Abstract
Electronic health records contain patient-level data collected during and for clinical care. Data within the electronic health record include diagnostic billing codes, procedure codes, vital signs, laboratory test results, clinical imaging, and physician notes. With repeated clinic visits, these data are longitudinal, providing important information on disease development, progression, and response to treatment or intervention strategies. The near universal adoption of electronic health records nationally has the potential to provide population-scale real-world clinical data accessible for biomedical research including genetic association studies. For this research potential to be realized, high quality research-grade variables must be extracted from these clinical data warehouses. We describe here common and emerging electronic phenotyping approaches applied to electronic health records as well as current limitations of both the approaches and the biases associated with these clinically-collected data that impact their use in research.
Key Concepts
Electronic Medical Record
Electronic medical records (EMRs) are digital versions of a patient’s medical record. Medical records, whether electronic or on paper, contain the medical and clinical data collected in a provider’s office on a patient based on his or her visits to a health care provider. According to United States (US) Code of Federal Regulations (45 CFR § 160.103), health care providers are defined as any provider of health care as well as any other organization that furnishes, bills, or is paid for health care in the course of business. Health care providers collect patient-level data for several broad areas of the EMR (Table 1).
Table 1. Examples of patient level data collected during the course of clinical care and recorded in the electronic medical record.
| Demographic Data | Lists | Health History | Medical Encounter Data | Reports |
|---|---|---|---|---|
|
|
|
|
|
EMR data contain both unstructured and structured data. Unstructured data, such as the clinical free text, is not organized in a specific manner. In contrast, structured data use a controlled vocabulary rather than narrative text, setting limits on how the data are recorded in the clinic, resulting in consistency of that structured health record data within a health care provider. Structured EMR data also lend themselves to straightforward digital searchability across the EMR for specific information.
Electronic Health Record
Electronic health records (EHRs) are often described interchangeably with EMRs despite subtle and important differences. The term EHR is used to encompass a broader set of information than that collected in standard clinical care. Thus, the term “health” replaces “medical” to reflect this other information collected at the point of clinical care. Further, while EMRs contain and store clinical data from visits to a specific provider or clinic, EHRs are designed to include data outside the clinic or provider that originally collected and stored the data. These data can be linked across health insurance providers in addition to health care providers, including information on prescriptions filled. External sources of collected information can also be linked to health care provider EHRs, including health data from other clinicians involved in the care of the patient, patient-provided data on lifestyle and environmental exposures, and state-based data that track controlled substance prescriptions [prescription drug monitoring systems (PDMP)(Manasco, Griggs et al. 2016)]. Perhaps most importantly, data within EHRs are meant to move with the patient over the patient’s lifetime.
International Classification of Disease Codes
International Classification of Disease (ICD) codes are an important part of the structured data within EHRs. These codes provide important information about patients, and can include patient diagnoses, procedures, factors influencing health status, complications of procedures, and causes of morbidity and mortality. ICD codes have long been an important component of epidemiologic research (e.g., (Calle, Rodriguez et al. 2003)) and disease surveillance (e.g., (Lewis, Pavlin et al. 2002)), and their role is predicted to increase as more and more EHRs are deployed and expanded (Birkhead, Klompas et al. 2015). Also emerging from the widespread availability of EHRs is the prominent role of ICD codes play in new genomic discovery and precision medicine research. The two ICD versions available for computable phenotyping in most US EHRs are the ICD 9th Revision Clinical Modification (ICD-9-CM) and the 10th Revision clinical Modification (ICD-10-CM).
Common EHR algorithm performance metrics
As described in the Commentary section, the data within the EHR can be used to identify individuals who have specific phenotypes by using computational algorithms. The performance of the algorithm can be described using a variety of metrics, the uses of which depend on the goal of the algorithm. Common algorithm performance metrics include the calculation of positive predictive value (PPV) and negative predictive value (NPV). The PPV is the proportion of patients that have the disease identified by the algorithm and confirmed with manual chart review (true positives) among patients with the disease identified by the algorithm (true positives and false positives). The NPV is the proportion of patients that are classified by the algorithm as non-case or control and confirmed by manual chart review (true negatives) among all non-cases or controls identified by the algorithm (true negatives and false negatives). The fraction of algorithm-identified cases that are relevant and accurate is the precision whereas recall is sensitivity, the fraction of relevant cases identified by the algorithm. Precision and recall can be measured together through an F-measure (2 × [(precision × recall)/(precision + recall)]).
Commentary
History of EMRs and EHRs
In general in the US, the information and data contained within the medical record is owned by the patient, while the physical medical record is owned by the provider and the office or facility in which it was created. The majority of states have no specific legislation regarding ownership of medical records(Policy 2015), and the Health Insurance Portability and Accountability Act (HIPAA)(1996), which defines regulations for provider-collected patient data, does not define who has ownership. As of 2015, 20 states have legislation that declares providers or hospitals as owners of the medical records, and only one (New Hampshire) declares patients as owners(Policy 2015). Under HIPAA, these data are considered protected health information (PHI), and privacy of these data is maintained by the covered entities defined as health plans, health care clearinghouses, and health care providers, through data security measures and limited data sharing(Kayaalp 2017).
EMRs offer several advantages over paper medical records. They provide increased readability, accessibility, and accuracy compared with their paper counterparts. The centralized location of EMRs increases efficiency and speed when accessing information, decreases storage costs, and supports the sharing of these records within a health care institution that may be located in multiple geographic locations. For a health care provider that provides both primary and specialty care, the result of this centralized data repository for each patient is a centralized longitudinal data history of a patient across health and disease, which can theoretically help improve patient care and patient safety(Sittig and Singh 2012). EMRs have also made it easier to share health histories and status with individual patients, most often through web-based patient portals. In general, for research, the expansion of EMR data has made patient- and population-level analyses possible.
One of the earliest EHRs, the Veterans Health Information Systems and Technology Architecture (VistA), was developed first as an EMR by the US Department of Veterans Affairs (VA) in the 1970s(Brown, Lincoln et al. 2003, Allen 2017). VistA began as support for laboratory and pharmacy computing developed by VA software engineers in close collaboration with physicians as an open, modular, and decentralized system. This vendor-free, open-source system of independently developed software packages from different VA sites has since evolved as an EHR system that serves all VA sites nationwide under the umbrella of a graphical user interface (the Computerized Patient Record System or CPRS) that allows for the integration of the numerous existing programs contained within VistA(Brown, Lincoln et al. 2003, Fihn, Francis et al. 2014). The VistA/CPRS EHR currently boasts higher ratings for connectivity (across four domains: with diagnostic devices, practice management, reference and hospital labs, and for supporting referrals), usefulness as a clinical tool, and overall user satisfaction compared with vender-dependent systems(Murff and Kannry 2001, Gue 2016). The enhanced capabilities of EHRs was quickly realized soon after Hurricane Katrina in 2005 when all paper medical records in the city of New Orleans were lost while the VA EHRs were preserved and available for health care support of VA evacuees(Brown, Fischetti et al. 2007).
Despite the existence of VistA and other vender-independent and –dependent EHRs in the 1980s and 1990s, adoption of these paperless systems across the US was slow. In 2008, <10% of US non-federal acute care hospitals had a basic EHR, defined by ten required functions that include capabilities in electronic clinical information (patient demographics, physician notes, nursing assessments, problem lists, medication lists, and discharge summaries), computerized provider order entry (medications), and results management (view lab reports, view radiology reports, and view diagnostic test results)(Henry, Pylypchuk et al. 2016). By 2015, almost 84% of non-federal acute care hospitals reported having a basic EHR(Henry, Pylypchuk et al. 2016). The substantial expansion that occurred in EHR usage between 2008 and 2015 in US hospitals can be attributed to the Health Information Technology for Economic and Clinical Health (HITECH) Act of 2009(Adler-Milstein and Jha 2017), which as part of the American Recovery and Reinvestment Act (ARRA) provided financial incentives intended to accelerate the adoption of EHRs with the ultimate goal of improving healthcare. Among office-based physicians, EHR adoption has doubled since 2008 albeit at a slower pace compared with non-federal acute care hospitals(Mennemeyer, Menachemi et al. 2016, Technology 2016).
With the widespread adoption of EHRs is the anticipation that patient and heath care organization data will be analyzed in real-time, resulting in better clinical care and patient safety(King, Patel et al. 2014, Bae, Rask et al. 2018). In the US, EHR incentive Medicare and Medicaid payments are made when eligible professionals and hospitals demonstrate “meaningful use” of the health care organization’s EHR((CMS) and HHS 2010, Marcotte, Seidman et al. 2012), which is defined as broad objectives that “improve quality, safety, efficiency, and reduce health disparities; engage patients and family; improve care coordination, and population and public health; and maintain privacy and security of patient health information(HealthIt.gov 2015).” Attaining meaningful use requires that EHRs meet the broad objectives now further defined as specific tasks and capabilities, including electronic reporting of quality of care(Blumenthal and Tavenner 2010), in three stages of increasing benchmarks. Early data suggest these additional incentives and regulations would positively affect quality of care(2011), but early post-incentive analysis is inconclusive(Enriquez, de Lemos et al. 2015, Kern, Edwards et al. 2015). Nevertheless, it is ultimately anticipated that the adoption EHRs and meaningful use functions will spur the evolution of learning health care systems that link and leverage patient and population data to provide an up-to-date and comprehensive knowledge base that can be readily accessed at the point of clinical care for treatment and disease prevention(Medicine 2012, Flum, Alfonso-Cristancho et al. 2014, Kaggal, Elayavilli et al. 2016, Breitenstein, Liu et al. 2018).
Using EHR data for research
The linkage and leverage of patient and population data in EHRs can also be readily accessed for research purposes. Epidemiological cohort and cross-sectional studies, such as the Framingham Heart Study(Dawber, Meadors et al. 1951, Mahmood, Levy et al. 2014), the National Health and Nutrition Examination Surveys (NHANES)((CDC) and (NCHS) 2012), and the Women’s Health Initiative (WHI)(Group 1998), have been long-standing important repositories of health-related data for research. Collectively, these respective decades-long studies were major contributors to establishing hypertension, hypercholesterolemia, and smoking as risk factors for cardiovascular disease(Dawber, Moore et al. 1957, Kannel, Dawber et al. 1961), to providing data essential for pediatric growth charts(Kuczmarski, Ogden et al. 2000), and to establishing a link between post-menopausal hormone replacement therapy and cardiovascular disease and breast cancer risk(Rossouw, Anderson et al. 2002, Force, Grossman et al. 2017, Lewis and Wellons 2017), among other scientific accomplishments. While invaluable, gold-standard cohort study designs can be expensive and time consuming in the collection of data. Population-based cross-sectional studies are further hampered by the lack of longitudinal data on participants, thereby limiting the scientific questions that can be addressed by these data. These among other limitations have encouraged the use of existing EHR real-world clinical data for biomedical research(Kohane 2011). EHRs offer longitudinal data on health and treatment outcomes for millions of patients, and can be used both retrospectively, cross-sectionally, and prospectively, depending on the epidemiologic study design applied when the EHR-based research is conducted.
EHR-based research is not limited to traditional academic endeavors. Because EHR data are collected in real time, these data can be captured for in-house analysis and research in a database independent of clinical operations. These research databases are known as a research data warehouse, a clinical data warehouse, an integrated data warehouse, an integrated data repository, or an enterprise data warehouse(Chen and Sarkar 2014). Data warehouses can be used by individual health care systems to evaluate the efficiency and effectiveness of clinical services and treatments, which may help identify potential improvement opportunities for patient safety and satisfaction. For research purposes, data warehouses can be augmented by incorporating other data not relevant to the EHR but relevant for research, such as payer claims databases and disease registries(Chen and Sarkar 2014).
Despite the relative ease of access and potential for research, data in EHRs suffer from several immediate limitations. First and foremost, EHR data are collected for clinical care and billing purposes, not for biomedical research. These data are prone to biases and error(Hersh, Weiner et al. 2013). Consequently, extensive and sophisticated quality control, phenotyping, and extraction strategies are required for EHRs to be effectively used for research. Herein we describe basic strategies emerging from the field of biomedical informatics(Sarkar 2010) for computable phenotyping using EHR data as well as trends in the field with an emphasis on present and future precision medicine research efforts.
Data in electronic health records
In addition to the “home-grown” EHR varieties such as the VA’s VistA, there are hundreds of private developers offering EHRs(Technology 2017). Example developers or vendors include Cerner, Allscripts, and Epic Systems Corporation. Epic Systems Corporation to date is the most widely adopted EHR in the US participating in the incentive Medicare and Medicaid programs(Koppel and Lehmann 2015, Technology 2017). Although there is no single EHR format, EHRs regardless of vender store similar patient data, including medical history, laboratory test results, diagnostic test results, problem list, clinical notes, and treatment notes(Spooner and Pesaturo 2013). Both EHR format and content are driven, in part, by the fundamentals of the practice of medicine as well as medical record documentation requirements for reimbursement, resulting in somewhat consistent and predictable approaches that can be exploited in computable phenotyping.
International Classification of Disease Codes
The basis for disease coding is ancient. Some of the earliest documented examples of disease classification include Sri Lankan exorcism rituals that use sanni demon masks worn by sick persons that depict the group of disease with which the person is afflicted(Bailey and de Silva 2006). Other early examples include disease classifications documented from ancient Greece and Egypt. Within Europe, early 15th century death certificates and 18th century work in creating a classification system for diseases(Moriyama, Loy et al. 2011, Organization 2012) are often attributed as the basis for disease coding. Modern classification codes have direct roots to the development of the International List of Causes of Death, which was presented at the International Statistical Institute and adopted by the American Public Health Association in the late 19th century. The International List of Causes of Death, also known as the Bertillon Classification of Causes of Death, was regularly revised in ten year intervals starting in 1900, the first of which resulted in ICD-1(Moriyama, Loy et al. 2011, Organization 2012). As of the sixth revision (ICD-6), the World Health Organization (WHO) leads ICD revisions, a role that continues to this day.
Today’s ICD codes have evolved from local rudimentary surveillance systems of plague-induced deaths to a truly international standard for reporting disease and health conditions. These modern statistics, like their predecessors, are used in monitoring the prevalence and incidence of diseases (surveillance) which informs both population and patient safety. Unlike their predecessors, modern codes also provide statistics towards factors that influence health status and causes of death, both of which are important data repositories for research. Within the US, modern ICD codes are also a required standard for billing and clinical purposes, an administrative use of codes that also influences the quality of data available for research.
ICD-9 was developed in the 1970s and then further revised in the US by the National Center for Health Statistics (NCHS) within the Department of Health and Human Services to meet the needs of clinicians and payers such as the Centers for Medicare and Medicaid Services (CMS)(Topaz, Shafran-Topaz et al. 2013). ICD-9-CM has nearly 15,000 codes (3,824 procedure codes and 14,025 diagnostic codes) and was mandated for use in the US from 1979 until 1999 for death certificates and until 2015 for medical records and eventually EHRs. ICD-9-CM was also a major element for defining diagnosis-related groups (DRGs), an inpatient hospital coding system developed in the early 1980s and adopted by CMS in 1983 to curb US Medicare spending through a prospective payment system (PPS)(Mayes 2006).
ICD-10, the 10th version of ICD, was released by WHO in 1990, and many countries apart from the US transitioned to these codes in the late 1990s. Transition to ICD-10 in the US was delayed until 2015 due to the extensive development and adoptions of clinical modifications(Chute, Huff et al. 2012, Topaz, Shafran-Topaz et al. 2013). The number of codes ballooned from nearly 15,000 codes in ICD-9-CM to 71,924 procedure and 69,823 diagnosis codes in ICD-10-CM(Statistics 2015). Furthermore, ICD-10 coding in the US is now split into “clinical modification” and “procedural classification system” (PCS), the latter of which was developed by CMS and is to be used only for inpatient settings. As of October 1, 2015, CMS requires ICD-10-CM/PCS codes for medical diagnoses and inpatient hospital procedures.
Regardless of version, the anatomy of modern ICD codes is uniform and somewhat similar across 9 and 10 code sets. Both consist of a string of characters, ranging from 3–5 (ICD-9-CM) or 3–7 (ICD-10-CM). For ICD-9-CM codes, the first character is numeric or alpha while characters 2–5 are numeric. In contract, the first ICD-10-CM code character is alpha, character 2 is numeric, and the remaining characters can be alpha or numeric(Statistics 2015). The expansion of permissible characters in ICD-10-CM allows coders to include information about laterality (position 6) and stage of disease (position 7), as shown in Figure 1. This expansion increases the granularity of the data, useful for research among other purposes.
Figure 1. Anatomy of International Classification of Diseases (ICD) codes.
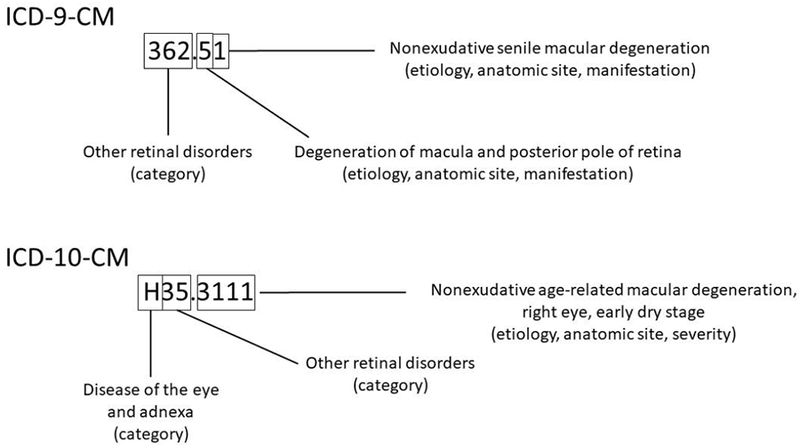
ICD-9-CM and ICD-10-CM codes are given for dry (nonexudative) age-related macular degeneration. Note that the expansion of characters in ICD-10-CM codes (3–7) compared with ICD-9-CM codes (3–5) allows for both laterality (6th position) and staging (7th position). 362.51 (362.50 is “macular degeneration (senile), unspecified”; 362.51 is “dry”). H35.3111 (Nonexudative age-related macular degeneration, dry age-related; right eye, early dry stage).
One will note that ICD-10-CM contains three times the number of codes of ICD-9-CM. While the two versions of ICD are structured as illustrated in Figure 1, there is not a one-to-many connection from ICD-9-CM to ICD-10-CM. For EHRs with a mix of legacy ICD-9-CM codes and newer ICD-10-CM codes, shifting from one code in ICD-9 to a code or codes in ICD-10, or vice-versa, became an immediate challenge. CMS and NCHS have made General Equivalence Mappings (GEMSs) to assist in mapping between the two versions. GEMS links any code in question to all valid alternatives in the other coding system, thus providing forward and backward mappings. Despite the existence of GEMs and its updates, bridging relevant codes across the two mappings has remained a challenge, prompting alternative mapping proposals such as a network of mappings instead of awkward one-to-one mappings(Boyd, Li et al. 2013).
The earliest uses of ICD codes in genomic studies used codes alone or in combination with laboratory values and medications to define cases and controls for genetic association studies to replicate known genotype-phenotype associations from published candidate gene and genome-wide association studies(Wood, Still et al. 2008, Ritchie, Denny et al. 2010). ICD codes have since been used extensively for phenotyping in genome-wide association studies(Crawford, Crosslin et al. 2014, Hoffmann, Keats et al. 2016, Dumitrescu, Ritchie et al. 2017), fine-mapping studies(Restrepo, Farber-Eger et al. 2015), whole exome sequencing studies(Abul-Husn, Manickam et al. 2016, Dewey, Murray et al. 2016), and phenome-wide association studies (PheWAS)(Bush, Oetjens et al. 2016). ICD codes are also being used to extract data on environmental exposures and lifestyle/behavioral variables for gene-environment and genetic association studies(Wiley, Shah et al. 2013).
Later, we will describe EHR-based phenotyping that outlines the use of data contained within the EHR, including ICD codes, to define a phenotype of interest (Phenotype Algorithm Development). For now, it is important to note that ICD codes can be used alone or in combination with other ICD codes and EHR data in the development of algorithms for computable phenotyping. While there are numerous examples of high-quality phenotypes derived from ICD codes, it is always important to consider that these data, and the entirety of EHRs, were collected for clinical purposes, including billing, and not for research purposes. Numerous studies have highlighted errors in billing codes that, while stemming from diverse sources or causes(O’Malley, Cook et al. 2005), adversely impact research and other secondary uses of codes. These limitations inform the strategies used in electronic phenotyping as detailed below.
Medication data
Data on medications is an essential part of the clinical narrative and a required element of an EHR qualifying for meaningful use. These required data provide multiple research opportunities, notably in pharmacogenomics and precision medicine research. It is well known that clinical trials are limited in sample size and, with few exceptions (e.g., (Group, Link et al. 2008)), are typically too small for powerful genetic association studies of rare outcomes such as adverse drug events. Clinical trials are also frequently limited in ethnic and racial diversity, thereby limiting the genetic ancestry represented in the trials(National Academies of Sciences 2016). The number of patients that can be monitored for a much longer time within an EHR is far more expansive, providing deep data on larger number of patients compared with clinical trials. And, depending on the health care provider, EHRs may have with a greater range of genetic ancestry within the patient population being served. Further, patients using health care providers may be on a multiple drugs and a range of drugs, and that information can provide important knowledge for drug-drug interactions(Rinner, Grossmann et al. 2015).
Studies accessing EHR medication data will often require medication name, dose, frequency, and duration data, and would likely be more successful with information on how consistently patients are taking medications. EHRs can vary on how medication data are collected, and how and when a medication list is updated for a given patient, leading to errors in information on what drugs are being taken and at what doses, and what prescriptions have been discontinued. The vast majority of EHRs are not linked to pharmacy dispensing information systems(Keller, Kelling et al. 2015) which provide information on prescriptions filled. Consequently, extraction of seemingly straightforward medication data is remarkably complex, as the required information is buried within the medication lists, medication orders, and even clinical narrative (the “free text” of the EHR) which altogether offer varying levels of completeness, uniformity, and availability(Laper, Restrepo et al. 2016).
Standardized medication name, dosage, and route information, is arguably the first challenge to address in conducting EHR-based studies involving medication use. The same medication, defined by identical formulation, can be associated with several brand names and generic names, resulting in multiple terms and text descriptions that convey the same information. To help remedy this known problem, the National Library of Medicine (NLM) developed RxNorm, a standardize nomenclature for clinical drugs. RxNorm provides RxNorm Concept Unique Identifiers (RxCUIs) that are linked to normalized descriptions of each drug from various sources(Medicine 2004). Developed in 2002 by NLM as one of the terminologies in the Unified Medical Language System (UMLS), RxNorm was built upon existing drug information used in pharmacy management and drug interaction software. RxNorm was released as an independent terminology in 2004 and is currently updated weekly and monthly(Nelson, Zeng et al. 2011). RxNorm provides normalized information for generic and branded medications, as well as drug packs that contain multiple drugs that need to be consumed in a specific sequence. This normalization is critical within the health care setting for efficient and unambiguous communication of drug information across multiple computer systems. RxCUIs are linked to information including the full name of a drug, the ingredient(s), and dose form(Fung, McDonald et al. 2008) thereby providing a link to equivalent drugs that would have otherwise been considered unique drugs based on the provided descriptions. RxNorm and RxCUIs can be coupled with text mining tools such as MedEx(Xu, Stenner et al. 2010) to extract research-grade drug data.
Drug information organized through RxNorm is helpful and can be used to determine if the patient has likely had a drug exposure. However, these EHR-extracted data are still limited in usability for several reasons. Even when prescriptions are given to patients, determining if the patients are taking the drugs, at the expected dose, at prescribed intervals is not systematically recorded in the EHR. If health payer plan data are available and accessible to research, these additional data can be used to determine if prescriptions were filled. PDMPs are state-wide level systems that collect data on designated substances, both prescribed and over-the-counter. These systems are in place to allow access to controlled substances; to identify, deter, and prevent drug abuse and drug diversion; to identify individuals addicted to prescription drugs; and to provide information for states on drug usage. Not all states have these systems, and not all of these systems allow access for research purposes. Furthermore, the provided information only includes prescriptions filled and does not include over-the-counter drugs, supplements, or herbal remedies which may or may not be reported by patients and recorded in the EHR. Finally, these systems also do not include information on foods or beverages, such as red grapefruit, alcohol, and caffeine known to alter drug response and in some cases lead to adverse drug reactions.
In general, little information on patient drug response is available in the EHR unless there is a significant adverse event that results in the need for medical care. These missing data are a particular challenge for pharmacogenomics research, a field that focuses on identifying how the genetic architecture impacts the pharmacokinetics (absorption, distribution, metabolism, and excretion or ADME) and pharmacodynamics of the drug. Candidate gene and genome-wide associations studies have identified at least 20 genes whose variants affect 80 medications(Whirl-Carrillo, McDonagh et al. 2012), resulting in changes to Food and Drug Administration (FDA) labeling and, in some cases, clinical care(Relling and Evans 2015, Relling, Krauss et al. 2017). Identifying cases of adverse events for pharmacogenomics research accessing EHRs is not straightforward as information about an adverse event may be only recorded in detail in the clinical free-text beyond billing codes related to treatment a patient had for an adverse event (e.g., (Wiley, Moretz et al. 2015)). If the side effect or adverse event was mild, it may never be reported by the patient and yet may be a reason the patient stops taking a drug without informing their physician. Mild events may also result in medication switching without detailed documentation in the clinical free text, a strategy used to identify potential cases of statin intolerance or statin-induce myopathy(Zhang, Plutzky et al. 2013, Preiss and Sattar 2015). Relevant drug dosing data can be obtained from the EHR using a variety of approaches including extracting dosing data from the medications list, extracting dosing data from clinical free text including free text from specialty clinics (e.g., warfarin clinics) using MedEx(Xu, Stenner et al. 2010) or similar extraction tools, and accessing intra-operative data such as electronic anesthesia records(Levin, Joseph et al. 2017).
Clinical Laboratory Measures and Vital Signs
Both clinical laboratory measures and vital signs are used to assess the current health of the patient as well as to screen for preventable or emerging conditions that require intervention. These data are also important quantitative traits or outcomes for genomic discovery studies(Crawford, Crosslin et al. 2014, Verma, Leader et al. 2016, Verma, Lucas et al. 2017). Examples of clinical laboratory measures include glucose, hemoglobin A1c, lipids, and blood cell counts. Vital signs include systolic and diastolic blood pressure, body temperature, respiration, pulse or heart rate, height, and weight. Secondary to ICD codes, laboratory measures and vital signs represent some of the most readily available structured data within the EHR.
While the extraction of clinical laboratory measures and vital signs from EHRs is relatively straightforward, their use and interpretation in research is not. First and foremost, patients are not uniformly screened for even a small subset of available clinical laboratory measures, and this introduces known and unknown biases into any extracted EHR data(Beaulieu-Jones, Lavage et al. 2018). Second, even though these data exist in EHR structured fields, a fraction represent transcription errors or errors in unit assignment (including missing units). As an example, height and weight are typically measured in meters and kilograms, respectively, and used to calculate body mass index (BMI), an important metric used to assess healthy versus unhealthy weight. In addition to representing a health condition (e.g., underweight, normal weight, overweight, obese, and morbidly obese), BMI is one of the most important covariates in association studies. This nearly ubiquitous variable in the EHR, however, requires data cleaning efforts that leverage repeated measures available in the EHR to overcome transcription errors and unit errors(Goodloe, Farber-Eger et al. 2017) as well as imputation for missing data (in this case, most likely height)(Bailey, Milov et al. 2013). These strategies may not be feasible for less common clinical laboratory measures(Beaulieu-Jones, Lavage et al. 2018).
Other data quality issues that are not unique to but can be exacerbated by EHRs and clinical practice are related to variability in the data collection process. While timing of the measurement is recorded, it is not uniform across patients. Also highly variable is the setting in which the measurement was taken, which is not well documented in the EHR. As an example, variability in blood pressure measurements based on timing and exposure to environmental stimuli, including that of patients seeing health care professionals (i.e., the white coat syndrome), is well known. For clinical laboratory measures, fasting status is not uniformly documented. For both clinical laboratory measures and vital signs, the clinical context in which they were collected may have an impact on the quality and usability of these data depending on the research questions or objectives. That is, inpatient data and data collected during surgery, trauma, or other acute injury may not be appropriate for the research question or objective compared with outpatient data. Depending on the EHR, these data may be indistinguishable and require data cleaning approaches such as classifying repeated measures within a specified time frame as “inpatient data” as opposed to repeated measures collected on different outpatient visits. Finally, EHRs do not thoroughly document all possible meta-data associated with clinical laboratory measures (such as laboratory assays or brands) and vital signs (such as the equipment), all of which can evolve over time with new technological developments.
In addition to some of the data cleaning approaches referenced above, data harmonization across EHRs for similar clinical laboratory measures with different labels can be facilitated by logical observation identifiers names and codes (LOINC). LOINC are unique numerical identifiers maintained by the Regenstrief Institute that distinguish relevant differences between laboratory measures(Forrey, McDonald et al. 1996, McDonald, Huff et al. 2003). The LOINC database currently has more than 71,000 different unique codes. Adoption of LOINC improves communication in integrated health systems and also facilitates research by providing a means to standardize seemingly disparate data collected across clinical laboratories.
Other structured data
Other structured data available within the EHR vary from health care provider to health care provider. Example of such data are those collected in optometry and ophthalmology specialty clinics(Peissig, Schwei et al. 2017). These specialty clinics offer additional structured data beyond ICD-9-CM/10-CM codes related to ocular disease and traits, and access to these data had already enabled genomic discovery for common and debilitating ocular diseases(Bauer, Cha et al. 2018). Other examples include modifications made to EHRs for smoking status and treatment either in response to specialty clinic requests or to comply with Meaningful Use regulations(Schindler-Ruwisch, Abroms et al. 2017). Additional modifications to EHRs are expected in the near future as many research groups recognize that important exposure, lifestyle, and behavioral variables (e.g., social determinants of health(Adler and Stead 2015, The National Academies of Sciences 2017)) are poorly represented in clinical notes.
Unstructured data
In addition to the structured data described above, the EHR contains semi-structured and unstructured data. These data include any EHR data that do not conform to a pre-defined model or organizational structure. An example of semi-structured data is the problems list, which is populated by the health care provider without constraints, leading to variable representation of similar concepts. An example of unstructured data is the “free text” of clinical notes and reports. The kinds of data that are semi-structured or unstructured can vary from health care provider to health care provider.
Extracting structured data from unstructured text poses several challenges. Manual extraction of key concepts is considered “gold-standard” compared with automated extraction approaches; however, this strategy is not scalable for routine or real-time monitoring of patients(Kaggal, Elayavilli et al. 2016) and for the purposes of large-scale research. A basic text-mining strategy designed to search for and extract key words is relatively straightforward to develop and implement, but this approach is limited(Farber-Eger, Goodloe et al. 2017). For example, when tasked with searching and extracting mentions of type 2 diabetes from the clinical notes, the strategy should take into account the many words, acronyms, and abbreviations that represent this condition as well as include common misspellings (e.g., T2D, type 2 diabetes, diabetes, type II diabetes, diabetes mellitus). Simple key word searches also do not account for sentence context, including negation (e.g., “does not have type 2 diabetes”) and hedge phrases(Hanauer, Liu et al. 2012) (e.g., “possible type 2 diabetes”). In general, unstructured data are highly heterogeneous and do not strictly conform to punctuation or grammar rules.
The access of these unstructured data while addressing the scalability limitations of manual chart review as well as the data quality limitations of simple text searches usually requires some form of natural language processing (NLP). Broadly defined, NLP is an approach that extracts structured natural language data from unstructured text in a high-throughput manner(Ohno-Machado 2011) using rule-based and/or probabilistic methods(Joshi 1991, Hirschberg and Manning 2015, Wong, Plasek et al. 2018). NLP tasks can include sentence boundary detection, tokenization (using punctuation and spaces to split text roughly into words), part-of-speech assignment to individual words, morphological decomposition of compound words, shallow parsing or chunking of phrases, and problem-specific segmentation(Jensen, Jensen et al. 2012). Higher level NLP tasks can include spelling and/or grammatical error identification and recovery as well as named entity recognition and concept mapping(Nadkarni, Ohno-Machado et al. 2011). These NLP tasks can be coupled with mapping tasks such as mapping disease concepts to the Systemized Nomenclature of Medicine – Clinical Terms (SNOMED CT) or to another Unified Medical Language System(Jensen, Jensen et al. 2012).
Many open source and proprietary tools have been developed or modified recently that implement NLP and applied to clinical free text(Nadkarni, Ohno-Machado et al. 2011, Ohno-Machado 2011). Common open source tools include the Apache clinical Text Analysis and Knowledge Extraction System (cTAKES)(Savova, Masanz et al. 2010), the Health information Text Extraction system (HiTEx)(Goryachev, Sordo et al. 2006), Medical Language Extraction and Encoding System (MedLEE), and MetaMap(Aronson and Lang 2010). Also popular are indexing information retrieval approaches like that performed by the Electronic Medical Record Search Engine (EMERSE)(Hanauer, Mei et al. 2015). Although not as sophisticated as some NLP approaches, medical indexing can be a fast, intuitive, and inexpensive option for searching free-text clinical notes.
Portable document format files
Apart from free-text clinical notes and problems lists, semi-structured and unstructured data can also theoretically be extracted from portable document format (PDF) files. PDFs available in the EHR are of two types: 1) scanned and 2) native. A scanned PDF is a document produced outside the EHR, such as a faxed report, that is then electronically captured as an image. Searchable PDFs are native files that contain text.
Although native PDF files are preferred, EHRs can have considerable legacy collections of scanned PDF files. A scanned PDF file is not easily searchable using automated methods such as NLP or indexing described above because the file does not contain text. Optical character recognition (OCR) can be applied to convert images of typed or hand-written text into machine-encoded text; however, this approach can require extensive image training data. Alternatively, data within PDF files can be manually extracted and entered into the EHR as structured or semi-structured data. The former approach is not yet widely used(Rasmussen, Peissig et al. 2012) and the latter approach is not easily scalable.
In contrast to scanned PDF files, text from a native PDF file can be retrieved using automated approaches such as NLP. The challenge here is that native PDF files contain header, footer, and other meta data, as well as tables or figures, all of which can present a challenge for NLP approaches designed to extract concepts from clinical narratives. There are open source and commercial solutions available that extract raw text from these files such as the freely available PDFBox Tool from Apache(Apache). The raw text files can then be further processed using NLP, machine learning, or other approaches(Bui, Del Fiol et al. 2016, Hassanpour and Langlotz 2016).
Imaging data
Imaging data are regularly captured within health care systems and are ordered as recommended screenings (e.g., mammography) or diagnostics (e.g., kidney stones). Common imaging data available in the EHR include x-rays, computerized tomography (CT) scans, magnetic resonance imaging (MRI) scans, ultrasounds, echocardiography, positron emission tomography (PET), medical photography, and endoscopy, to name a few. Although not strictly considered medical imaging, tests that do not produce images such as electrocardiograms (ECGs) are visually represented as the measured parameter versus time or as maps.
There is much interest in automating data extraction from all of these images for research as these data often contain quantitative or additional measures related to specific diagnoses that can meaningfully augment structured diagnostic code data. Also, images can be used for research beyond why the images were collected. As an example, CT scans ordered to visualize kidney stones for diagnostic purposes can be used to obtain measures of visceral and subcutaneous adipose tissue for research purposes(Cha, Veturi et al. 2018).
Future trends in EHR data collection
While the EHR already has a variety of data useful for research, there are other sources of data that can improve the use of EHR data for research, as well as contribute to improving learning health care systems and contributing to precision medicine. These data can include patient-reported data through structured digital surveys that can be used to augment lifestyle, behavioral, and family history data often missing from the EHR(Murray, Giovanni et al. 2013, McClung, Ptomey et al. 2018) or to provide key information on efficacy of treatment, such as pain management and asthma control. Health systems now have patient portals that can be used by patients for appointment scheduling, messaging providers, and maintaining prescription lists. Many patient portals also provide limited views of the EHR to the patient. Increasingly, patient portals are being recognized as a convenient mechanism to acquire additional patient data through structured digital surveys as well as through emerging wearable technologies(Shameer, Badgeley et al. 2017). Note that not all patients have access to the internet or are digitally inclined(Graetz, Gordon et al. 2016, Perzynski, Roach et al. 2017), resulting in persistent and biased missing health-related data.
Other EHR data collection trends attempt to address poor documentation of social determinants of health such as socioeconomic status(Hollister, Restrepo et al. 2016, The National Academies of Sciences 2017). Zipcodes, census blocks, and geocoded addresses can be linked to various public repositories of community-level environmental data such as walkability maps, air pollution monitors, food desert maps(King and Clarke 2015, Pike, Trapl et al. 2017, Xie, Greenblatt et al. 2017). These Geographic Information Systems (GIS) have the potential to help identify patterns or associations between EHR conditions or disease status and exposures related to the physical, built, and social environments of the patients(Casey, Schwartz et al. 2016, Bush, Crawford et al. 2018).
Phenotype Algorithm Development
Many strategies and approaches have been developed to extract research-grade data from the EHR data described above for a variety of study designs, including genetic association studies. Here we describe the two most commonly applied approaches: high-throughput approaches accessing only structured data and more time-intensive rule-based approaches using a combination of structured, semi-structured, and unstructured data.
High-throughput phenotype algorithm development
A key goal of a high-throughput phenotype algorithm is the rapid classification of case or disease status across the entire data warehouse with little or no curation. Most high-throughput approaches access only structured data such as ICD-9-CM/ICD-10-CM billing codes for rapid case-status phenotyping. An emerging study design known as the phenome-wide association study (PheWAS) employs this approach when applied to EHRs(Bush, Oetjens et al. 2016). Case-status is determined using presence of billing codes, and this can be achieved using individual billing codes or applying pre-specified code groups sometimes known as phecodes. Once case-status is determined, tests of association can be performed between case/non-case status and genetic variants of interest. Since the first EHR-based PheWAS(Denny 2010), approaches to rapidly determine case-status have widened in scope to include other structured data such as clinical laboratory measures(Verma, Leader et al. 2016).
There are many limitations to rapid phenotyping approaches that only access structured data. For phenotyping that only considers billing codes, a major challenge is that presence of ICD-9-CM/ICD-10-CM codes may not accurately represent the disease status of the patient. In addition to diagnosis, billing codes can be used to indicate a potential diagnosis to explain the reason for an additional clinical test, visit, or procedure. For difficult-to-diagnose conditions, the presence of billing codes over the course of multiple clinic visits may be reflecting a diagnostic odyssey with multiple misdiagnoses before a final diagnosis is achieved. It is also important to note that absence of a billing code may not reflect absence of the disease. That is, EHR diagnosis data only exist if the individual patient is seen by a provider for that chief complaint. For example, the patient may have primary open-angle glaucoma (POAG), but does not have diagnosis codes for this disease as he or she is being seen by an allergist. Until the patient visits the ophthalmologist, POAG tests and diagnosis will not be added to this patient’s EHR. In addition, criteria for disease change over time, such as the criteria for preeclampsia as well as criteria for excessive gestational weight gain, and these changing criteria will ultimately change who within an EHR are considered to have, or not have, a given condition.
Given that patients with a billing code may not truly be a case, and those without a diagnosis code may not actually be a non-case, billing codes are often viewed as a source of sub-par phenotype data. To overcome these well-known short-comings, a standard approach in phenotyping using billing codes only is to increase the required number of code mentions across separate clinic visit dates(Leader, Pendergrass et al. 2015, Dumitrescu, Diggins et al. 2016). In this approach, single mentions of a code are not considered when classifying a patient as a case. Beyond stricter code mention requirements, previously-curated code groupings can be applied to determine case and non-case status.
Low-throughput phenotype algorithm development
Development of rule-based phenotype algorithms that access EHR data beyond straight-forward structured data requires substantial resources and time. A typical workflow includes the establishment of a team of physicians knowledgeable of the disease in question and informaticians knowledgeable of the EHR data architecture. Team discussions then identify the EHR data necessary to classify cases of the disease or condition and non-cases or controls free of the condition. Team discussions also clarify clinical patterns and practices that may be relevant, such as what and when laboratory test orders should be expected in the course of a patient’s diagnosis. After all the EHR data required for case and non-case status are identified, the team drafts a rule-based algorithm and sequential flow chart (Figure 2) where at each step of the chart, a patient’s EHR data are evaluated for the required criteria (e.g., presence of two mentions of the desired billing code) before moving to the next step of the flow chart. The flow chart will eventually end as “case”, “non-case” or “control”, or “excluded” as the final decision or designation. This draft algorithm is then applied to the EHR data warehouse or a subset of the data warehouse, and its performance is evaluated through limited manual chart reviews. If the performance is not acceptable, the algorithm is revised for another round of manual review. This process is repeated until the desired performance is achieved. In some cases, algorithm performance cannot be improved with further revisions, highlighting a major limitation of using clinically-collected data for research purposes.
Figure 2. Example rule-based algorithm flow chart used to assign case status of a generic disease.
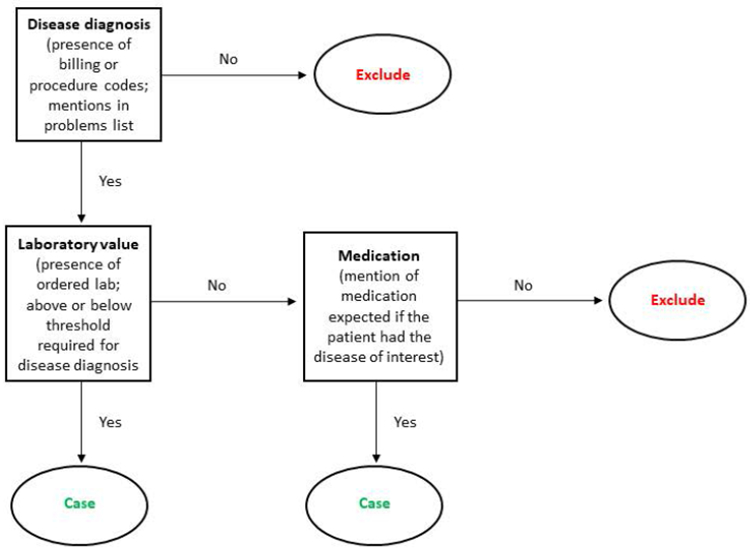
In this simple rule-based example, disease status is first considered using presence or absence of structured data such as ICD-9-CM/ICD-10-CM codes, procedure codes, or mentions of the disease in the problems list. If yes, the algorithm then requires a corroborating laboratory measure and threshold value associated with the disease or condition of interest. True cases of the disease may have the required code or problems list mention but might be missing the laboratory data. In these cases, the algorithm then asks if the patient with the required code or problems list mention has an EHR mention of a mediation associated with the disease or condition of interest. Rule-based algorithms in practice are more complex than shown here and can incorporate a combination of codes, temporal relationships between diagnosis, laboratory tests, or imaging and medication mentions, and natural language processing techniques to search the clinical free text for evidence of case status.
Measuring Algorithm Performance
PPV and NVP are dependent on prevalence of the disease in the population. Alternatively, sensitivity (true positive rate) and specificity (true negative rate) can be calculated by applying the algorithm to already curated or gold-standard data. For algorithms that employ NLP to extract research-grade variables from unstructured EHR data, common metrics include precision, recall, and F-measures. Finally, for EHRs linked to genome-wide data, known genotype-phenotype relationships can be used to assess algorithm performance (e.g., (Ritchie, Denny et al. 2010)) or to help calibrate algorithms(Halladay, Hadi et al. 2018).
The development of a high quality rule-based phenotype algorithm is considered low-throughput as the process can take months to years to complete. Given that substantial resources are expended for each algorithm, consortia such as the electronic Medical Records and Genomics (eMERGE) network(McCarty, Chisholm et al. 2011) are providing the algorithms, including any associated documents or pseudocode, to the larger scientific community through the Phenotype KnowledgeBase (PheKB)(Kirby, Speltz et al. 2016). Within the eMERGE network, algorithms are developed at one health care system and deployed and evaluated at other study sites representing other health care systems. Many algorithms have generalized well across different health care systems(Kirby, Speltz et al. 2016). For algorithms that do not perform well when applied to an EHR from a different health care system, modifications may be necessary to account for that health care system’s unique characteristics or clinical culture.
Future trends in electronic phenotyping
Most published electronic phenotype algorithms are rule-based, and none take full advantage of the data available in the EHR(Shivade, Raghavan et al. 2014). While the majority of phenotype algorithms have focused on the use of supervised methods, a trend in electronic phenotyping is the development of semi-supervised or unsupervised methods(Beaulieu-Jones and Greene 2016). Supervised methods as described above depend on expert knowledge that is then applied when classifying each patient as case, non-case/control, and/or excluded. In contrast, unsupervised methods use the patterns across the data of the EHR to identify patient groupings. As an example, a machine learning approach has been developed to identify new phenotype subgroups among patients with type 2 diabetes(Li, Cheng et al. 2015, Beaulieu-Jones 2017). Another approach has been to use multiple algorithmic methods to bring together multiple clinical lab measures(Bauer, Lavage et al. 2017). Even among supervised methods, the trend is increase speed and accuracy of electronic phenotyping by leveraging common data models and noisy labeled training data(Peissig, Santos Costa et al. 2014, Banda, Halpern et al. 2017).
A major trend in basic EHR infrastructure is the emphasis on interoperability, which has tremendous potential in ensuring that phenotype algorithms are portable across health care systems. With an estimated 1,100 EHR vendors as of 2017, interoperability is not yet common(Sittig and Wright 2015). In lieu of truly interoperable systems, administrators and investigators have adopted common data models, an approach that standardizes EHR and claims data that then can be easily shared with other health care systems that adopted the same or similar common data model. Several common data models exist, including the Observational Health Data Sciences and Informatics (OHDSI)’s Observational Medical Outcomes Partnership (OMOP)(OHDSI) and the National Patient-Centered Clinical Research Network (PCORnet)(PCORNet). It is important to note that the remapping of the data from the native EHR data structures to a common data model and a research data warehouse is a complex process. However, once implemented updating the data warehouse is an automated process requiring only monitoring and occasional updating. Analogous to epidemiologic variable harmonization efforts such as the consensus measures for Phentoypes and eXposures (PhenX) Toolkit(Hendershot, Pan et al. 2015), common data models have the potential to enable electronic phenotype necessary for downstream research.
Conclusions
Today’s ubiquity of EHRs makes population-scale research possible for clinical variables and clinical contexts that cannot be completely recapitulated or represented by controlled trials or epidemiologic studies. The major bottleneck in effective and efficient use of these data is the accurate extraction of research-grade variables, including case status. Electronic or computable phenotyping methods and approaches along with associated evaluation metrics are now emerging, the most common of which are described here. More sophisticated approaches are being developed, ensuring in part that the full potential of the EHR for precision medicine research, including genomic discovery, will be achieved as many envision.
Acknowledgements
This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland, UL1TR002548 and UL1 TR000439 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. This work was also supported by 1R01GM126249 and institutional funds. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- (ADME)
Absorption, distribution, metabolism, and excretion
- (ARRA)
American Recovery and Reinvestment Act
- (BMI)
Body mass index
- (CMS)
Centers for Medicare and Medicaid Services
- (CM)
Clinical Modification
- (cTAKES)
clinical Text Analysis and Knowledge Extraction System
- (CFR)
Code of Federal Regulations
- (CPRS)
Computerized Patient Record System
- (CT)
Computerized tomography
- (CUI)
Concept unique identifier
- (DRGs)
Diagnosis-related groups
- (ECGs)
Electrocardiograms
- (EHR)
Electronic health record
- (EMR)
Electronic medical record
- (eMERGE)
electronic Medical Records and Genomics
- (EMERSE)
Electronic Medical Record Search Engine
- (FDA)
Food and Drug Administration
- (GIS)
Geographic Information Systems
- (GEMSs)
General Equivalence Mappings
- (HiTEx)
Health information Text Extraction system
- (HIPAA)
Health Insurance Portability and Accountability Act
- (HITECH)
Health Information Technology for Economic and Clinical Health
- (ICD)
International Classification of Disease
- (LOINC)
Logical observation identifiers names and codes
- (MRI)
Magnetic resonance imaging
- (MedLEE)
Medical Language Extraction and Encoding System
- (NCHS)
National Center for Health Statistics
- (NHANES)
National Health and Nutrition Examination Surveys
- (NLM)
National Library of Medicine
- (NLP)
Natural language processing
- (NVP)
Negative predictive value
- (OHDSI)
Observational Health Data Sciences and Informatics
- (OMOP)
Observational Medical Outcomes Partnership
- (OCR)
Optical character recognition
- (PCORnet)
Patient-Centered Clinical Research Network
- (PheWAS)
Phenome-wide association study
- (PhenX)
Phentoypes and eXposures
- (PheKB)
Phenotype KnowledgeBase
- (PDF)
Portable document format
- (PPV)
Positive predictive value
- (PET)
Positron emission tomography
- (PDMP)
Prescription drug monitoring systems
- (POAG)
Primary open-angle glaucoma
- (PCS)
Procedural classification system
- (PPS)
Prospective payment system
- (PHI)
Protected health information
- (SNOMED CT)
Systemized Nomenclature of Medicine – Clinical Terms
- (UMLS)
Unified Medical Language System
- (US)
United States
- (VA)
Veterans Affairs
- (VistA)
Veterans Health Information Systems and Technology Architecture
- (WHI)
Women’s Health Initiative
- (WHO)
World Health Organization
Literature Cited
- (1996). Health Insurance Portability and Accountability Act (HIPPA). Public Law 104–191. t. Congress Public Law 104.-.
- (2011). “The Benefits Of Health Information Technology: A Review Of The Recent Literature Shows Predominantly Positive Results.” Health Affairs 30(3): 464–471. [DOI] [PubMed] [Google Scholar]
- (CDC), C. f. D. C. a. P. and N. C. f. H. S. (NCHS) (2012) National Health and Nutrition Examination Surveys (NHANES).
- (CMS), C. f. M. M. S. and HHS (2010). “Medicare and Medicaid pograms; electronic health record incentive program. Final rule.” Fed Regist 75(144): 44313–44588. [PubMed] [Google Scholar]
- Abul-Husn NS, et al. (2016). “Genetic identification of familial hypercholesterolemia within a single U.S. health care system.” Science 354(6319). [DOI] [PubMed] [Google Scholar]
- Adler-Milstein J and Jha AK (2017). “HITECH Act Drove Large Gains In Hospital Electronic Health Record Adoption.” Health Affairs 36(8): 1416–1422. [DOI] [PubMed] [Google Scholar]
- Adler NE and Stead WW (2015). “Patients in Context — EHR Capture of Social and Behavioral Determinants of Health.” New England Journal of Medicine 372(8): 698–701. [DOI] [PubMed] [Google Scholar]
- Allen A (2017). A 40-year ‘conspiracy” at the VA. Politico. [Google Scholar]
- Apache. “Apache PDFBox -- A Java PDF Library.” Retrieved 09/20/2018, from https://pdfbox.apache.org/.
- Aronson AR and Lang FM (2010). “An overview of MetaMap: historical perspective and recent advances.” J Am Med Inform Assoc 17(3): 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J, et al. (2018). “The Impact of Electronic Medical Records on Hospital-Acquired Adverse Safety Events: Differential Effects Between Single-Source and Multiple-Source Systems.” American Journal of Medical Quality 33(1): 72–80. [DOI] [PubMed] [Google Scholar]
- Bailey LC, et al. (2013). “Multi-Institutional Sharing of Electronic Health Record Data to Assess Childhood Obesity.” PLoS ONE 8(6): e66192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MS and de Silva HJ (2006). “Sri Lankan sanni masks: an ancient classification of disease.” BMJ 333(7582): 1327–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda JM, et al. (2017). “Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHSDI) data network.” AMIA Jt Summits Transl Sci Proc 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- Bauer CR, et al. (2018). Electronic health records elucidate the relationships between genetics, the anatomy of the eye, and disease. [Google Scholar]
- Bauer CR, et al. (2017). “Opening the door to the large scale use of clinical lab measures for association testing: exploring different methods for defining phenotypes.” Pac Symp Biocomput 22: 356–367. [DOI] [PubMed] [Google Scholar]
- Beaulieu-Jones B (2017). “Machine learning for structured clinical data.” [Google Scholar]
- Beaulieu-Jones BK and Greene CS (2016). “Semi-supervised learning of the electronic health record for phenotype stratification.” Journal of Biomedical Informatics 64: 168–178. [DOI] [PubMed] [Google Scholar]
- Beaulieu-Jones BK, et al. (2018). “Characterizing and managing missing structured data in electronic health records: data analysis.” JMIR Med Inform 6(1): e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead GS, et al. (2015). “Uses of Electronic Health Records for Public Health Surveillance to Advance Public Health.” Annual Review of Public Health 36(1): 345–359. [DOI] [PubMed] [Google Scholar]
- Blumenthal D and Tavenner M (2010). “The “Meaningful Use” Regulation for Electronic Health Records.” New England Journal of Medicine 363(6): 501–504. [DOI] [PubMed] [Google Scholar]
- Boyd AD, et al. (2013). “The discriminatory cost of ICD-10-CM transition between clinical specialties: metrics, case study, and mitigating tools.” Journal of the American Medical Informatics Association 20(4): 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenstein MK, et al. (2018). “Electronic Health Record Phenotypes for Precision Medicine: Perspectives and Caveats From Treatment of Breast Cancer at a Single Institution.” Clinical and Translational Science 11(1): 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SH, et al. (2007). “Use of Electronic Health Records in Disaster Response: The Experience of Department of Veterans Affairs After Hurricane Katrina.” American Journal of Public Health 97(Supplement_1): S136–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SH, et al. (2003). “VistA—U.S. Department of Veterans Affairs national-scale HIS.” International Journal of Medical Informatics 69(2): 135–156. [DOI] [PubMed] [Google Scholar]
- Bui DDA, et al. (2016). “PDF text classification to leverage information extraction from publication reports.” Journal of Biomedical Informatics 61: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, et al. (2018). “Integrating community-level data resources for precision medicine research.” Pac Symp Biocomput 23: 618–622. [PubMed] [Google Scholar]
- Bush WS, et al. (2016). “Unravelling the human genome-phenome relationship using phenome-wide association studies.” Nat Rev Genet 17(3): 129–145. [DOI] [PubMed] [Google Scholar]
- Calle EE, et al. (2003). “Overweight, Obesity, and Mortality from Cancer in a Prospectively Studied Cohort of U.S. Adults.” New England Journal of Medicine 348(17): 1625–1638. [DOI] [PubMed] [Google Scholar]
- Casey JA, et al. (2016). “Using Electronic Health Records for Population Health Research: A Review of Methods and Applications.” Annual Review of Public Health 37(1): 61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha EDK, et al. (2018). “Using adipose measures from healht provider based imaging data for discovery.” Journal of Obesity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES and Sarkar IN (2014). Mining the Electronic Health Record for Disease Knowledge Biomedical Literature Mining. Kumar VD and Tipney HJ. New York, NY, Springer New York: 269–286. [DOI] [PubMed] [Google Scholar]
- Chute CG, et al. (2012). “There Are Important Reasons For Delaying Implementation Of The New ICD-10 Coding System.” Health Affairs 31(4): 836–842. [DOI] [PubMed] [Google Scholar]
- Crawford DC, et al. (2014). “eMERGEing progress in genomics---the first seven years.” Frontiers in Genetics 5: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, et al. (1951). “Epidemiological approaches to heart disease: the Framingham Study.” Am J Public Health Nations Health 41(3): 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, et al. (1957). “Coronary heart disease in the Framingham study.” Am J Public Health Nations Health 47(4 Pt 2): 4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC (2010). “PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations.” Bioinformatics 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, et al. (2016). “Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study.” Science 354(6319). [DOI] [PubMed] [Google Scholar]
- Dumitrescu L, et al. (2016). “Testing population-specific quantitative trait associations for clinical outcome relevance in a biorepository linked to electronic health records: LPA and myocardial infarction in African Americans.” Pac Symp Biocomput 21: 96–107. [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu L, et al. (2017). “Genome-wide study of resistant hypertension identified from electronic health records.” PLoS ONE 12(2): e0171745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez JR, et al. (2015). “Modest Associations Between Electronic Health Record Use and Acute Myocardial Infarction Quality of Care and Outcomes.” Results From the National Cardiovascular Data Registry 8(6): 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber-Eger E, et al. (2017). “Extracting country of origin from electronic health records for gene-envrionment studies as part of the Epidemiologic Architecture for Genes Linked to Environment“AMIA Jt Summits Transl Sci Proc. [PMC free article] [PubMed] [Google Scholar]
- Fihn SD, et al. (2014). “Insights From Advanced Analytics At The Veterans Health Administration.” Health Affairs 33(7): 1203–1211. [DOI] [PubMed] [Google Scholar]
- Flum DR, et al. (2014). “Implementation of a “real-world” learning health care system: Washington state’s Comparative Effectiveness Research Translation Network (CERTAIN).” Surgery 155(5): 860–866. [DOI] [PubMed] [Google Scholar]
- Force UPST, et al. (2017). “Hormone therapy for the primary prevention of chronic conditions in postmenopausal women: US Preventive Services Task Force Recommendation Statement.” JAMA 18(22): 2224–2233. [DOI] [PubMed] [Google Scholar]
- Forrey AW, et al. (1996). “Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results.” Clinical Chemistry 42(1): 81–90. [PubMed] [Google Scholar]
- Fung KW, et al. (2008). “RxTerms -- a drug interface terminology derived from RxNorm.” AMIA Annu Symp Proc 6: 227–231. [PMC free article] [PubMed] [Google Scholar]
- Goodloe R, et al. (2017). “Reducing clinical noise for body mass index measures due to unit and transcription errors in te electronic health record.” AMIA Jt. Summits Transl Sci Proc. [PMC free article] [PubMed] [Google Scholar]
- Goryachev S, et al. (2006). “A suite of natural language processing tools developed for the I2B2 project.” AMIA Annu Symp Proc 2006: 931. [PMC free article] [PubMed] [Google Scholar]
- Graetz I, et al. (2016). “The Digital Divide and Patient Portals: Internet Access Explained Differences in Patient Portal Use for Secure Messaging by Age, Race, and Income.” Medical Care 54(8): 772–779. [DOI] [PubMed] [Google Scholar]
- Group SC, et al. (2008). “SLCO1B1 Variants and Statin-Induced Myopathy −− A Genomewide Study.” New England Journal of Medicine 359(8): 789–799. [DOI] [PubMed] [Google Scholar]
- Group TW, s. H. IS (1998). “Design of the Women’s Health Inititiative clinical trail and observational study.” Control Clin Trials 19(1): 61–109. [DOI] [PubMed] [Google Scholar]
- Gue D (2016). VistA retains top spot in most recent Medscape EHR survey. Medshere. 2018. [Google Scholar]
- Halladay CW, et al. (2018). “Genetically-guided algorithm development and sample size optimization for age-related macular degeneration cases and controls in electronic health records for the VA Million Veteran Program.” [PMC free article] [PubMed] [Google Scholar]
- Hanauer DA, et al. (2012). “Hedging their mets: the use of uncertainty terms in clinical documents and its potential implications when sharing the documents with patients.” AMIA Annu Symp Proc 2012: 321–330. [PMC free article] [PubMed] [Google Scholar]
- Hanauer DA, et al. (2015). “Supporting information retrieval from electronic health records: A report of University of Michigan’s nine-year experience in developing and using the Electronic Medical Record Search Engine (EMERSE).” Journal of Biomedical Informatics 55: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour S and Langlotz CP (2016). “Information extraction from multi-institutional radiology reports.” Artificial Intelligence in Medicine 66: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HealthIt.gov (2015, 02/06/2015). “Meaningful use definition & objectives.” from https://www.healthit.gov/providers-professionals/meaningful-use-definition-objectives.
- Hendershot T, et al. (2015). Using the PhenX Toolkit to Add Standard Measures to a Study Curr Protoc Hum Genet Current Protocols in Human Genetics, John Wiley & Sons, Inc; 86: 1.21.21–21.21.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J, et al. (2016). Adoption of electronic health record systems among non-federal acute care hospitals: 2008–2015 ONC Data Brief. Washington, DC, Office of the National Coordinator for Health Information Technology. [Google Scholar]
- Hersh WR, et al. (2013). “Caveats for the use of operational electronic health record data in comparative effectiveness research.” Med Care 51(8 Suppl 3): S30–S37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg J and Manning CD (2015). “Advances in natural language processing.” Science 349(6245): 261–266. [DOI] [PubMed] [Google Scholar]
- Hoffmann TJ, et al. (2016). “A Large Genome-Wide Association Study of Age-Related Hearing Impairment Using Electronic Health Records.” PLoS Genetics 12(10): e1006371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister BM, et al. (2016). “Development and performance of text-mining algorithms to extract socioeconomic status from de-identified electronic health records.” Pac Symp Biocomput 22: 230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PB, et al. (2012). “Mining electronic health records: towards better research applications and clinical care.” Nat Rev Genet 13(6): 395–405. [DOI] [PubMed] [Google Scholar]
- Joshi AK (1991). “Natural Language Processing.” Science 253(5025): 1242–1249. [DOI] [PubMed] [Google Scholar]
- Kaggal VC, et al. (2016). “Toward a Learning Health-care System – Knowledge Delivery at the Point of Care Empowered by Big Data and NLP.” Biomedical Informatics Insights 8s1: BII.S37977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, et al. (1961). “Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study.” Ann Intern Med 55: 33–50. [DOI] [PubMed] [Google Scholar]
- Kayaalp M (2017). “Patient privacy in the era of big data.” Balkan Med J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller ME, et al. (2015). “Enhancing practice efficiency and patient care by sharing electronic health records.” Perspect Health Inf Manag 12(ib). [PMC free article] [PubMed] [Google Scholar]
- Kern LM, et al. (2015). “The Meaningful Use of Electronic Health Records and Health Care Quality.” American Journal of Medical Quality 30(6): 512–519. [DOI] [PubMed] [Google Scholar]
- King J, et al. (2014). “Clinical Benefits of Electronic Health Record Use: National Findings.” Health Services Research 49(1pt2): 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KE and Clarke PJ (2015). “A disadvantaged advantage in walkability: findings from socioeconomic and geographical analysis of national built environment data in the United States.” Am J Epidemiol 181(1): 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JC, et al. (2016). “PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability.” Journal of the American Medical Informatics Association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohane IS (2011). “Using electronic health records to drive discovery in disease genomics.” Nat Rev Genet 12(6): 417–428. [DOI] [PubMed] [Google Scholar]
- Koppel R and Lehmann CU (2015). “Implications of an emerging EHR monoculture for hospitals and healthcare systems.” J Am Med Inform Assoc 22(2): 465–471. [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, et al. (2000). “CDC growth charts: United States.” Adv Data 8(314): 1–27. [PubMed] [Google Scholar]
- Laper SM, et al. (2016). “The challenges in using electronic health records for pharmacogenomics and precision medicine research.” Pac Symp Biocomput 21: 369–380. [PMC free article] [PubMed] [Google Scholar]
- Leader JB, et al. (2015). “Contrasting association results between existing PheWAS phenotype definition methods and five validated electornic phentoypes.” AMIA Annu Symp Proc 2015: 824–832. [PMC free article] [PubMed] [Google Scholar]
- Levin MA, et al. (2017). “iGAS: A framework for using electronic intraoperative medical records for genomic discovery.” Journal of Biomedical Informatics 67: 80–89. [DOI] [PubMed] [Google Scholar]
- Lewis CE and Wellons MF (2017). “Menopausal hormone therapy for primary prevention of chronic disease.” JAMA 318(22): 2187–2189. [DOI] [PubMed] [Google Scholar]
- Lewis MD, et al. (2002). “Disease outbreak detection system using syndromic data in the greater Washington DC area.” American Journal of Preventive Medicine 23(3): 180–186. [DOI] [PubMed] [Google Scholar]
- Li L, et al. (2015). “Identification of type 2 diabetes subgroups through topological analysis of patient similarity.” Science Translational Medicine 7(311): 311ra174–311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood SS, et al. (2014). “The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective.” Lancet 383(9921): 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasco AT, et al. (2016). “Characteristics of state prescription drug monitoring programs: a state-by-state survey.” Pharmacoepidemiology and Drug Safety 25(7): 847–851. [DOI] [PubMed] [Google Scholar]
- Marcotte L, et al. (2012). “Achieving meaningful use of health information technology: a guide for physicians to the EHR incentive programs.” Arch Intern Med 172(9): 731–736. [DOI] [PubMed] [Google Scholar]
- Mayes R (2006). “The origins, development, and passage of Medicare’s revolutionary prospective payment system.” Journal of the History of Medicine and Allied Sciences 62(1): 21–55. [DOI] [PubMed] [Google Scholar]
- McCarty C, et al. (2011). “The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies.” BMC Medical Genomics 4(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung HL, et al. (2018). “Dietary Intake and Physical Activity Assessment: Current Tools, Techniques, and Technologies for Use in Adult Populations.” American Journal of Preventive Medicine 55(4): e93–e104. [DOI] [PubMed] [Google Scholar]
- McDonald CJ, et al. (2003). “LOINC, a Universal Standard for Identifying Laboratory Observations: A 5-Year Update.” Clinical Chemistry 49(4): 624–633. [DOI] [PubMed] [Google Scholar]
- Medicine I. o. (2012). Best care at lower cost: the path to continuously learning health care in America. Washington, DC, National Academies Press. [PubMed] [Google Scholar]
- Medicine U. N. L. o. (2004, 2018). “RxNorm.” 2018, from https://www.nlm.nih.gov/research/umls/rxnorm/.
- Mennemeyer ST, et al. (2016). “Impact of the HITECH Act on physicians’ adoption of electronic health records.” Journal of the American Medical Informatics Association 23(2): 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama IM, et al. (2011). History of the statistical classification of diseases and causes of dealth. Rosenberg HM and Hoyert DL. Hyattsville, MD. [Google Scholar]
- Murff HJ and Kannry J (2001). “Physician satisfaction with two order entry systems.” J Am Med Inform Assoc 8(5): 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MF, et al. (2013). “Comparing Electronic Health Record Portals to Obtain Patient-Entered Family Health History in Primary Care.” Journal of General Internal Medicine 28(12): 1558–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni PM, et al. (2011). “Natural language processing: an introduction.” J Am Med Inform Assoc 18(5): 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, E., and Medicine (2016). Strategies for ensuring diversity, inclusion, and meaninful participation in clinical trials: Proceedings of a workshop. Washington, DC, The National Academies Press. [PubMed] [Google Scholar]
- Nelson SJ, et al. (2011). “Normalized names for clinical drugs: RxNorm at 6 years.” Journal of the American Medical Informatics Association 18(4): 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley KJ, et al. (2005). “Measuring Diagnoses: ICD Code Accuracy.” Health Services Research 40(5p2): 1620–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OHDSI. “OMOP Common Data Model.” Retrieved 09/26/2018, 2018, from https://www.ohdsi.org/data-standardization/the-common-data-model/.
- Ohno-Machado L (2011). “Realizing the full potential of electronic health records: the role of natural language processing.” J Am Med Inform Assoc 18(5): 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W. H. (2012, 2012). “History of the development of the ICD.” Retrieved 02/13/2018, 2018, from http://www.who.int/classifications/icd/en/HistoryOfICD.pdf.
- PCORNet. “PCORnet Common Data Model (CDM).” Retrieved 09/26/2018, 2018, from https://pcornet.org/pcornet-common-data-model/.
- Peissig P, et al. (2017). “Prototype development: context-driven dynamic XML ophthalmologic data capture application.” JMIR Med Inform 5(3): e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peissig PL, et al. (2014). “Relational machine learning for electronic health record-driven phenotyping.” Journal of Biomedical Informatics 52: 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perzynski AT, et al. (2017). “Patient portals and broadband internet inequality.” Journal of the American Medical Informatics Association 24(5): 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike SN, et al. (2017). “Examining the food retail choice context in urban food deserts, Ohio 2015.” Prev Chronic Dis 14: E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policy GWU s. H. H. L. a. (2015). “Who Owns Medical Records: 50 State Comparison.” Health Information & the Law. Retrieved 01/16/2018, 2018, from http://www.healthinfolaw.org/comparative-analysis/who-owns-medical-records-50-state-comparison.
- Preiss D and Sattar N (2015). “Classification of reported statin intolerance.” Current Opinion in Lipidology 26(1): 65–66. [DOI] [PubMed] [Google Scholar]
- Rasmussen LV, et al. (2012). “Development of an optical character recognition pipeline for handwritten form fields from an electronic health record.” Journal of the American Medical Informatics Association 19(e1): e90–e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling M, et al. (2017). “New Pharmacogenomics Research Network: An Open Community Catalyzing Research and Translation in Precision Medicine.” Clinical Pharmacology & Therapeutics 102(6): 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV and Evans WE (2015). “Pharmacogenomics in the clinic.” Nature 526: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo NA, et al. (2015). “Extracting primary open-angle glaucoma from electronic medical records for genetic association studies.” PLoS ONE 10(6): e0127817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinner C, et al. (2015). “Effects of Shared Electronic Health Record Systems on Drug-Drug Interaction and Duplication Warning Detection.” BioMed Research International 2015: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, et al. (2010). “Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record.” Am J Hum Genet 86(4): 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw JE, et al. (2002). “Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women’s Healht Initiative Randomized Controlled Trial.” JAMA 288(3): 321–333. [DOI] [PubMed] [Google Scholar]
- Sarkar IN (2010). “Biomedical informatics and translational medicine.” Journal of Translational Medicine 8(1): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savova GK, et al. (2010). “Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation, and applications.” J Am Med Inform Assoc 17(5): 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler-Ruwisch JM, et al. (2017). “A content analysis of electronic health record (EHR) functionality to support tobacco treatment.” Transl Behav Med 7(2): 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer K, et al. (2017). “Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams.” Briefings in Bioinformatics 18(1): 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivade C, et al. (2014). “A review of approaches to identifying patient phenotype cohorts using electronic health records.” J Am Med Inform Assoc 21(2): 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig DF and Singh H (2012). “Electronic Health Records and National Patient-Safety Goals.” New England Journal of Medicine 367(19): 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig DF and Wright A (2015). “What makes an EHR “open” or interoperable?” J Am Med Inform Assoc 22(5): 1099–1101. [DOI] [PubMed] [Google Scholar]
- Spooner LM and Pesaturo KA (2013). The medical record Fundamental skills for patient care in pharmacy practice. Lauster CD and Srivastava SB. Burlington, MA, Jones & Bartlett Learning. [Google Scholar]
- Statistics NC f. H (2015, 10/01/2015). “International Classification of Diseases, (ICD-10-CM/PCS) Transition -- Background.” Retrieved 02/14/2018, 2018, from https://www.cdc.gov/nchs/icd/icd10cm_pcs_background.htm.
- Technology O o. t. N. C. f. H. I. (2016, 12/2016). “Office-based physician electronic health record adoption.” Health IT Quick-Stat #50. Retrieved 01/23/2018, 2018, from dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php.
- Technology O o. t. N. C. f. H. I. (2017, July 2017). “Certified health IT developers and editions reported by health care professionals participating in Medicare EHR incentive program.” Health IT Quick-Stat #30. Retrieved 02/07/2018, 2018, from https://dashboard.healthit.gov/quickstats/pages/FIG-Vendors-of-EHRs-to-Participating-Professionals.php.
- The National Academies of Sciences, E., and Medicine (2017). “Recommended Social and Behavioral Domains and Measures for Electronic Health Records.” Retrieved 08/12/2017, from http://nationalacademies.org/HMD/Activities/PublicHealth/SocialDeterminantsEHR.aspx.
- Topaz M, et al. (2013). “ICD-9 to ICD-10: evolution, revolution, and current debates in the United States.” Perspect Health Inf Manag 10(Spring): 1:d. [PMC free article] [PubMed] [Google Scholar]
- Verma A, et al. (2016). “Integrating clinical laboratory measures and ICD-9 code diagnoses in phenome-wide association studies.” Pac Symp Biocomput 21: 168–179. [PMC free article] [PubMed] [Google Scholar]
- Verma SS, et al. (2017). “Identifying genetic associations with variability in metabolic health and blood count laboratory values: diving into the quantitative traits by leveraging longitudinal data from an EHR.” Pac Symp Biocomput 22: 533–544. [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo M, et al. (2012). “Pharmacogenomics Knowledge for Personalized Medicine.” Clinical Pharmacology & Therapeutics 92(4): 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LK, et al. (2015). “Phenotyping adverse drug reactions: statin-induced myotoxicity.” AMIA Jf. Summits Transl Sic Proc 2015: 466–470. [PMC free article] [PubMed] [Google Scholar]
- Wiley LK, et al. (2013). “ICD-9 tobacco use codes are effective identifiers of smoking status.” Journal of the American Medical Informatics Association 20(4): 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, et al. (2018). “Natural Language Processing and Its Implications for the Future of Medication Safety: A Narrative Review of Recent Advances and Challenges.” Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 38(8): 822–841. [DOI] [PubMed] [Google Scholar]
- Wood GC, et al. (2008). “Association of chromosome 9p21 SNPs with cardiovascular phentoypes in morbid obesity using electronic health record data.” Genomic Med 2(1–2): 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, et al. (2017). “Enhancing electronic health record data with geospatial information.” AMIA Jt. Summits Transl Sci Proc 2017: 123–132. [PMC free article] [PubMed] [Google Scholar]
- Xu H, et al. (2010). “MedEx: a medication information extraction system for clinical narratives.” Journal of the American Medical Informatics Association 17(1): 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. (2013). “Discontinuation of statins in routine care settings: a cohort study.” Ann Intern Med 158(7): 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]


