286–296. [DOI] [PubMed] [Google Scholar]
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM, Saumweber T, Huser A, Eschbach C, Gerber B, et al. (2017). The complete connectome of a learning and memory centre in an insect brain. Nature 548, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- Fukuda T, and Hoog C (2010). The Mouse Cohesin-Associated Protein PDS5B Is Expressed in Testicular Cells and Is Associated with the Meiotic Chromosome Axes. Genes 1, 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Waites CL, and Ziv NE (2006). Synapse development: still looking for the forest, still lost in the trees. Cell and tissue research 326, 249–262. [DOI] [PubMed] [Google Scholar]
- Gordon MD, and Scott K (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, and Kyriacou CP (2014). A Drosophila RNAi collection is subject to dominant phenotypic effects. Nature methods 11, 222–223. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Busto GU, Schutte SS, Cervantes-Sandoval I, O’Dowd DK, and Davis RL (2016). MiR-980 Is a Memory Suppressor MicroRNA that Regulates the Autism-Susceptibility Gene A2bp1. Cell reports 14, 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KP, and Littleton JT (2015). Transmission, Development, and Plasticity of Synapses. Genetics 201, 345–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawasli AH, Benavides DR, Nguyen C, Kansy JW, Hayashi K, Chambon P, Greengard P, Powell CM, Cooper DC, and Bibb JA (2007). Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nature neuroscience 10, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M (2003). Mushroom body memoir: from maps to models. Nature reviews Neuroscience 4, 266–275. [DOI] [PubMed] [Google Scholar]
- Iobbi C, Korte M, and Zagrebelsky M (2017). Nogo-66 Restricts Synaptic Strengthening via Lingo1 and the ROCK2-Cofilin Pathway to Control Actin Dynamics. Cerebral cortex 27, 2779–2792. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell reports 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038. [DOI] [PubMed] [Google Scholar]
- Kern JV, Zhang YV, Kramer S, Brenman JE, and Rasse TM (2013). The kinesin-3, unc-104 regulates dendrite morphogenesis and synaptic development in Drosophila. Genetics 195, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, and Jalink K (2011). A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PloS one 6, e19170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CW, Hirst E, Alexandre C, and Vincent JP (2003). Segment boundary formation in Drosophila embryos. Development 130, 5625–5635. [DOI] [PubMed] [Google Scholar]
- Lee J, Yokota T, and Yamashita M (2002). Analyses of mRNA expression patterns of cohesin subunits Rad21 and Rec8 in mice: germ cell-specific expression of rec8 mRNA in both male and female mice. Zoological science 19, 539–544. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, and Luo L (1999). Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126, 4065–4076. [DOI] [PubMed] [Google Scholar]
- Lee T, and Luo L (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- Lee YS (2014). Genes and signaling pathways involved in memory enhancement in mutant mice. Molecular brain 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, and Silva AJ (2009). The molecular and cellular biology of enhanced cognition. Nature reviews Neuroscience 10, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehalle D, Mosca-Boidron AL, Begtrup A, Boute-Benejean O, Charles P, Cho MT, Clarkson A, Devinsky O, Duffourd Y, Duplomb-Jego L, et al. (2017). STAG1 mutations cause a novel cohesinopathy characterised by unspecific syndromic intellectual disability. Journal of medical genetics [DOI] [PubMed]
- Lei M, Shafique A, Shang K, Couttas TA, Zhao H, Don AS, and Karl T (2017). Contextual fear conditioning is enhanced in mice lacking functional sphingosine kinase 2. Behavioural brain research 333, 9–16. [DOI] [PubMed] [Google Scholar]
- Lepeta K, Lourenco MV, Schweitzer BC, Martino Adami PV, Banerjee P, Catuara-Solarz S, de La Fuente Revenga M, Guillem AM, Haidar M, Ijomone OM, et al. (2016). Synaptopathies: synaptic dysfunction in neurological disorders - A review from students to students. Journal of neurochemistry 138, 785–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Ponnusamy R, Widagdo J, Choi JA, Ge W, Probst C, Buckley T, Lou M, Bredy TW, Fanselow MS, et al. (2017). MicroRNA-mediated disruption of dendritogenesis during a critical period of development influences cognitive capacity later in life. Proceedings of the National Academy of Sciences of the United States of America 114, 9188–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, and Krantz ID (2009). Cornelia de Lange syndrome, cohesin, and beyond. Clinical genetics 76, 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, and Wu MN (2012). Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Current biology : CB 22, 2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A (2014). Cohesin in cancer: chromosome segregation and beyond. Nature reviews Cancer 14, 389–393. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, and Davis RL (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 17651768. [DOI] [PubMed] [Google Scholar]
- Mullegama SV, Klein SD, Mulatinho MV, Senaratne TN, Singh K, Center UCG, Nguyen DC, Gallant NM, Strom SP, Ghahremani S, et al. (2017). De novo loss-of-function variants in STAG2 are associated with developmental delay, microcephaly, and congenital anomalies. American journal of medical genetics Part A 173, 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Haginoya K, Osaka H, Kubota K, Ishiyama A, Hiraide T, Komaki H, Sasaki M, Miyatake S, Nakashima M, et al. (2015). De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. Journal of human genetics 60, 739–742. [DOI] [PubMed] [Google Scholar]
- Pack-Chung E, Kurshan PT, Dickman DK, and Schwarz TL (2007). A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nature neuroscience 10, 980–989. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, and Nasmyth K (2008). Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Developmental cell 14, 239251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech U, Pooryasin A, Birman S, and Fiala A (2013). Localization of the contacts between Kenyon cells and aminergic neurons in the Drosophila melanogaster brain using SplitGFP reconstitution. The Journal of comparative neurology 521, 3992–4026. [DOI] [PubMed] [Google Scholar]
- Pech U, Revelo NH, Seitz KJ, Rizzoli SO, and Fiala A (2015). Optical dissection of experience-dependent pre- and postsynaptic plasticity in the Drosophila brain. Cell reports 10, 2083–2095. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, and Schmitz J (2008). The cohesin complex and its roles in chromosome biology. Genes & development 22, 3089–3114. [DOI] [PubMed] [Google Scholar]
- Pinto MJ, and Almeida RD (2016). Puzzling out presynaptic differentiation. Journal of neurochemistry 139, 921–942. [DOI] [PubMed] [Google Scholar]
- Rizzoli SO (2014). Synaptic vesicle recycling: steps and principles. The EMBO journal 33, 788–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoli SO, and Betz WJ (2005). Synaptic vesicle pools. Nature reviews Neuroscience 6, 57–69. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, and Luo L (2008). piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Developmental cell 14, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, and Davis RL (2008). Cyclic AMP imaging sheds light on PDF signaling in circadian clock neurons. Neuron 58, 161–163. [DOI] [PubMed] [Google Scholar]
- Valdeolmillos A, Villares R, Buesa JM, Gonzalez-Crespo S, Martinez C, and Barbero JL (1998). Molecular cloning and expression of stromalin protein from Drosophila melanogaster: homologous to mammalian stromalin family of nuclear proteins. DNA and cell biology 17, 699–706. [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, and Davis RL (2015). Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics 199, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel O, Henkel AW, Stroebel AM, Jung J, Tischbirek CH, Ebert K, Kornhuber J, Rizzoli SO, and Groemer TW (2011). Systematic heterogeneity of fractional vesicle pool sizes and release rates of hippocampal synapses. Biophysical journal 100, 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, and Shafer OT (2012). Analysis of functional neuronal connectivity in the Drosophila brain. Journal of neurophysiology 108, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, and Tully T (1994). Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


