Abstract
Advances in single-cell biology have enabled measurements of >40 protein features on millions of immune cells within clinical samples. However, the data analysis steps following cell population identification are susceptible to bias, time-consuming, and challenging to compare across studies. Here, an ensemble of unsupervised tools was developed to evaluate four essential types of immune cell information, incorporate changes over time, and address diverse immune monitoring challenges. The four complementary properties characterized were: 1) systemic plasticity, 2) change in population abundance, 3) change in signature population features, and 4) novelty of cellular phenotype. Three systems immune monitoring studies were selected to challenge this ensemble approach. In serial biopsies of melanoma tumors undergoing targeted therapy, the ensemble approach revealed enrichment of double-negative (DN) T cells. Melanoma tumor resident DN T cells were abnormal and phenotypically distinct from those found in non-malignant lymphoid tissues, but similar to those found in glioblastoma and renal cell carcinoma. Overall, ensemble systems immune monitoring provided a robust, quantitative view of changes in both the system and cell subsets, allowed for transparent review by human experts, and revealed abnormal immune cells present across multiple human tumor types.
Keywords: systems immunology, immunotherapy, mass cytometry, double-negative T cells
INTRODUCTION
The immune system is a complex network of cells and tissue types, and it is increasingly important to simultaneously track cell subsets and understand the system as a whole. Longitudinal monitoring of changes in the immune system has provided insight into drug response and disease progression (1–3). Differences in response to perturbation can stratify clinical outcome (4, 5) and indicate mechanism of action (6). Challenges to the immune system, such as vaccination, infection, surgical intervention, or the emergence of a malignancy, can elicit detectable changes above the relatively stable basal state of each individual (7, 8). Cytomic approaches that can characterize all the cells in a system, like high-dimensional flow and mass cytometry, are an area of active development in immunology (9). A common framework for data analysis will allow researchers using these cytomic approaches to compare and contrast immune systems from diverse research areas, including tumor immunology and treatment response (4, 10, 11), blood cancer (12, 13), bone marrow failure (14, 15), and human immune variation and autoimmunity (3, 7, 16).
Systematically monitoring the cells of the immune system and their features is especially powered when serial samples from an individual can be used as comparison points. However, this approach generates vast amounts of data. Simultaneously tracking entire systems and the parts that comprise them can become overwhelming in the context of clinical research, where cells of interest can be rare (4, 10), and the entire system can change quickly in unanticipated ways (15). Thus, in cytomic, system-level studies, the data analysis strategy is as important as the experiment design (4, 17, 18), and it is vital to track known reference populations and place new observations in the context of prior knowledge (19, 20). Tools exist for visualizing and gating (21–24), supervised population and biomarker analysis (25, 26), and describing cell population identity (19) within high-dimensional data sets. In contrast, a great need exists for automated analysis for the steps immediately after population identification (gating). Tools for cellular clustering are especially numerous, specific for each data type, and have been extensively addressed in prior work (27). This ensemble aims to quantify changes in the whole system, prior to cellular clustering, and to characterize how cellular populations change after the appropriate clustering tool has been applied to the system.
The growth and success of high-dimensional single-cell technologies relies on the ongoing development of data analysis tools needed to parse large amounts of data and place results into context (18–20). It remains relatively rare for researchers to choose data analysis approaches that explicitly incorporate time or other changes. Across longitudinal studies, four dynamic elements stand out as key features of cellular systems: 1) plasticity or stability of the system as a whole, 2) changes in abundance of cell subsets, 3) emergence of unexpected features on cell subsets, and 4) emergence of novel or unexpected cell types. The central goal here was to simultaneously employ distinct tools focused on these different data types as an ensemble and to organize this copious information into an automated “first pass” analysis that could be easily interpretable by an immunologist and that would highlight cell subsets for in-depth review. The components selected for the ensemble toolkit here included the Earth Mover’s Distance algorithm (28), t-distributed stochastic neighbor embedding (t-SNE) (21), and Marker Enrichment Modeling (MEM) (19).
To validate and challenge computational analysis tools, it is valuable to explore immunology problems representing differing levels of prior knowledge, amounts of change, and abundances of target cell types. Here, the first challenge (Dataset 1) comprised data from melanoma patients undergoing anti–PD-1 therapy with pembrolizumab. Dataset 1 was chosen for the relevance of this therapeutic strategy and because the dataset includes an unusual case from a patient with a distinct, unexpected immune system trajectory (15). This example was chosen to include an outlier case, which is unusual enough that training datasets would not normally include an example of it. Dataset 2 comprised of a previously published data set of peripheral blood from acute myeloid leukemia (AML) patients undergoing chemotherapy (12). Dataset 2 was chosen as an example of dynamic cellular populations shifting dramatically over therapy. This dataset was also included to represent the challenge of tracking and characterizing treatment-refractory leukemic blasts, which did not converge on a single phenotype and instead, shifted into different phenotypic compartments, none of which matched the phenotype of healthy cells (12). Dataset 3 included serial melanoma tumor biopsies from patients treated with dabrafenib, a BRAFV600 inhibitor (BRAFi), and trametinib, a MEK inhibitor (MEKi). A challenge of Dataset 3 was to apply multiple tools in a system that was relatively less well-studied and included a diverse set of individuals and mass cytometry panels. By providing both high-level and detailed views of cellular systems changing over time in human patients, the ensemble approach revealed knowledge about immune system interactions in these three study types with contrasting changes and challenges.
MATERIALS AND METHODS
Study Design
Dataset 1:
In the case of peripheral blood collected from melanoma patients receiving pembrolizumab, the purpose of the study was to identify biological characteristics of melanoma occurring prior to treatment and at different time points following therapy for patients being treated with immune-based therapies. Each patient gave consent to an Institutional Review Board (IRB)- approved research protocol. To be included in this study, patients met the following inclusion criteria: 1) pathologically proven diagnosis of melanoma, 2) 18 years of age or older, 3) treated with immune-based therapies, and 4) willing to have several serial blood draws. Patients not receiving immune-based therapy or unwilling or unable to provide consent were not included. There was no blinding or randomization process in this study. Blood specimens were obtained from patients during the time of scheduled phlebotomy for routine clinical laboratory analysis. Peripheral blood draws were done on the day of therapy start and 21 days (± 10 days), 84 days (± 21 days), and 180 days (± 21 days) following initiation of therapy.
Cohort 2:
This cohort has been previously published and includes peripheral blood from acute myeloid leukemia (AML) patients undergoing chemotherapy (12). Briefly, patients consented to a blood draw through an IRB-approved research protocol. Peripheral blood was drawn pre-treatment at the time of diagnosis, every 2–3 days within the first two weeks after the start of chemotherapy, at the two-week time point, and at the time of recovery, if applicable.
Dataset 3:
In the case of tumors sampled sequentially from melanoma patients treated with targeted therapy, the objective was to identify biomarkers of response and resistance to B-RAF and MEK targeted therapy in melanoma. Patients with advanced, operable BRAF mutation-positive melanoma will received GSK-2118436 (BRAF inhibitor) for two weeks, followed by the combination of GSK-2118436 and GSK-1120212 (MEK inhibitor) for two weeks, followed by surgical resection of the disease. Tumor biopsies were obtained prior to start of therapy and 2 weeks after combined GSK-2118436 and GSK-1120212 (29). To be included in this study, patients met the following inclusion criteria: 1) signed written, informed consent, 2) between the ages of 18 and 90, 3) patients with locally or regionally advanced melanoma being considered for resection of the lesion(s) for local-regional control and potential cure, 4) BRAF V-600 mutation-positive by snapshot molecular analysis, 5) measurable disease, 6) all prior treatment related toxicities CTCAE ≤ Grade 1 at the time of enrollment, 6) adequate baseline organ function, 8) women of childbearing potential with a negative serum pregnancy test within 14 days of first dose of study or men with female partner of childbearing potential must have had either had a prior vasectomy or agree to use effective contraception, and 9) able to swallow and retain oral medication. There was no blinding or randomization process in this study.
Human Tissue Sample Collection and Preservation
All human samples were obtained in accordance with the Declaration of Helsinki following protocols approved by Vanderbilt University Medical Center IRB. The patient information for unpublished samples can be found in Supplementary Table S1. Healthy donor tonsil, adenoid, and blood were collected as “non-human subjects”, without gender or age information. Upon single-cell isolation, all cells were cryopreserved in 88% fetal calf serum plus 12% DMSO. Cells from human samples were collected and isolated as follows:
Peripheral Blood
Peripheral blood mononuclear cells (PBMCs) were collected, isolated, and cryopreserved from approximately 20 mL of freshly drawn blood as previously described (15). Briefly, peripheral blood was drawn into sodium heparin anticoagulant, and PBMCs were isolated by centrifugation after layering on top of a Ficoll-Paque PLUS (GE Healthcare Bio-Sciences) gradient.
Solid Tissue
Melanoma tumors, glioblastoma tumors, and non-malignant human adenoid and tonsil tissue were resected from patients with consent and in accordance with the Declaration of Helsinki. All solid tissue samples were dissociated into live, single-cell suspensions, and samples were cryopreserved using a previously documented protocol (30, 31). Solid tissue samples were first manually dissociated using a scalpel. The minced tissue was then incubated in RPMI 1640 (Corning/Mediatech, Corning, NY) plus 10% FBS, collagenase II (1 mg/mL; Sigma Aldrich; Darmstadt, Germany), and DNase (0.25 mg/mL; Sigma Aldrich) for 1 hour in a 37° C incubator with 5% CO2. Cells were then strained through a 70 μm and 40 μm filter prior to cryopreservation.
Renal cell carcinoma samples were processed and stored as described by Siska et al (32). Briefly, malignant tissue was removed from patients and processed by mechanical dissociation and enzymatic digestion. Single cell suspensions were achieved by passing dissociated tissue through a 70 μm strainer. Finally, red blood cells were lysed, and the single cell suspension was cryopreserved.
Human Induced Pluripotent Stem Cells (iPSCs)
Reprogramming of human neonatal foreskin fibroblast cells (strain BJ; ATCC no. CRL2522) was induced by transduction with CytoTune Sendai virus (Life Technologies). All experiments were performed under the supervision of the Vanderbilt Institutional Human Pluripotent Cell Research Oversight (VIHPCRO) Committee. Induced pluripotent stem cells were grown in feeder-free conditions in plates coated with Matrigel (BD Biosciences) and maintained in mTESR1 media (Stem Cell Technologies) at 37°C with 5% CO2. iPSCs were generated in the lab of Vivian Gama, Ph.D. and were passaged 30–35 times prior to staining by mass cytometry. The iPSCs were first characterized by live staining with Tra1–60 or Tra1–81 antibodies that recognize undifferentiated iPSCs. Every 30 passages, iPSCs were characterized by gene expression analysis using the TaqMan hPSC score card panel (Life Technologies) and karyotyping. Cells were checked daily for differentiation and were passaged every 3–4 days using Gentle dissociation solution (Stem Cell Technologies). iPSCs were treated with 0.5% EDTA prior to staining with mass cytometry antibody panel described below. iPSCs used in this study were mycoplasma negative.
Mass Cytometry
Thawed samples were first incubated with cisplatin (25 μmol/L, Enzo Life Sciences). After incubation with cisplatin, cells were washed in PBS containing 1% BSA. Staining occurred in 50 μL PBS/1% BSA for 30 minutes at room temperature using the antibodies listed in Supplementary Table S2. Cells were then washed twice with PBS/1% BSA and fixed with a final concentration of 1.6% paraformaldehyde (PFA, Electron Microscopy Sciences). Cells were washed again, using PBS, and then resuspended in iced cold methanol to permeabilize. Cells were incubated at −20° C overnight before being washed twice in PBS and stained with iridium DNA intercalator (Fluidigm Sciences). Purified, carrier free antibodies were purchased from the listed provider and labeled with the listed metal using the protocol provided by Fluidigm. Stained samples were collected at Vanderbilt University Flow Cytometry Shared Resource on a CyTOF 1.0 mass cytometer (Fluidigm Sciences). All events were normalized prior to analysis using Fluidigm normalization beads.
CyTOF data preprocessing
Data (FCS files) were collected and stored in the online, analysis platform Cytobank. Data analysis was performed in Cytobank and statistical programming environment R (version 3.4.0) via R Studio.
Earth Mover’s Distance
The Earth Mover’s Distance (EMD) was calculated between each pair of populations using the “transport” library for R (28, 33) (https://cran.r-project.org/web/packages/transport/citation.html). The parent population (e.g. live CD45+ events) were gated in Cytobank, followed by the creation of a viSNE map in Cytobank. A viSNE analysis with two output dimensions was performed, equally sampling 5000 events per file, with 1000 iterations, perplexity equal to 30, and theta equal to 0.5. The events with their viSNE axes were then downloaded from Cytobank, and the Earth Mover’s Distance (EMD) was calculated between each pair of files using the “transport” library for R. The “wpp” object was used to represent each set of points in the two viSNE axes, and the “wasserstein” function was called on each pair of point sets to produce a distance matrix. Each point was assigned unit weight.
Because calculating a matrix with the EMD between each set of 5000 events from the viSNE analysis is computationally expensive, four optimizations were performed. (1) Each file was further down-sampled to 1000 out of the original 5000 events per file in the viSNE analysis. Each event was still assigned unit weight, and each point set, therefore, still had an equal total mass of 1000. (2) The “shortsimplex” method was used for the “wasserstein” function in the “transport” library, which accepted no other parameters besides the pair of weighted point sets (34). (3) Each population was automatically assigned a zero EMD compared to itself, and EMD scores already computed across the diagonal were simply copied because EMD is a metric. (4) The “parallel” library was used to parallelize the computation of each row of the matrix, in addition to the above, using the number of cores detected from the “detectCores” function in the “parallel” library. EMD values computed by ‘emdist’ were compiled in a CSV file and used to create a heatmap, in R, for visualization. Statistical comparisons of EMD values between groups were done in Excel using a Student’s t- test. CSV file and heatmap are each produced as an output.
Change in population equation
The frequency of immune populations was determined in Cytobank and exported into CSV files prior to re-organization. For Cohorts 1 and 3, populations were identified by traditional biaxial gating. For Dataset 2, populations were identified by first running a viSNE on nucleic acid expressing events from all patients at all time points and then running a SPADE on the t-SNE axes. Fifteen nodes (15) were identified with 5% down sampling. The following equation was used to determine the change in frequency for all data sets where FREQt is equal to the frequency of a population at a given time point and FREQpre is the frequency of that same population prior to the start of therapy. The addition of 0.01 to both the numerator and the denominator is to account for the appearance of new populations over the course of therapy.
Change in frequency = ln((FREQt + 0.01)/( FREQpre + 0.01))
R was used to conduct a paired Student t test to compare samples from the same patient at different time points of treatment. R script provided by Carr, et al. was used to create boxplots in R (7). In the case of Dataset 1, a Bonferroni correction was used for multiple hypothesis testing.
MEM
MEM creates a quantitative label of cell identity for given populations (19), and the MEM equation is implemented in R. MEM labels were either created for the indicated populations using the bulk, non-population as the reference, except, where indicated, when iPSCs or hematopoietic stem cells were stained and run on mass cytometry as a respective common reference (19). Median MEM labels were created by taking the median MEM score of each marker for each population. Standard deviation is shown. ΔMEM scores are calculated by subtracting the MEM score of the pre-therapy sample from the MEM score of the indicated time point.
Similarity of MEM labels
Root mean square deviation (RMSD) and hierarchical clustering were used to compare MEM labels, as previously described (19). The MEM vectors for each non-reference population were calculated over phenotype channels which were shared across all non-reference populations and the single reference population. Each MEM vector contained the population’s MEM score, calculated for each of the common phenotype channels, in reference to the single reference population. The MEM RMSD between pairs of non-reference populations was then calculated using the Euclidean distance between these MEM vectors.
Heatmaps representing population similarity were generated from each distance matrix using the “heatmap.2” function of the “gplots” library for R. The distance matrix was normalized by the maximum non-normalized distance d_max between any pair of populations, then multiplied by 100, then subtracted from 100. The result was that zero entries in the original distance matrix would receive a similarity score of 100, whereas the pair of populations with greatest distance in the original distance matrix would receive a similarity score of 0. Thus, two populations with the exact same enrichment score would have 100% similarity (19). To compare populations, median RMSD scores were compared using a two-tailed, Student t test.
Code availability
Original data sets were provided as FCS files in Flow Repository. Software for calculating EMD and displaying it as a heat map is available as Supplementary Software. Software for generating MEM scores is available in the Supplementary Software of Diggins et al. (19) (http://mem.vueinnovations.com/ ).
Data availability
Dataset 1
Peripheral blood from melanoma patients treated with anti–PD-1 and healthy peripheral blood controls is available as FCS files in Flow Repository. SNaPshot genotyping was done in the clinic on tumors resected from each patient. Forty-eight mutations in NRAS, BRAF, KIT, CTNNB1, and GNAQ were monitored (35).
Dataset 2
Peripheral blood from AML patients treated with chemotherapy was generated by CyTOF analysis as described by Ferrell et al. (12) and is available as FCS files in Flow Repository (http://flowrepository.org/id/FR-FCM-ZZMC). Patient characteristics and treatment details are available in Ferrell et al.
Dataset 3
Serially biopsied melanoma tumors from patients treated with BRAFi and MEKi were generated in separate mass cytometry experiments. Patients MP-034, MP-029, MP-031, MP-032, MP-055, and MP-059 were stained with the mass cytometry panel described in Supplementary Table S2. Patients MP-019, MP-023, MP-054, MP-052, and MP-062 were stained with the panel described by Doxie et al. (36). FCS files are available in Flow Repository. SNaPshot genotyping was done as described above.
Additional Data
Data for comparison across cancer types were generated by us in separate mass cytometry studies characterizing untreated melanoma tumors, glioblastoma tumors, and non-malignant tonsil and adenoid. FCS files are available in Flow Repository. Renal cell carcinoma tumors RC-29, RC-37, and RC-52 were published by Siska et al. (32), and Staphylococcal enterotoxin B (SEB)-stimulated PBMCs were published by Nicholas et al. (37). Briefly, PBMCs were stimulated with SEB (final concentration 1 μg/mL in in 200 μL) at a cell concentration of 10 × 106 cells/mL. Cells were incubated at 37° C for 16 hours before being washed twice with PBS and stained for mass cytometry.
Data sets used in the RMSD heatmap are described by Diggins et al. (19).
RESULTS
Application of Earth Mover’s Distance to characterize the peripheral immune system during therapy
We sought to investigate how the overall composition of circulating immune populations changes over the course of anti–PD-1 therapy. To do this we generated viSNE maps, which reduce the dimensionality of the data by plotting cells in two dimensions on the basis of their high-dimensional similarity. Changes in the distribution, or topography, of these maps, are indicative of changes in the abundance or profile of individual cell populations. Quantifying changes that exist between viSNE maps relies on either deconstructing the system into populations or qualitative assessment of the overall “shape” of the map. Approaches for quantifying differences in multidimensional cytometry distributions include Quadratic Form (38) and Earth Mover’s Distance (EMD) (28). Here, we used EMD to quantify the similarity of two viSNE maps, where differences were quantified as the amount of work required to make the images like another. In this instance, “work” was defined as the number of units, or cells, times the distance moved. To quantify differences in viSNE maps, all samples must be run together on the same axes. It is imperative that measures be taken to reduce batch effects, like the use of normalization beads here.
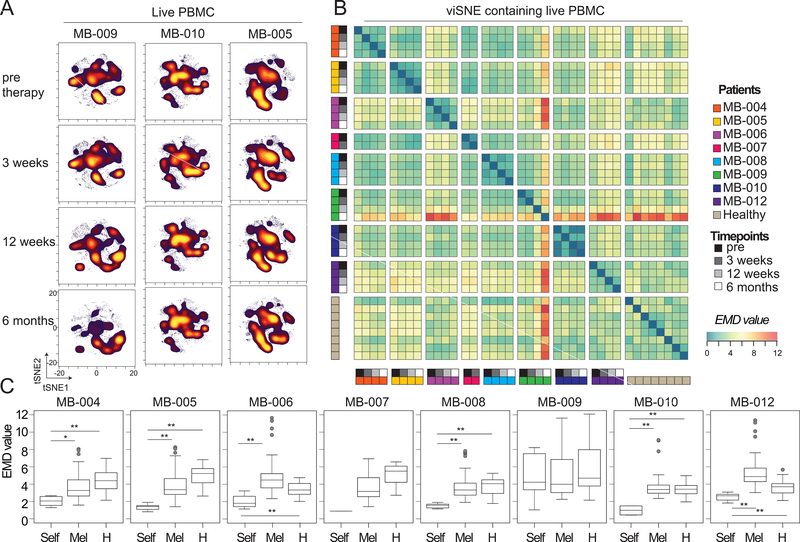
Blood was drawn from melanoma patients immediately prior to therapy, and 3 weeks, 12 weeks, and 6 months after the start of anti–PD-1 therapy (Dataset 1). Mass cytometry was used to characterize PBMCs from each patient at each time point (Supplementary Table S1 and Supplementary Table S2). To monitor and quantify the phenotypic plasticity of the peripheral immune system as a whole, the EMD algorithm was used to quantify differences between viSNE maps as described above (21, 28, 33). One viSNE map was created for eight patients analyzed at four clinical timepoints and eight healthy controls (Fig. 1A, Supplementary Table S1). EMD was then used to quantify the differences between each viSNE map, and the numerical results were displayed in a heat map (Fig. 1B). Low EMD scores indicated that the maps were similar, whereas larger EMD scores indicated divergent maps. To determine whether the peripheral immune systems of each patient remained stable or had increased phenotypic plasticity over the course of anti-PD-1 therapy, intra-patient EMD values, those generated by comparing viSNE maps within a single patient over therapy, were compared to inter-patient EMD values, those generated by comparing viSNE maps across patients. In 7 out of 8 melanoma patients receiving anti–PD-1 therapy, the intra-patient EMD score was lower than the inter-patient EMD score, indicating that each patient’s peripheral blood immune system was more similar to itself than to that of any of any other patient, regardless of any ongoing therapy response (Fig. 1C). The exceptional patient, MB-009, did not conform to this pattern (median intra-patient EMD value±standard deviation: 4.22±2.69, versus median inter-patient EMD value: 3.99±2.48). This patient was diagnosed with myeloid dysplastic syndrome 8 months after starting anti–PD-1 therapy and was known to have an expansion of mature and blasting myeloid cells and a decrease in all other major cell types in the periphery (15). Thus, combining EMD and viSNE allowed for an automated approach to quantify stability and plasticity of a system over the course of therapy.
Figure 1. EMD quantifies phenotypic plasticity of the system over therapy and identifies an outlier patient.
Peripheral blood mononuclear cells (PBMCs) from melanoma patients undergoing anti–PD-1 therapy and healthy donors were characterized by mass cytometry. Equal numbers of live events from each sample were run together on a viSNE map. (A) Representative live leukocyte viSNE plots are shown for three patients at all collection points during therapy. (B) EMD was calculated, pairwise, for all samples. Heat indicates magnitude of EMD value. (C) Median EMD was calculated for each patient from pairwise EMD between samples from that same patient (light grey), between that patient and all other pembrolizumab samples (white), and between that patient and all healthy donors (dark grey). N=6, 104, and 32 respectively (with exception of MB-007 where N=1, 52, and 16, respectively). *p<0.001, **p<0.0001. The whiskers of the boxplot extend to the most extreme data point which is no more than 1.5x the interquartile range from the box. See also Supplementary Tables S1 and S2.
Ensemble analysis revealed decreases in PD-1+ T cells during anti–PD-1 therapy
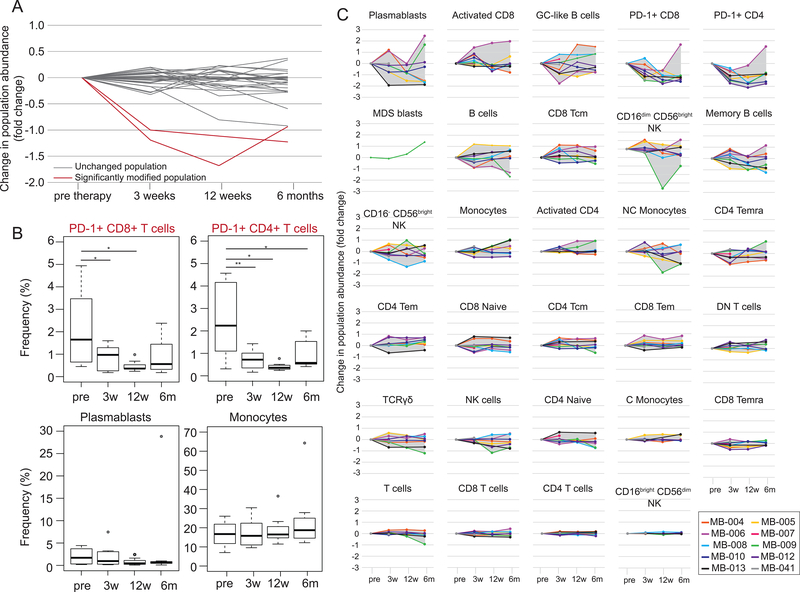
Although viSNE and EMD provided a quantitative, first-glance at system stability, it is possible that shifts in abundance or phenotype of small, biologically relevant populations may be overlooked when using a single summary statistic. To avoid missing small but crucial cell subsets, cells should be broken into smaller populations manually or computationally and compared. The cells were next manually split into classical immune populations using traditional, biaxial gating (Supplementary Fig. S1). Although biaxial gates were used for cell population identification, it is possible to integrate other automated clustering tools, like Phenograph (23), VorteX (39), and FlowSOM (40) into the ensemble at this step. The change in frequency of these populations was calculated as a fold-change over pre-therapy, allowing for visualization of all patients and populations on one graph.
Twenty-eight cell populations were identified, and the change in frequency of those populations compared to the pre-therapy time point was calculated (Fig. 2A-B, Supplementary Fig. S2). Two populations showed significant changes in frequency. CD4+PD-1+ and CD8+PD-1+ T cells had a significant change in frequency [N=10; p<0.0001 (pre- vs. 3 weeks) and p=0.0011 (pre- vs. 12 weeks) for CD4+PD-1+ T cells, and p=0.00057 (pre- vs. 12 weeks) for CD8+PD-1+ T cells] over the course of therapy, in which both decreased (Fig. 2A-C). PD-1 gates were drawn with non-malignant tonsil as a reference and considerations discussed and explored by Nicholas et al. (37). This decrease in relative frequency is most likely the result of receptor occupancy by pembrolizumab, as opposed to a biological decrease in these cell populations (41, 42).
Figure 2. Frequency tracking of populations identifies a loss of detectable PD-1+ T cells.
Established immune populations were manually gated and population frequencies were determined over the course of anti–PD-1 therapy. (A) Population frequencies at 3 weeks, 12 weeks, and 6 months post start of anti–PD-1 therapy were normalized to the pre-therapy frequency. Each line represents a change in frequency for one population. Significantly changing populations, compared to pre-therapy, are shown in red. (B) Change in population frequency is shown for individual patients for each population median shown in (A). Each time point after the start of therapy was compared to the pre-therapy time point using a two tailed, paired t test. With a Bonferroni correction, significantly different populations had a p value<0.0018. P values are indicated for populations with significant changes. (C) Boxplots of population frequency are shown for each significantly changing population (top) and for two populations that did not significantly change (bottom). P values were derived from an uncorrected, two tailed, paired t-test. *p<0.05, **p<0.01. See also Supplementary Figures S1 and S2 and Supplementary Tables S1 and S2.
Marker Enrichment Modeling identified signature features of PD-1+ T cells in tumor and blood
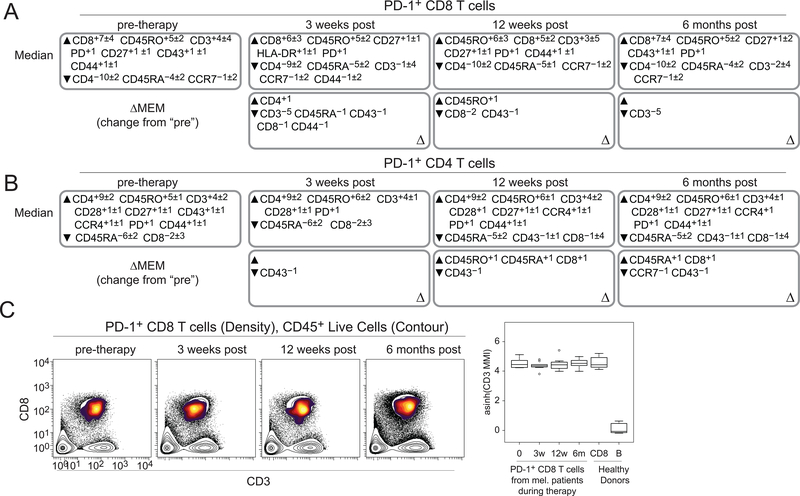
The next step in the ensemble systems immune analysis pipeline was to automatically quantify enrichment of measured parameters and determine how those enriched parameters changed during treatment. MEM was used to identify signature features of each population at each time point following established methods (19). MEM scores can range from +10 (maximum enrichment) through 0 (no enrichment) to −10 (maximum lack) and here, are reported as the median±standard deviation in MEM value for the cell population. Enrichment quantifications for each population are a vector to be used for additional computational analysis. In this ensemble workflow, vectors describing the high-dimensional phenotype of populations are compared using root-mean-square deviation (RMSD) (19).
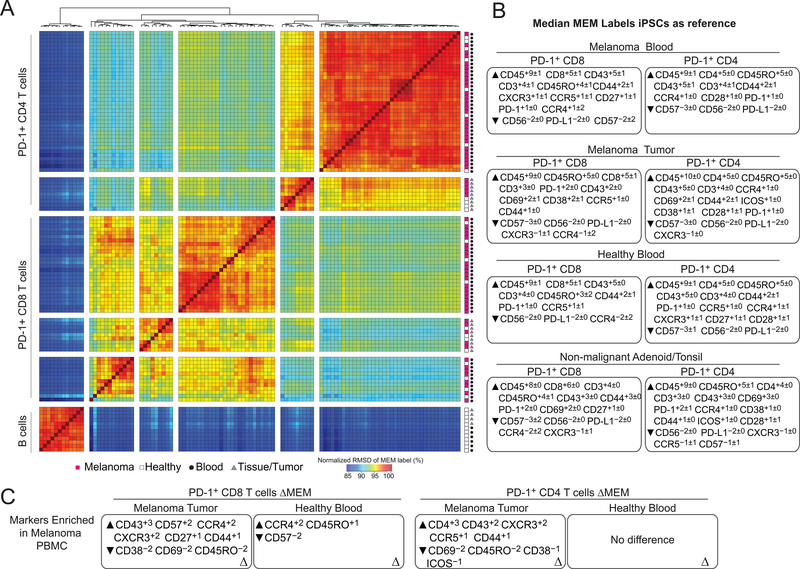
PD-1+ CD8+ and CD4+ T cells from pre-therapy samples were enriched for canonical identity makers CD3, and CD8 and CD4, respectively (Fig. 3A-B, top). To indicate changes in enrichment patterns, changes in MEM score (ΔMEM) were calculated by subtracting the median MEM score for each parameter at the pre-therapy time point from the indicated time point after the start of therapy. After 3 weeks and 6 months of anti–PD-1 therapy, PD-1+CD8+ T cells lost enrichment, but not expression, for CD3 (ΔMEM of –5; median±standard deviation, CD3 MMI 40.9±11 (3 weeks) and 48.3±14.1 (6 months)) (Fig. 3A, bottom; Fig. 3C). This was not the case for PD-1+CD4+ T cells (Fig. 3B, bottom). Given a loss of enrichment of the TCR CD3 subunit on these peripheral blood PD-1+CD8+ T cells, it was informative to determine their novelty by comparing them to other subsets, including PD-1+CD8+ T cells from tumors or healthy donors. To assess this population, MEM labels were created for PD-1+CD4+ and PD-1+CD8+ T cells gated from (1) blood from melanoma patients during anti–PD-1 therapy, (2) human melanoma tumors, (3) blood from healthy donors, and (4) tonsil or adenoid tissue from healthy donors. IPSCs, analyzed by mass cytometry, were used as a control cell reference for MEM calculations. Similarity in MEM labels was then compared using RMSD (Fig. 4A-B). B cells gated from healthy donor blood and tonsils were also included for contrast because of their distinctly different enrichment profiles. As expected, B cells clustered separately from the T-cell populations. PD-1+CD4+ T cells from the blood of healthy donors and melanoma patients receiving anti–PD-1 therapy clustered together, whereas PD-1+CD4+ T cells from melanoma tumors and healthy donor tonsils formed a different cluster (Fig. 4A). Similarly, PD-1+CD8+ T cells from melanoma tumors clustered with those from healthy donor tonsils, whereas PD-1+CD8 T cells from healthy donor blood and melanoma blood formed two, intermixed clusters. These results indicated PD-1+ T cells found in the blood were distinct from those found in the tumor or healthy tonsil.
Figure 3. Marker enrichment modeling identifies signature features of populations over the course of therapy.
Tissue-specific MEM labels were created for each cell population, from each patient, at each time point. (A) Median MEM labels are shown for PD-1+CD8+ T cells at each time point during therapy (top). ΔMEM labels were calculated by subtracting the median pre-therapy MEM scores from the median MEM scores at each time point. ΔMEM labels indicate the change in MEM value compared to pre-therapy (bottom). (B) Median MEM labels are shown for PD-1+CD4+ T cells at each time point during therapy (top). ΔMEM labels are shown for PD-1+CD4+ T cells from each time point during therapy (bottom). MEM values are represented as the median MEM value±standard deviation. (C) Biaxial plots of CD3 and CD8 are shown for PD-1+CD8+ T cells from representative melanoma patients (MB-004) undergoing anti–PD-1 therapy (left). Density: PD-1+CD8+ T cells, Contour: live CD45+ cells. Transformed (arcsinh5) CD3 median metal intensity (MMI) is shown for PD-1+CD8+ T cells at each time point during therapy [pre-therapy (0), n=10; 3 weeks (3w), n=10; 12 weeks (12w), n=7; 6 month (6m), n=8; healthy, n=8]. Healthy PBMC donor CD8+ T and B cells are shown for reference. See also Supplementary TablesS1 and S2.
Figure 4. MEM reveals that PD-1+ T cells from blood differ from those in the tumor.
(A) MEM labels were compared for each of the 112 populations (PD-1+ CD4+ and CD8+ T cells and B cells) from three human tissues. Populations were defined using traditional biaxial gates as in Supplementary Fig. S1. Tissue type and source are indicated in the bottom left. (B) Median MEM labels are shown for PD-1+ CD4+ and CD8+ T cells from each tissue type. MEM values are shown±standard deviation. (C) ΔMEM scores show the difference in median MEM scores between PD-1+CD8+ T cells (left) or PD-1+CD4+ T cells (right) in the peripheral blood and those found in the tumor or blood of healthy donors. See alsoSupplementary TablesS1 and S2.
Median MEM labels, which display enrichment scores for each measured feature, are shown for each tissue’s PD-1+ T-cell populations (Fig. 4B). PD-1+CD8+ T cells from the blood of melanoma patients were enriched for CD43 protein expression and trafficking markers, like CCR4 and CXCR3, and specifically lacked activation markers CD38 and CD69 compared to PD-1+CD8+ T cells in melanoma tumors (Fig. 4C, left). PD-1+CD4+ T cells were enriched for CD4 (+3) compared to their melanoma tumor counterparts. No difference in enrichment between PD-1+CD4+ T cells in the peripheral blood of melanoma patients and healthy donors (Fig. 4C, right). PD-1+ T cells from the blood of melanoma patients and healthy donors were, therefore, phenotypically similar. Thus, MEM, as part of this ensemble, automated quantitative comparisons of high-dimensional data from different tumor and donor types.
Increased CD4+ T-cell frequency following chemotherapy in AML patients
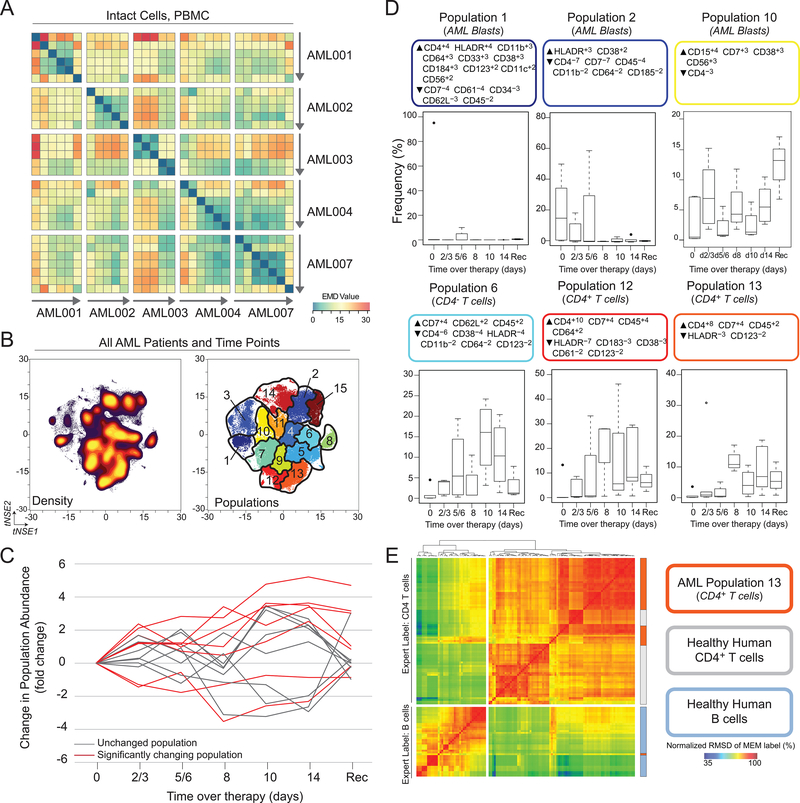
The ensemble systems immune monitoring pipeline was applied to a previously published dataset of peripheral blood from acute myeloid leukemia (AML) patients undergoing chemotherapy (Dataset 2, (12)) in order to describe and dissect system wide changes. Five AML patients were consented for peripheral blood draws over the course of chemotherapy, which were then characterized by mass cytometry. All PBMCs, including blasts and non-blasts, were identified using the gating scheme published by Ferrell et al. (12). EMD on t-SNE revealed lack of intra-patient stability in 3 of 5 patients, indicated by no significant difference between intra- and inter-patient EMD values (Fig. 5A, Supplementary Fig. S3). The remaining two patients showed intra-patient stability, most likely due to the presence of leukemic blasts throughout treatment. Unlike in Dataset 1, where cellular populations were identified by traditional biaxial gates, fifteen populations were defined automatically using the SPADE algorithm to cluster on t-SNE axes (Fig. 5B), as previously described (17, 24). These populations were then fed into the remaining three steps of the ensemble analysis pipeline.
Figure 5. Ensemble immune analysis and automated gating identifies loss of peripheral blasts and increase in non-malignant immune cells in AML patients undergoing chemotherapy.
PBMCs from AML patients undergoing chemotherapy were characterized by mass cytometry. Equal numbers of live events from each sample were run together on a viSNE map. (A) EMD was calculated, pairwise, for all samples. Heat indicates magnitude of EMD value. (B) Populations were identified (right) by SPADE of cell density on t-SNE axes (left). (C) Frequency of populations identified in (B) was normalized to the pre-therapy frequency and compared using a paired t test. Populations with time points that significantly change from pre-therapy are shown in red. (D) Boxplots are shown for each significantly changing populations. Each population is labeled with a MEM label and an expert given name derived from the MEM label. (E) MEM labels from Population 13 were compared with the 80 populations (CD4+ T cells and B cells) using RMSD (36). See also Supplementary Fig. S3 and S4.
Of the fifteen populations identified, seven showed significant changes at some point during chemotherapy (Fig. 5C, Supplementary Fig. S4). MEM was used to label the automatically characterized signature features on each population (Fig. 5D, Supplementary Fig. S4). Two populations of blasts were defined by HLA-DR enrichment and were observed to decrease over the course of chemotherapy. In contrast, CD4– T cells and two subsets of CD4+ T cells were observed to expand, relative to other populations, following chemotherapy. Population 13 was enriched for CD4 (+5), CD7 (+4), and CD45 (+2), while specifically lacking expression of HLA-DR (–3) and CD123 (–2). Combined with the expression of CD3 (Supplementary Fig. S5), population 13 is likely a population of T cells. Next, the similarity of population 13 to CD4+ T cells from healthy donors was assessed. To do this, a common reference population of hematopoietic stem cells was used to create MEM labels for population 13 and for previously published healthy donor CD4+ T cells and B cells (19). RMSD was used to compare MEM labels and determine whether population 13 shared a phenotype similar to that of previously characterized CD4+ T cells. Population 13 clustered with CD4+ T cells from healthy donors, suggesting that they are phenotypically similar (Fig. 5E). Taken together, these results represent an automated way to dissect immune responses to therapy and suggest that chemotherapy resulted in a relative increase of a non-malignant population of CD4+ T cells.
Loss of activated T cells and expansion of CD4–CD8– T cells in melanoma tumors with MEKi and BRAFi
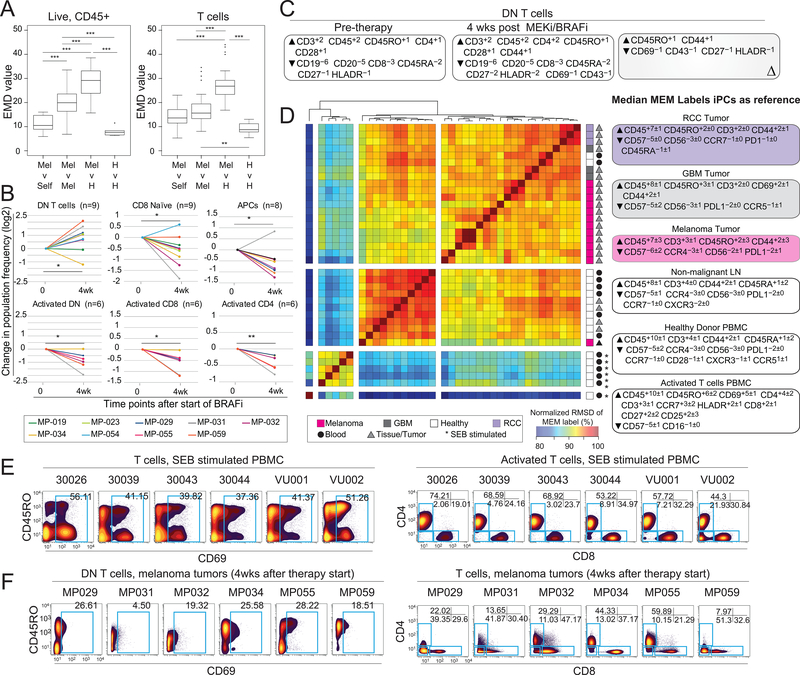
Samples from a cohort of melanoma patients with BRAFV600E mutations treated with targeted therapies dabrafenib and trametinib (n=11; Dataset 3) were characterized by mass cytometry, and the data were analyzed by the ensemble systems immune monitoring pipeline. Combining EMD and viSNE to quantify stability of the immune compartment showed that each patient remained more similar to itself than to other melanoma tumors or healthy tonsils over the course of therapy. However, EMD run on t-SNE axes created from analysis of T cells revealed that the T-cell compartment did not have lower intra-patient EMD values compared to inter-patient EMD values (Fig. 6A). Thus, significant immune plasticity followed therapy. Immune populations were defined using traditional biaxial gates, and the change in frequency for those populations was calculated. After 4 weeks of treatment, a statistically significant change in frequency of 5 immune populations was observed. One population of interest included double-negative (DN) T cells lacking expression of CD4 or CD8 (DN T cells) that comprised 7.23±17.18% in pre-therapy tumors, 26.27±16.36% in tumors 4 weeks post-therapy, and 3.57±1.52% of a healthy lymph node (median±standard deviation, Fig. 6B, Supplementary Fig. S6–7). Pre-therapy MEM labels showed that DN T cells were enriched for CD3 (+2), CD45 (+2), CD45RO (+1), CD4 (+1), and CD28 (+1) but specifically lacked CD8 (–3) and CD45RA (–2) when compared to all cells found within the melanoma tumors. ΔMEM scores indicated that, over the course of therapy, DN T cells became more enriched for CD45RO (+1) and CD44 (+1) but lost enrichment of CD69 (–1), CD43 (–1), CD27 (–1), and HLA-DR (–1) (Fig. 6C).
Figure 6. Ensemble immune analysis identifies expansion of CD8 and CD4 double-negative T cells in tumors from patients treated with BRAF and MEK inhibitors.
Single-cell suspensions of melanoma tumor biopsies from before and after treatment with BRAF and MEK inhibitors were characterized by mass cytometry. (A) Equal numbers of live events (left) or T cells (right) from each sample were run together on a viSNE map. EMD was calculated, pairwise, for all samples. Boxplots show median, pairwise EMD values for listed comparisons. Unpaired student t test. *p<0.05. **p<0.01, ***p<0.001. (B) Changes in population frequency is shown for individual patients for each significantly changing population. Each time point after the start of therapy was compared to the pre-therapy time point using a two-tailed, paired t test. *p<0.05. **p<0.01. (C) Median MEM labels are shown for double-negative (DN) pre-therapy and 4 weeks after therapy. A ΔMEM score shows the difference in median MEM scores DN T cells before and after the start of therapy. (D) MEM labels from tumor resident DN T cells were compared with SEB-stimulated T cells from peripheral blood using RMSD. Median MEM scores for each tissue are shown on the right. (E) Biaxial plots of all T cells from SEB-stimulated PBMCs (left) and activated T cells (CD69+, plots on right). (F) Biaxial plots of DN T cells from tumors of patients treated for 4 weeks with BRAFi and MEKi (left) and all T cells from (right). See also Supplementary Fig. S6 and S7, Supplementary Tables S1 and S2.
Given that T cells can downregulate expression of CD4 or CD8 if activated (43), DN T cells from melanoma tumors were compared to acutely activated peripheral blood T cells stimulated through the TCR by Staphylococcal enterotoxin B (SEB) (37). Additionally, we sought to understand whether similar T cells were found in other cancers, such as glioblastoma (GBM) and renal cell carcinoma (RCC) (Figure 6D). SEB-stimulated T cells formed their own cluster, apart from all other DN T cells, and shared 87.2% (±2.88%, n=78 comparisons) similarity with DN T cells from melanoma tumors. In contrast, DN T cells from melanoma, GBM, and RCC clustered together, with a similarity score of 93.6% (±1.06%, n=78 comparisons). Of the T cells activated by SEB, close to half were CD69+ (41.26±7.41%), and most retained CD4 or CD8 co-receptor expression (Fig. 6E). In contrast, DN T cells from post-treatment melanoma tumors contained fewer CD69+ cells (22.45±8.76%) (Fig. 6F). Thus, DN T cells from melanoma, GBM, and RCC consisted of a phenotypically distinct population of DN T cells not observed in resting non-diseased blood and tonsils or acutely stimulated blood. Taken together, these data suggest that an unusual population of T cells emerged in tumors from melanoma patients treated with BRAFi and MEKi and that this DN T-cell population occurs in multiple types of tumors.
DISCUSSION
The manuscript describes an analysis suite designed to be a common starting point for immunologists tracking cells over time, provides three reference datasets for testing tools designed to discover and characterize cell subsets, and reveals unexpected cells in the context of cancer therapies. This ensemble data analysis strategy was designed specifically for systems immune monitoring in longitudinal, clinical studies, but it could be applied for any system with changes. We envision adapting it to study experimental perturbations to map signaling networks and drug responses (44). Although we expect the algorithms in the ensemble will change and improve over time, the four cellular properties identified should be considered essential features for immune monitoring with any single-cell platform. Cancer immune monitoring strategies must expect the unexpected and be prepared for novel phenotypes (9, 45, 46). The first property, systems plasticity, automatically quantifies the state of a system compared to baseline. In the cohort described here, the peripheral blood of melanoma patients remained stable over the course of therapy. By monitoring systems plasticity with EMD and t-SNE, a patient who experienced bone marrow failure (15) was identified without gating or clustering. In this case, quantifying systems plasticity allowed for the unbiased identification of a patient experiencing large, biologically relevant changes during therapy. Caveats exist for using EMD to quantify differences between viSNE maps. All samples must be embedded on the same viSNE map in order to be appropriately compared, and precautions, like barcoding and bead normalization, must be taken to ensure that differences in viSNE maps are attributed to biological differences and not batch effects. In addition to EMD, the quadratic form (38) could be used to quantify differences in t-SNE axes. Looking forward, we envision automating a process in which t-SNE and a plasticity test like EMD are first run on the system as a whole, as described here, and then run iteratively on increasingly refined populations in order to pinpoint the populations undergoing the greatest change or stability.
The remaining components of the ensemble work together to describe changes or stability revealed in the first step, systems plasticity. To accomplish this, the system is divided into subpopulations and the frequency, signature features, and novelty of each subpopulation are quantified. Identification of cells into groups can be accomplished in whichever way is most appropriate, or in multiple ways, prior to ensemble analysis (22–24, 27). For example, FlowSOM (40) could be used to identify cellular populations, and those populations could be fed into the remaining parts of the ensemble. Because this ensemble toolkit does not rely on known populations, it remains independent from methods of automated population identification (17, 27) and can be used to compare and communicate analysis results from teams relying on computational approaches, immunologists, and bioinformatics experts. The ensemble approach was especially adept at capturing shifts over time and was able to identify a shift in relative abundance of a subset of T lymphocytes, their phenotype and intra-patient recovery of these phenotypes, all of which could have substantial implications for maintenance of remission and clinical outcomes (47, 48). A human immune monitoring strategy using this ensemble toolkit might provide new insight into the immune system’s interaction with leukemia remission and relapse.
This ensemble approach was robust across multiple, contrasting studies in quantifying changes and stability in immune system cells. This approach also provided a detailed analysis of the abundance and tractable quantitative phenotype of populations that comprised each system. We found a population of CD4–CD8– DN T cells observed to have a common phenotype in three human tumor types, melanoma, renal cell carcinoma, and glioblastoma that had a phenotype distinct from that of resting CD4–CD8– DN and SEB-activated T cells from healthy individuals. This result is consistent with reports characterizing the phenotypic similarity of T cells infiltrating melanoma and colon cancer mouse tumor models (49) and provides evidence that common changes to immune cell mechanisms are shared across human tumor types and play a role in response to targeted therapy.
The ensemble toolkit detected known biological occurrences, as well as identified potential mechanisms for further study. The ensemble toolkit detected an overall loss of PD-1+ T cells in the peripheral blood over the course of therapy. This has previously been described and attributed to receptor occupancy by the drug itself (41, 50). Previous work revealed an expanded population of Ki67+ T cells in the peripheral blood during immunotherapy (10, 51). Although our comparable study did not measure Ki67 in the blood and, thus, could not be compared directly, we did observe a trend towards increased activated CD4+ and CD8+ T cells, defined by expression of HLA-DR, at 3 weeks after the start of therapy, and CD4+ effector memory T (TEM) cells at all time points after the start of therapy. In addition to capturing known clinical events, the ensemble tool kit identified a novel population of DN T cells present in unexpectedly high frequencies in melanoma tumors before and after treatment with BRAFi and MEKi compared to healthy lymphoid tissue. Expanded DN T cell populations have previously been reported in metastatic lymph nodes of melanoma patients (52). However, their deep phenotype or frequency in response to inhibitor therapy has not yet been described. Previous work has described the expansion of a regulatory CD3+ T-cell population lacking both CD4 and CD8 after TCR and cytokine stimulation (53–55). MEK inhibitors can support antitumor T-cell function by blunting TCR-induced apoptosis (56). Therefore, it is possible that high frequencies of DN T cells after treatment with MEKi and BRAFi indicate an accumulation of T cells derived from tumor resident T cells, although additional mechanistic studies are required.
Overall, the melanoma tumor dataset (Dataset 3) highlights the complexity of tumor-associated T cells in human malignancies and provides further evidence that phenotypically diverse populations of tumor resident T cells can be found across multiple, distinct tumor types (49). The data presented here will join a common immunology reference set (19, 57, 58) that can be mined further to characterize and understand changes in immune and cancer cell populations in diverse disease settings and build a reference of cellular identity.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Holly Crandall and Kate Carson for their help with consenting patients and collecting patient samples.
Funding: NIH/NCI R00 CA143231 (J.M.I.), F31 CA199993 (A.R.G.), U01 CA196405 (D.D.M. and J.M.I.), R25 GM062459 (D.B.D.), T32 CA009592 (D.B.D.), K23 HL138291 (P.B.F.), K23 CA204726 (D.B.J.), the Vanderbilt-Ingram Cancer Center (VICC, P30 CA68485), a Vanderbilt University Discovery Grant (J.M.I. and N.L.), a VICC Provocative Question award (M.C.K. and J.M.I.), and VICC Ambassadors (J.M.I. and A.R.G.).
Footnotes
Conflict of interest disclosure: J.M.I. is co-founder and a board member of Cytobank Inc. and received research support from Incyte Corp and Janssen.
REFERNCES
- 1.Greenplate AR, Johnson DB, Ferrell PB Jr., Irish JM Systems immune monitoring in cancer therapy. Eur J Cancer. 2016;61:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodin P, Davis MM. Human immune system variation. Nature reviews Immunology. 2017;17:21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudilliere B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Science translational medicine. 2014;6:255ra131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaczorowski KJ, Shekhar K, Nkulikiyimfura D, Dekker CL, Maecker H, Davis MM, et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proceedings of the National Academy of Sciences of the United States of America. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer cell. 2008;14:335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, Wouters C, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nature immunology. 2016;17:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irish JM. Beyond the age of cellular discovery. Nature immunology. 2014;15:1095–7. [DOI] [PubMed] [Google Scholar]
- 10.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017;168:487–502 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrell PB Jr., Diggins KE, Polikowsky HG, Mohan SR, Seegmiller AC, Irish JM. High-Dimensional Analysis of Acute Myeloid Leukemia Reveals Phenotypic Changes in Persistent Cells during Induction Therapy. PloS one. 2016;11:e0153207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–28. [DOI] [PubMed] [Google Scholar]
- 14.Kordasti S, Costantini B, Seidl T, Perez Abellan P, Martinez Llordella M, McLornan D, et al. Deep-phenotyping of Tregs identifies an immune signature for idiopathic aplastic anemia and predicts response to treatment. Blood. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenplate AR, Johnson DB, Roussel M, Savona MR, Sosman JA, Puzanov I, et al. Myelodysplastic Syndrome Revealed by Systems Immunology in a Melanoma Patient Undergoing Anti-PD-1 Therapy. Cancer immunology research. 2016;4:474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roederer M, Quaye L, Mangino M, Beddall MH, Mahnke Y, Chattopadhyay P, et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diggins KE, Ferrell PB Jr., Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saeys Y, Gassen SV, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nature reviews Immunology. 2016;16:449–62. [DOI] [PubMed] [Google Scholar]
- 19.Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, Irish JM. Characterizing cell subsets using marker enrichment modeling. Nature methods. 2017;14:275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science (New York, NY). 2015;349:1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology. 2013;31:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr., Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE). Proceedings of the National Academy of Sciences of the United States of America. 2014;111:202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arvaniti E, Claassen M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nat Commun. 2017;8:14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2016;89:1084–96. [DOI] [PubMed] [Google Scholar]
- 28.Orlova DY, Zimmerman N, Meehan S, Meehan C, Waters J, Ghosn EE, et al. Earth Mover’s Distance (EMD): A True Metric for Comparing Biomarker Expression Levels in Cell Populations. PloS one. 2016;11:e0151859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson AS, Crandall H, Dahlman K, Kelley MC. Preliminary results from a prospective trial of preoperative combined BRAF and MEK-targeted therapy in advanced BRAF mutation-positive melanoma. J Am Coll Surg. 2015;220:581–93 e1. [DOI] [PubMed] [Google Scholar]
- 30.Leelatian N, Doxie DB, Greenplate AR, Mobley BC, Lehman JM, Sinnaeve J, et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom. 2017;92:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leelatian N, Doxie DB, Greenplate AR, Sinnaeve J, Ihrie RA, Irish JM. Preparing Viable Single Cells from Human Tissue and Tumors for Cytomic Analysis. Curr Protoc Mol Biol. 2017;118:25C 1 1–C 1 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siska PJ, Beckermann KE, Mason FM, Andrejeva G, Greenplate AR, Sendor AB, et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Japp AS, Hoffmann K, Schlickeiser S, Glauben R, Nikolaou C, Maecker HT, et al. Wild immunology assessed by multidimensional mass cytometry. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2017;91:85–95. [DOI] [PubMed] [Google Scholar]
- 34.Gottschlich C, Schuhmacher D. The Shortlist Method for fast computation of the Earth Mover’s Distance and finding optimal solutions to transportation problems. PloS one. 2014;9:e110214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovly CM, Dahlman KB, Fohn LE, Su Z, Dias-Santagata D, Hicks DJ, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PloS one. 2012;7:e35309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doxie DB, Greenplate AR, Gandelman JS, Diggins KE, Roe CE, Dahlman KB, et al. BRAF and MEK inhibitor therapy eliminates Nestin-expressing melanoma cells in human tumors. Pigment Cell Melanoma Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, et al. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2016;89:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernas T, Asem EK, Robinson JP, Rajwa B. Quadratic form: a robust metric for quantitative comparison of flow cytometric histograms. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2008;73:715–26. [DOI] [PubMed] [Google Scholar]
- 39.Samusik N, Good Z, Spitzer MH, Davis KL, Nolan GP. Automated mapping of phenotype space with single-cell data. Nature methods. 2016;13:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, et al. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2015;87:636–45. [DOI] [PubMed] [Google Scholar]
- 41.Das R, Verma R, Sznol M, Boddupalli CS, Gettinger SN, Kluger H, et al. Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. Journal of immunology (Baltimore, Md : 1950). 2015;194:950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. The New England journal of medicine. 2012;2012:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viola A, Salio M, Tuosto L, Linkert S, Acuto O, Lanzavecchia A. Quantitative contribution of CD4 and CD8 to T cell antigen receptor serial triggering. The Journal of experimental medicine. 1997;186:1775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nature biotechnology. 2012;30:858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. The Journal of clinical investigation. 2015;125:3356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeMets DL, Ellenberg SS. Data Monitoring Committees - Expect the Unexpected. The New England journal of medicine. 2016;375:1365–71. [DOI] [PubMed] [Google Scholar]
- 47.Behl D, Porrata LF, Markovic SN, Letendre L, Pruthi R, Hook C, et al. Absolute lymphocyte count recovery after induction chemotherapy predicts superior survival in acute myelogenous leukemia. Leukemia. 2006;20:29. [DOI] [PubMed] [Google Scholar]
- 48.Mackall CL, Fleisher TA, Brown MR, Andrich MP, Chen CC, Feuerstein IM, et al. Age, Thymopoiesis, and CD4+ T-Lymphocyte Regeneration after Intensive Chemotherapy. New England Journal of Medicine. 1995;332:143–9. [DOI] [PubMed] [Google Scholar]
- 49.Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, et al. Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell. 2017;170:1120–33 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallacchi V, Vergani E, Camisaschi C, Deho P, Cabras AD, Sensi M, et al. Transcriptional profiling of melanoma sentinel nodes identify patients with poor outcome and reveal an association of CD30(+) T lymphocytes with progression. Cancer research. 2014;74:130–40. [DOI] [PubMed] [Google Scholar]
- 53.Fischer K, Voelkl S, Heymann J, Przybylski GK, Mondal K, Laumer M, et al. Isolation and characterization of human antigen-specific TCR alpha beta+ CD4(−)CD8- double-negative regulatory T cells. Blood. 2005;105:2828–35. [DOI] [PubMed] [Google Scholar]
- 54.Priatel JJ, Utting O, Teh HS. TCR/self-antigen interactions drive double-negative T cell peripheral expansion and differentiation into suppressor cells. Journal of immunology (Baltimore, Md : 1950). 2001;167:6188–94. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D, Yang W, Degauque N, Tian Y, Mikita A, Zheng XX. New differentiation pathway for double-negative regulatory T cells that regulates the magnitude of immune responses. Blood. 2007;109:4071–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ebert PJ, Cheung J, Yang Y, McNamara E, Hong R, Moskalenko M, et al. MAP Kinase Inhibition Promotes T Cell and Anti-tumor Activity in Combination with PD-L1 Checkpoint Blockade. Immunity. 2016;44:609–21. [DOI] [PubMed] [Google Scholar]
- 57.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–49 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong MT, Ong DE, Lim FS, Teng KW, McGovern N, Narayanan S, et al. A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity. 2016;45:442–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Dataset 1
Peripheral blood from melanoma patients treated with anti–PD-1 and healthy peripheral blood controls is available as FCS files in Flow Repository. SNaPshot genotyping was done in the clinic on tumors resected from each patient. Forty-eight mutations in NRAS, BRAF, KIT, CTNNB1, and GNAQ were monitored (35).
Dataset 2
Peripheral blood from AML patients treated with chemotherapy was generated by CyTOF analysis as described by Ferrell et al. (12) and is available as FCS files in Flow Repository (http://flowrepository.org/id/FR-FCM-ZZMC). Patient characteristics and treatment details are available in Ferrell et al.
Dataset 3
Serially biopsied melanoma tumors from patients treated with BRAFi and MEKi were generated in separate mass cytometry experiments. Patients MP-034, MP-029, MP-031, MP-032, MP-055, and MP-059 were stained with the mass cytometry panel described in Supplementary Table S2. Patients MP-019, MP-023, MP-054, MP-052, and MP-062 were stained with the panel described by Doxie et al. (36). FCS files are available in Flow Repository. SNaPshot genotyping was done as described above.