Abstract
Tryptophan-2,3-dioxygenase (TDO2), a rate-limiting enzyme in the tryptophan catabolism pathway, is induced in triple-negative breast cancer (TNBC) by inflammatory signals and anchorage-independent conditions. TNBC express extremely low levels of the microRNA-200 (miR-200) family compared to estrogen receptor-positive (ER+) breast cancer. In normal epithelial cells and ER+ breast cancers and cell lines, high levels of the family member, miR-200c serve to target and repress genes involved in epithelial-to-mesenchymal transition (EMT). To identify mechanism(s) that permit TNBC to express TDO2 and other proteins not expressed in the more well-differentiated ER+ breast cancers, miRNA-200c was restored in TNBC cell lines. The data demonstrate that miR-200c targeted TDO2 directly resulting in reduced production of the immune-suppressive metabolite kynurenine. Furthermore, in addition to reversing a classical EMT signature, miR-200c repressed many genes encoding immune-suppressive factors including PD-L1/2, HMOX1, and GDF15. Restoration of miR-200c revealed a mechanism whereby TNBC hijacks a gene expression program reminiscent of that used by trophoblasts to suppress the maternal immune system to ensure fetal tolerance during pregnancy.
Implications:
Knowledge of the regulation of tumor-derived immune-suppressive factors will facilitate development of novel therapeutic strategies that complement current immunotherapy to reduce mortality for TNBC patients.
Keywords: Tryptophan-2,3-dioxygenase; TDO2; microRNA-200; triple-negative breast cancer; immune suppression; epithelial to mesenchymal transition
Introduction
Triple-negative breast cancer (TNBC), which lacks estrogen and progesterone receptors (ER and PR) and amplification of the growth factor receptor HER2 (human epidermal growth factor receptor 2), recurs more often and more rapidly than other subtypes, with a peak risk of recurrence at 1–3 years post-diagnosis (1). If metastatic at time of initial diagnosis, TNBC has a median survival of 13 months (2,3). This high metastatic potential can be facilitated by an oncogenic epithelial to mesenchymal transition (EMT) that enhances anchorage independence and invasiveness. Partial EMT or mixed populations potentiate metastasis in breast cancer preclinical models (4) and circulating tumor cells isolated from breast cancer patients were enriched for mesenchymal markers upon chemo-resistance (5). ER positive breast cancer cell lines maintain epithelial markers and undergo significant programmed cell death termed “anoikis” under anchorage independent conditions (6). In contrast, TNBC lines aberrantly express mesenchymal and neuronal proteins that provide survival signals (7,8) and metabolic changes (9,10) including increased tryptophan catabolism (11).
Increasing evidence supports a role for tryptophan catabolism in tumor progression, whereby the tryptophan catabolite kynurenine (Kyn) facilitates anchorage independent survival by binding to and activating the aryl hydrocarbon receptor (AhR) (11–13). While the enzymes indoleamine 2,3-dioxygenase 1 (IDO1) and IDO2 can mediate the rate limiting step of tryptophan catabolism, we reported that in TNBC the increased intracellular and secreted levels of Kyn critical for anchorage independent survival are mediated by tryptophan-2,3-dioxygenase (TDO2) activity (11). Kyn can also exert its effects in a paracrine fashion, by binding to AhR in CD8+ T-cells to suppress anti-tumor activity (12,14,15), and by expanding the regulatory T-cell population to exhaust the anti-tumor response (14).
Members of the microRNA-200 (miR-200) family are high in normal epithelial cells and the more well-differentiated ER+ breast cancers, but extremely low in TNBC, and miR-200c is the most differentially expressed between human ER+ versus TNBC cell lines (16–18). Considered the “guardians of the epithelial phenotype,” this miRNA family suppresses EMT by direct targeting of Zinc Finger E-Box Binding Homeobox 1 and 2 (ZEB1/2) and other transcriptional repressors of E-cadherin (19,20) as well as numerous mesenchymal, neuronal and embryonic stem cell genes (21,22). Loss or silencing of this miRNA family in TNBC permits aberrant production of non-epithelial proteins that facilitate multiple steps in the metastatic cascade.
Here we report that restoration of miR-200c to TNBC cell lines not only potently reversed a “pan-EMT gene signature” derived from multiple types of carcinomas, but also significantly decreased TDO2 and consequent production of Kyn. Furthermore, miR-200c repressed multiple genes encoding immune-suppressive factors known to be produced by early embryo cells called trophoblasts. Trophoblasts undergo a normal EMT that facilitates invasion into the uterus and suppresses the maternal immune system to ensure fetal tolerance. The commonalities between tumor cells and trophoblasts are hypothesized to be due to adaptations to similar selective pressures, including limited nutrients, oxygen, and the need to avoid immune recognition (23). Although the idea that “trophoblast mimicry” might facilitate tumor progression has been previously recognized (24,25), a global mechanism permitting tumor cells to co-opt this immune-suppressive program, had not been identified.
Materials and Methods
Cell Lines
All cell lines were authenticated by Short Tandem Repeat DNA Profiling (Promega; Fitchburg, WI) at the University of Colorado Cancer Center (UCCC) Tissue Culture Core, and tested for mycoplasma every three months. Only cells under five passages were used in this study. Hs578T and SUM159PT cells were purchased from the UCCC Tissue Culture Core and the rest directly from ATCC (Manassas, VA). SUM159PT (SUM159) cells were grown in Ham’s F-12 with 5% fetal bovine serum (FBS), penicillin/streptomycin (P/S), hydrocortisone, insulin, HEPES and L-glutamine supplementation. BT549 were cultured in RPMI Medium 1640 with 10% FBS, P/S and insulin. MDA-MB-453 (MDA-453), were grown in DMEM Medium with 10% FBS and P/S. MDA-MB-231 (MDA-231) were grown in MEM with 5% FBS, HEPES, L-glutamine, nonessential amino acids, and insulin. MCF7 cells were obtained from Dr. Kate Horwitz at the University of Colorado Anschutz Medical Campus and were grown in DMEM with 10% FBS and L-glutamine.
Reagents
Interlukin-1 beta (IL-1β) and tumor necrosis factor alpha (TNF-α) were purchased from Affymetrix eBioscience (Santa Clara, CA) and used at a final concentration of 10ng/μl. Negative, scrambled control (SCR) or miR-200c mimics were purchased from Ambion (Foster City, CA) and used at a final concentration of 50nM. miRNA mimic and luciferase reporter transfections were performed using either RNAi Max or Lipofectamine 3000 purchased from Thermo Fisher Scientific (Waltham, MA) and experiments were conducted following the supplied protocols. TDO2 cDNA plasmid (TDO2-OE) and corresponding empty plasmid control vector (EV) control were obtained from Sino Biological (Wayne, PA), catalog # HG13215-UT and CV011.
Inducible system for miR-200c expression
BT549 cells were transduced with the doxycycline (DOX) inducible lentiviral vector pTripZ (Dharmacon, Lafayette, CO) encoding the precursor sequence for miR-200c (pTripZ-200c). Control cells were transduced with pTripZ empty vector (pTripZ-EV, pooled population). Stable expression was selected using 1 μg/mL puromycin, and multiple pTripZ-200c clones were tested to identify which had low/absent background expression of miR-200c and at least 500-fold expression of miR-200c in the presence of 1 μg/mL doxycycline (DOX) inducer.
Sample Preparation/UHPLC-MS analysis of cell lines
Cells were counted, pelleted at 1500 g at 4ºC, and stored at −80°C until analysis. Prior to LC-MS analysis, samples were placed on ice and re-suspended with methanol:acetonitrile:water (5:3:2, v:v) at a concentration of 2 million cells per ml. Media samples were extracted with the same solution at a dilution of 1:25 (v/v). Suspensions were vortexed continuously for 30 min at 4°C. Insoluble material was removed by centrifugation at 10,000 g for 10 min at 4°C and supernatants were isolated for metabolomics analysis by UHPLC-MS. Briefly, the analytical platform employs a Vanquish UHPLC system coupled online to a Q Exactive mass spectrometer (Thermo Fisher Scientific). Samples were resolved over a Kinetex C18 column, 2.1 × 150 mm, 1.7 μm particle size (Phenomenex, Torrance, CA, USA) equipped with a guard column (SecurityGuard™ Ultracartridge – UHPLC C18 for 2.1 mm ID Columns – AJO-8782 – Phenomenex, Torrance, CA, USA) using an aqueous phase (A) of water and 0.1% formic acid and a mobile phase (B) of acetonitrile and 0.1% formic acid. Samples were eluted from the column using either an isocratic elution of 5% B flowed at 250 μl/min and 25ºC or a gradient from 5% to 95% B over 1 minute, followed by an isocratic hold at 95% B for 2 minutes, flowed at 400 μl/min and 30ºC. The Q Exactive mass spectrometer (Thermo Fisher Scientific) was operated independently in positive or negative ion mode, scanning in Full MS mode (2 μscans) from 60 to 900 m/z at 70,000 resolution, with 4 kV spray voltage, 15 shealth gas, 5 auxiliary gas. Calibration was performed prior to analysis using the Pierce™ Positive and Negative Ion Calibration Solutions (Thermo Fisher Scientific). Acquired data was then converted from .raw to .mzXML file format using Mass Matrix (Cleveland, OH, USA). Metabolite assignments, isotopologue distributions, and correction for expected natural abundances of deuterium, 13C, and 15N isotopes were performed using MAVEN (Princeton, NJ, USA). Graphs, heat maps and statistical analyses (either T-Test or ANOVA), metabolic pathway analysis, PLS-DA and hierarchical clustering was performed using the MetaboAnalyst 3.0 package (www.metaboanalyst.com). Hierarchical clustering analysis (HCA) was also performed through the software GENE-E (Broad Institute, Cambridge, MA, USA). XY graphs were plotted through GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA).
Quantitative RT-PCR
Total RNA was isolated using TRIZOL RNA Isolation (QIAGEN, Germantown, MD) according to the manufacturer’s instructions. cDNA was synthesized with the qScript cDNA SuperMix (QuantaBio, Beverly, MA). qRT-PCR was performed in an ABI 7600 FAST thermal cycler. SYBR Green quantitative gene expression analysis was performed using ABsolute Blue qRT-PCR SYBR Green Low ROX Mix (ThermoScientific) and the following primers: TDO2 forward 5’- CGGTGGTTCCTCAGGCTATC-3’ and reverse 5’- CTTCGGTATCCAGTGTCGGG-3’; IL6 forward 5’- AAGCCAGAGCTGTGCAGATGA-3’ and reverse 5’- AACAACAATCTGAGGTGCCCA; HMOX-1 forward 5’-CAGGCAGAGAATGCTGAGTTC-3’ and reverse; GDF-15 forward 5’-CTCAGGACGCTGAATGGCTCT-3’ and reverse 5’-GGGTCTTGCAAGGCTGAGCTG-3’; PD-L1 forward 5’-TATGGTGGTGCCGACTACAA-3’ and reverse 5’-TGGCTCCCAGAATTACCAAG-3’; ZEB1 forward 5’-TCCATGCTTAAGAGCGCTAGCT-3’ and reverse 5’-ACCGTAGTTGAGTAGGTGTATGCCA-3’; GAPDH forward 5’-GTCAGTGGTGGACCTGACCT-3’ and reverse 5’-AGGGGTCTACATGGCAACTG-3’; β-actin: FP- CTGTCCACCTTCCAGCAGATG RP-CGCAACTAAGTCATAGTCCGC. Taqman quantitative gene expression analysis for miR-200c and U6 was performed with primer-specific reverse transcriptase using the following TaqMan MicroRNA Assays (Applied Biosystems/ThermoFisher): miR-200c assay ID #002300; U6 assay ID #001973. All experiments were performed in biological triplicate.
Immunoblotting
Whole-cell protein extracts (50 μg) were denatured, separated on SDS-PAGE gels, and transferred to polyvinylidene fluoride membranes. After blocking in 5% BSA in Tris-buffered saline–Tween, membranes were probed overnight at 4°C. The following antibodies were used in this study: TDO2 (Sigma, St. Louis, MO, #HPA039611, 1:1000), HMOX-1 (Cell Signaling, Danvers, MA, #5061, 1:1000), ZEB1 (Sigma, #HPA027524, 1:2000), α-tubulin (Sigma, #T5168, 1:10,000), and β-Actin (Cell Signaling, #3700, 1:4000). Polyclonal antisera directed against GDF-15 was generated using the C-terminus peptide KTDTGVSLQTYDDLLA (Zymed, San Francisco, CA). Affinity purified GDF-15 antibody was prepared using the SulfoLink coupling resin according to the manufacturer’s instructions (ThermoFisher). Following secondary antibody incubation, results were detected using Western Lighting Chemiluminescence Reagent Plus (Perkin Elmer; Waltham, MA). Densitometry quantifications were performed using ImageJ (NIH, Bethesda, MD). First, all bands were normalized to their respective α-Tubulin or β-Actin loading control. The values shown in figures are reported as a fold change compared to either untreated cells or cells transiently transfected with scrambled mimic (SCR) or DOX.
Flow Cytometry
Single-cell suspensions of BT549 cells were prepared and stained using the anti PD-L1 (Clone MIH1, eBioscience) following manufacturer’s instructions, then fixed in 1% paraformaldehyde and stored at 4º C until analysis. Flow cytometry was conducted using the BD LSRFortessa cell analyzer (BD Biosciences, Franklin Lakes, NJ) and data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR).
Gene expression analysis
BT549-pTripZ-200c cells, plated in triplicate, were cultured in DOX for 48 hours. RNA was isolated using the RNeasy PLUS kit (Qiagen; Germantown, MD) and quality was measured on an Agilent 2100 Analyzer (Agilent Technologies; Santa Clara, CA), RIN (RNA integrity number) ranged from 9.6–10.0. Gene expression was measured using the Affymetrix Human Transcriptome 2.0 array hybridization performed by the University of Colorado Cancer Center Genomics and Microarray core facility and was deposited in the Gene Expression Omnibus (GEO) database as GSE108271. Data were analyzed by one-way ANOVA performed at the gene and probe (exon) levels using Partek Genomics Suite software (Partek; St. Louis, Missouri). Microarray data were normalized by Robust Multiarray Average method using Affymetrix Power Tools. Multiple probe sets for the same gene were collapsed using average expression. Genes with false-discovery rate < 10% and fold change > 1.2 were selected as differentially expressed between the two groups. Pathway analysis was performed using Gene Set Enrichment Analysis (GSEA) software and KEGG gene sets. Gene sets with P< 0.05 (after 1000 gene set permutations) were deemed to be enriched in each group.
Pan-carcinoma EMT signature analysis
To determine the EMT states of the untreated and DOX-treated BT549 cells, our gene array results were compared to the published pan-carcinoma EMT signatures from Mak et al (26). The “pan-carcinoma EMT signature” genes were extracted from the normalized gene expression data of the untreated and DOX-treated BT549 cells. Unsupervised clustering was performed using CLUSTER 3.0 with Spearman correlation with average linkage. Cluster was visualized using Java TreeView (SourceForge, Mountain View, CA).
Cloning of TDO2 3’UTR and Site-Directed Mutagenesis
The 3’UTR of TDO2 was amplified from the BAC plasmid RP11–401G24. Primers were designed to amplify the 3’UTR of TDO2 and to additionally add restriction enzyme cut sites for Sac1 (5’ end) and Xho1 (3’end): XhoI-TDO2: AGACCGCTCGAGAATCGTCTGCAAAATCTATG
SalI-TDO2: GAACGCGTCGACTACAGGGAGAAAGATTAATAC. Once amplified, the insert was cloned into the pmiR-GLO vector (Promega) using the corresponding restriction enzyme cut sites. The pmiR-GLO vector containing the 3’UTR of TDO2 was confirmed by gel electrophoresis and its sequence confirmed by the DNA Sequencing Core at the University of Colorado (Aurora, CO).
Luciferase reporter activity
Reporter activation was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Briefly, cells were lysed for 15 minutes at room temperature using 1× passive lysis buffer and centrifuged to eliminate cell debris. The supernatant was used immediately or diluted with 1× passive lysis buffer for determination of luciferase activity. For analysis of the activity of miR-200c mimic on the wild type or mutated TDO2 3’UTR, both were cloned into the pmiR-Glo vector and luciferase activity was normalized to the Renilla signal to control for differences in transfection efficiency.
Measurement of anchorage-independent growth
Soft-agar assays were performed in 0.5% bottom and 0.25% top-layer agar (Difco Agar Noble; BD Biosciences). Cells were embedded in the top-layer agar and media refreshed every 4 days.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5. Student t test, ANOVA with Tukey post-hoc test, and two-way ANOVA with Bonferroni multiple comparison test were used as noted. P values are denoted as follows: *, P< 0.05; **, P< 0.01; ***, P< 0.001; ****, P< 0.0001; ns, not significant.
Results
Restoration of miR-200c to TNBC cell lines leads to widespread reversal of EMT and an immune-suppressive signature that includes TDO2
BT549 breast cancer cells engineered to express miR-200c in a doxycycline (DOX)-inducible manner were treated with or without DOX for 48 hours. RNA was collected, gene expression analysis performed, and unsupervised hierarchical clustering of the top differentially expressed genes (FDR < 10% and fold change > 1.2) is shown in heatmap form (Figure 1A). Analysis of, miR-200c and known target ZEB-1 from BT549-TripZ-EV and BT549-TripZ-200c cells treated with or without DOX showed significantly increased miR-200c and a reduction in ZEB-1 in DOX treated BT549-TripZ-200c cells exclusively (Figure 1B). Western blot confirmed decreased ZEB-1 and increased E-Cadherin proteins in DOX treated BT549-TripZ-200c, but not BT549-TripZ-EV cells (Figure 1C). GSEA analysis revealed alterations in a number of pathways involved in immune modulation upon induction of miR-200c, including tryptophan metabolism, allograft rejection, complement and coagulation pathways, and cytokine/cytokine receptor signaling (Figure 1D, Supplemental Figure S2, Supplemental Table S1). The list of altered genes involved in these immune related pathways is available in Supplemental Table S2. For select genes encoding proteins with immune-suppressive functions, qRT-PCR was performed from RNA isolated from an independent set of BT549-Tripz-200c cells treated with or without DOX. This experiment demonstrated significantly decreased levels of TDO2, Programmed death-ligand 1 (PD-L1/CD274), heme oxygenase 1 (HMOX-1), and Growth Differentiation Factor 15 (GDF-15) in the presence of miR-200c (Figure 1E). Although GDF-15 was decreased by 1.7 fold, it does not appear in the heatmap because its p value fell slightly below the cutoff of FDR < 10% at FDR =12%, but we followed up on its regulation because of its known immune-modulatory functions. Comparison of genes altered by induction of miR-200c in BT549-TripZ-200c cells to a “pan-carcinoma EMT signature” derived from multiple types of carcinomas (26) revealed that induction of miR-200c increased 60% (15/25) of the epithelial signature and decreased 80% (41/51) of the mesenchymal signature (Supplemental Figure S1A).
Figure 1. Restoration of miR-200c to TNBC represses epithelial to mesenchymal transition and pathways/genes involved in immune suppressive functions.
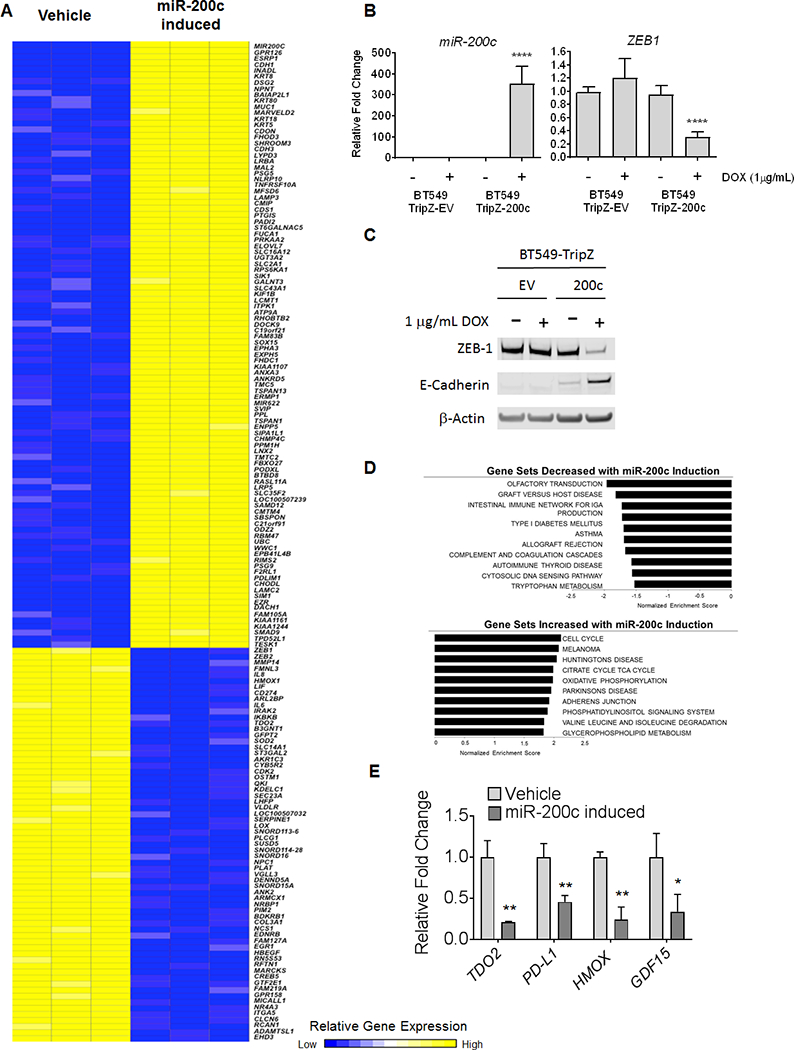
BT549-TripZ-200c cells containing the pre-miR-200c sequence in the DOX inducible pTripZ vector were treated in triplicate with 1 μg/mL Doxycycline (DOX) or vehicle for 48 hours. (A) Heat map of differentially expressed genes (adjusted p-value < 0.10 and fold-change > 1.2) between untreated (DOX-) and induced (DOX+) BT549-TripZ-200c cells in triplicate. (B) In both BT549-TripZ-200c and BT549 cells with empty pTripZ vector (BT549-TripZ EV), miR-200c and ZEB1 were measured by qRT-PCR and normalized to U6 and 18S respectively, and presented as fold change of DOX+ cells relative to untreated counterpart cells. ****P<0.0001, unpaired t-test. (C) Western blot for ZEB-1 and E-Cadherin proteins in BT549-TripZ-EV cells and BT549-TripZ-200c minus or plus DOX (48 hrs). (D) Gene Set Enrichment Analysis of pathways enriched in DOX- (no miR-200c) versus DOX+ (miR-200c induced). **P<0.01, *P<0.05, unpaired t-test. (E) Select transcripts measured by qRT-PCR from RNA isolated from independent BT549–200c cells DOX- and DOX+ for 48 hours.
Restoration of miR-200c in an immune-competent mouse model of triple-negative mammary carcinoma led to decreased metastatic outgrowth in the lungs (GSE62230) (27). While the immune system was not examined in that study, genes altered by restoration of miR-200c to human TNBC cells compared to those altered by miR-200c in the mouse mammary carcinoma found a significant number of overlapping genes, including TDO2, HMOX-1, PD-L1/CD274, and Inhibitor of Nuclear Factor Kappa B Kinase Subunit Beta (IKBKB) among others (Supplemental Figure S1B).
miR-200c directly targets TDO2 and decreases TNBC kynurenine production
To further validate the decrease in TDO2 upon miR-200c restoration observed in the initial gene expression findings, four TNBC cell lines (BT549, Hs578T, MDA-453, and SUM159) were transiently transfected with either miR-200c mimic or scrambled control mimic (SCR). Decreased TDO2 expression was detected in all four cell lines following miR-200c transfection (Figure 2A). Western blot of whole cell lysates showed decreased TDO2 and ZEB1 in three TNBC cell lines tested (Figure 2B). A predicted miR-200bc/429 binding site was identified in the TDO2 3’UTR (nucleotide position 5’−395–400-3’) using TargetScan. To test whether miR-200c directly binds to and targets TDO2, the entire 3’UTR of TDO2 was inserted downstream of luciferase in the pmiR-GLO reporter plasmid (Figure 2C). When this reporter was transfected into BT549 cells with miR-200c mimic or SCR, miR-200c mimic significantly decreased luciferase activity. This effect was reversed upon mutation of the predicted binding site for miR-200c in the TDO2 3’UTR (Figure 2D). To test the impact of miR-200c and subsequent reduction in TDO2 protein on tryptophan catabolism, Kyn levels were measured by UHPLC-MS following miR-200c induction in the BT549 TripZ DOX-inducible system. Intracellular and secreted Kyn levels were significantly reduced with DOX induction of miR-200c in the BT549 cells and the same effect was observed following addition of miR-200c mimic to the Hs578T and MDA-453 TNBC cell lines (Figure 2E).
Figure 2. Restoration of miR-200c to TNBC decreases TDO2 by direct targeting of TDO2 at a binding site in the 3’UTR, resulting in reduction of intracellular and secreted kynurenine.
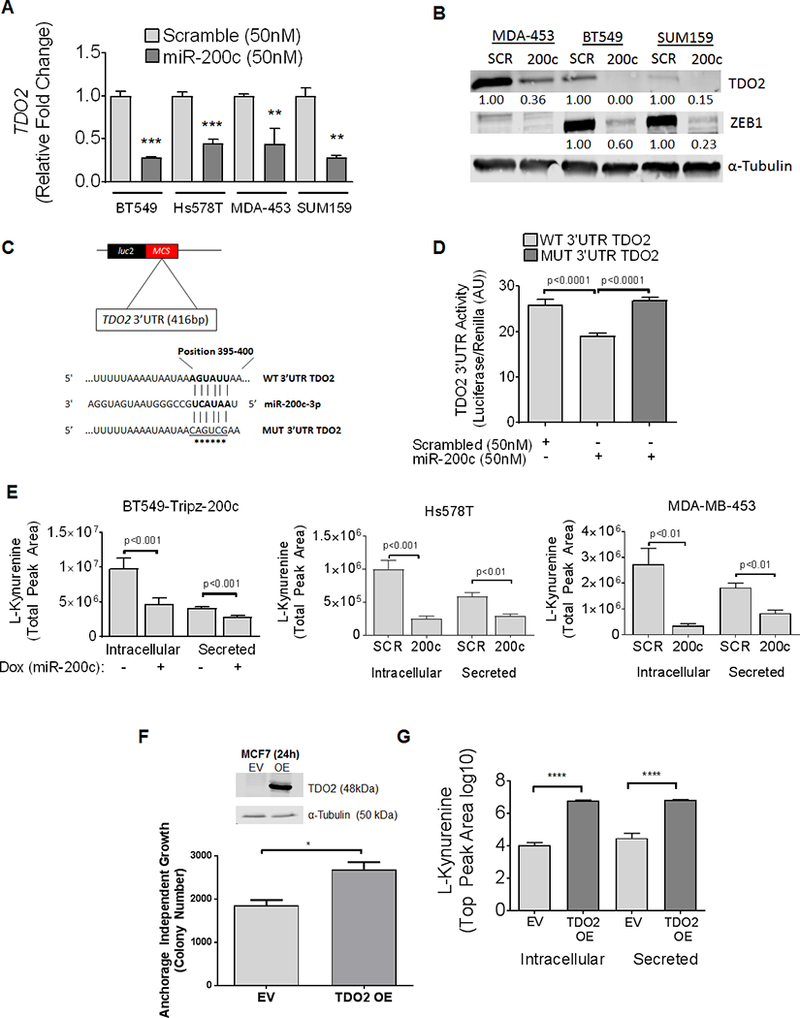
(A) Relative TDO2 mRNA levels determined by qRT-PCR in multiple TNBC cell lines following transfection with either negative SCR control mimic or miR-200c mimic for 48 hours. ***P<0.001, **P<0.01, unpaired t-test. (B) TDO2 and ZEB1 in multiple TNBC lines following transfection with negative control or miR-200c mimic for 72 hours. (C) Schematic of TDO2 3’UTR depicting predicted binding site of miR-200c at position 395–400 cloned into pmiR-Glo-luciferase vector with site-directed mutations indicated by asterisks. (D) Luciferase activity of the WT TDO2 3’UTR or mutated TDO2 3’UTR containing pmiR-Glo following transfection of BT549 cells with either negative control mimic or miR-200c mimic for 48 hours. ****P<0.0001, one-way ANOVA analysis. (E) Relative Kyn levels in BT549 cells as determined by UPLC-MS expressing a stable DOX-inducible lentiviral empty vector pTripZ vector or miR-200c-pTripZ vector after 72 hours of DOX induction (left). Relative Kyn levels measured in Hs578T and MDA-453 following transfection with SCR control or miR-200c mimic 72 hours prior (middle, right). (F) MCF7 cells transfected with expression vector for TDO2 (OE) or empty vector (EV) and immunoblot probed with TDO2 detecting antibody 24 hours after transfection (top). MCF7 cells with TDO2 OE or EV control were grown in soft-agar and colony number determined after 11 days (bottom). (G) Intracellular and secreted Kyn measured by UPLC-MS in MCF7-EV or MCF7-TDO2-OE cells after 72 hours. Data is shown on a log10 scale.
Exogenous TDO2 in ER+ breast cancer cells increased anchorage independent growth
TDO2 mRNA and protein are very low or undetectable in ER+ breast cancer cells and these cells do not survive well under anchorage independent conditions compared to TNBC lines (11). To determine whether increased TDO2 promotes anchorage independent survival, MCF7 cells were transfected with either TDO2-expression vector (TDO2 OE) lacking the 3’UTR or empty vector (EV) for 24 hours and plated in soft agar. After 11 days, colonies were stained and quantified to assess anchorage independent growth. MCF7-TDO2 OE demonstrated significantly enhanced growth on soft agar compared to MCF7 EV cells (Figure 2F). Western blot of whole-cell lysate collected prior to growth on soft agar confirmed TDO2 overexpression in the MCF7-TDO2 OE cells as compared to MCF7-EV (Figure 2F). Further, to functionally confirm the activity of the exogenous TDO2, we confirmed by UHPLC-MS that MCF7-TDO2 OE cells had increased intracellular and secreted Kyn levels compared to MCF7-EV cells (Figure 2G).
Restoration of miR-200c decreases TDO2, HO-1, and GDF-15 even in the presence of inflammatory stimuli
TDO2 is significantly upregulated in TNBC cells in forced-suspension culture as a result of increased Nuclear Factor kappa B (NFκB) activity (11). Given these findings, we examined whether miR-200c would repress NFκB-mediated stimulation of TDO2 expression. BT549-TripZ-200c inducible cells were treated with or without DOX in the presence or absence of the NFκB activating cytokines Interleukin 1 Beta (IL-1β) and Tumor Necrosis Factor alpha (TNF-α). Both TDO2 mRNA and protein were significantly upregulated following treatment with the NFκB activating cytokines, but suppressed in the presence of DOX, indicating that miR-200c blocks TDO2 expression even in the context of inflammatory stimuli (Figure 3A). As a positive control we examined Interleukin-6 (IL-6), a known NFκB target, which significantly increased following treatment with IL-1β and TNF-α (Figure 3B). IL-6 was also decreased with restoration of miR-200c in the initial array results and this was confirmed in an independent experiment shown in Figure 3B. While miR-200c levels significantly increased following treatment with DOX, they were not significantly affected by IL-1β and TNF-α treatment (Figure 3C).
Figure 3. Restoration of miR-200c abrogates expression of immunomodulatory gene and protein expression in TNBC cells.
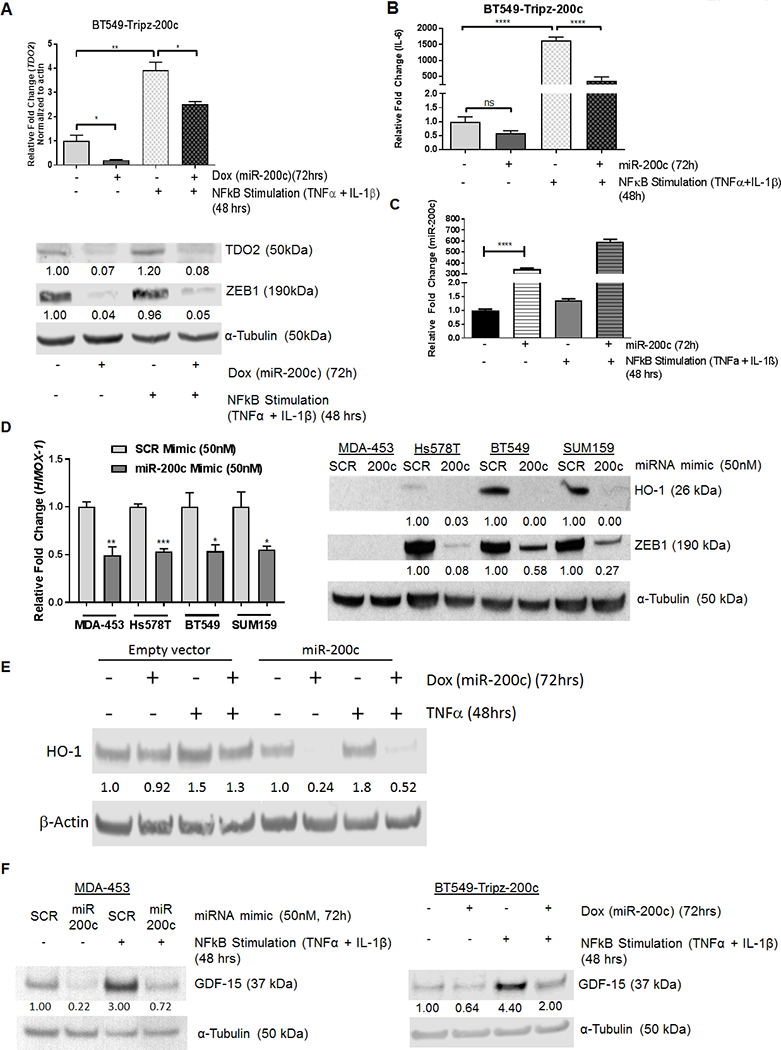
(A) Relative TDO2 mRNA levels in BT549-TripZ-200c cells after 72 hours of no treatment, DOX, NFκB stimulation (TNF-α, IL-1β (10 ng/μL each)) for the last 48 hrs, or the combination of DOX and NFκB stimulation (top). Relative TDO2 levels were normalized to β-actin. ***P<0.001, **P<0.01 by unpaired t-test. Immunoblot for TDO2 and ZEB1with α-Tubulin as a loading control in BT549-TripZ-200c cells after 72 hours of no treatment, DOX, NFκB stimulation (TNFα and IL-1β), or the combination of DOX and NFκB stimulation (bottom). Fold changes are indicated compared to vehicle treated after normalizing to loading control.(B) IL6 mRNA levels in BT549-TripZ-200c cells after 72 hours of no treatment, DOX, NFκB stimulation(TNF-α, IL-1β), or the combination of DOX and NFκB activation. Relative IL6 levels were normalized to B-actin. ***P<0.001, **P<0.01 by unpaired t-test. (C) Relative miR-200c in BT549-TripZ-200c cells after 72 hours of no treatment, DOX alone, NFκB stimulation (TNF-α, IL-1β), or the combination of DOX plus NFκB activation. Relative miR-200c levels were normalized to U6 snRNA. (D) HMOX-1 qRT-PCR in multiple TNBC cell lines following transient transfection with either negative control scrambled (SCR) mimic or miR-200c mimic after 48 hours (left) and immunoblot for HO-1 (encoded by HMOX-1) in TNBC lines following transient transfection with SCR mimic or miR-200c mimic for 72 hours (right). Fold changes were calculated over SCR after normalizing to the α-Tubulin loading control. (E) Immunoblot for HO-1 protein in in BT549-Tripz-EV cells as compared to BT549-Tripz-200c cells treated minus or plus DOX (72 hrs) with or without TNF-α for the last 48 hrs. (F) Immunoblot for GDF-15 in MDA-453 cells following transfection with SCR control or miR-200c mimic for 72 hrs with or without NFκB stimulation for the last 48 hrs (left) and in BT549-TripZ-200c cells after vehicle versus DOX for 72 hrs with or without NFκB stimulation for the last 48 hrs (right). Fold change was calculated over vehicle treated cells after normalization to α-Tubulin loading control.
We further investigated miR-200c decreased multiple other genes encoding immunomodulatory proteins (Figures 1A, Supplemental Figure S2). HMOX-1, which encodes heme oxygenase 1 (HO-1), was reduced by restoration of miR-200c in the microarray analysis (fold change −2.13; P=9.40E-05). Following transient transfection of BT549, Hs578T, MDA-453, and SUM159 TNBC lines with miR-200c mimic HMOX-1 was significantly reduced in all lines (Figure 3D, left). HO-1 protein (encoded by HMOX-1) was also decreased by miR-200c (Figure 3D, right). Since TNFα increased HO-1 protein levels, but IL-1β decreased them (Supplemental Figure S3), we examined HO-1 in empty vector containing BT549-TripZ-EV cells versus miR-200c containing BT549-TripZ-200c cells treated with or without DOX, then plus or minus TNF-α. It was clear that even in the face of TNFα stimulation, miR-200c suppressed production of HO-1 (Figure 3E).
GDF-15, which encodes an anti-inflammatory cytokine also known as Placental TGF-Beta (PTGFB) due to its high level of expression in placenta, decreased in BT549-TripZ-200c cells following induction of miR-200c in the array analysis (fold change −1.7); however, the p value fell slightly below the cutoff of P < 0.1, with P=0.12. We examined whether NFκB stimulation affected GDF-15 levels and whether miR-200c would limit this process. BT549-TripZ-200c inducible cells were treated with or without DOX in the presence or absence of the NFκB stimulating cytokines IL-1β and TNF-α for 48 hours. GDF-15 protein was upregulated by 4.4 fold following cytokine treatment, but the effect was suppressed in the presence of DOX induction of miR-200c (Figure 3F right). The same was true in MDA-453 cells, in which the NFκB activating cytokines significantly upregulated GDF-15, but addition of miR-200c mimic repressed GDF-15 baseline and cytokine induced levels (Figure 3F left).
Restoration of miR-200c decreases expression of PD-L1/2
PD-L1 was recently identified as a direct target of miR-200c in lung cancer and MCF7 breast cancer cells (28,29). Since PD-L1 was significantly decreased in BT549–200c cells following DOX induction in the array analysis (Figure 1A), we treated BT549-TripZ-200c cells with Interferon gamma (IFNγ) to induce PD-L1 expression, and confirmed that miR-200c significantly reduced the IFNγ-mediated upregulation of PD-L1 (Figure 4A). Additionally IFNγ-induced PD-L2 expression was significantly reduced in the presence of miR-200c (Figure 4B). Reduction in PD-L1 ligand following restoration of miR-200c was confirmed by flow cytometry (Figure 4C). Figure 4D illustrates a model how miR-200c suppression of TDO2-dependent tryptophan catabolism and resulting Kyn production, along with suppression of GDF-15 and HO-1, might affect T-cells in a non-contact dependent fashion in addition to reducing the contact-dependent immune suppressors, PD-L1 and 2.
Figure 4. miR-200c regulates PD-L1 and 2 and other immunosuppressive factors in Triple-Negative Breast Cancer.
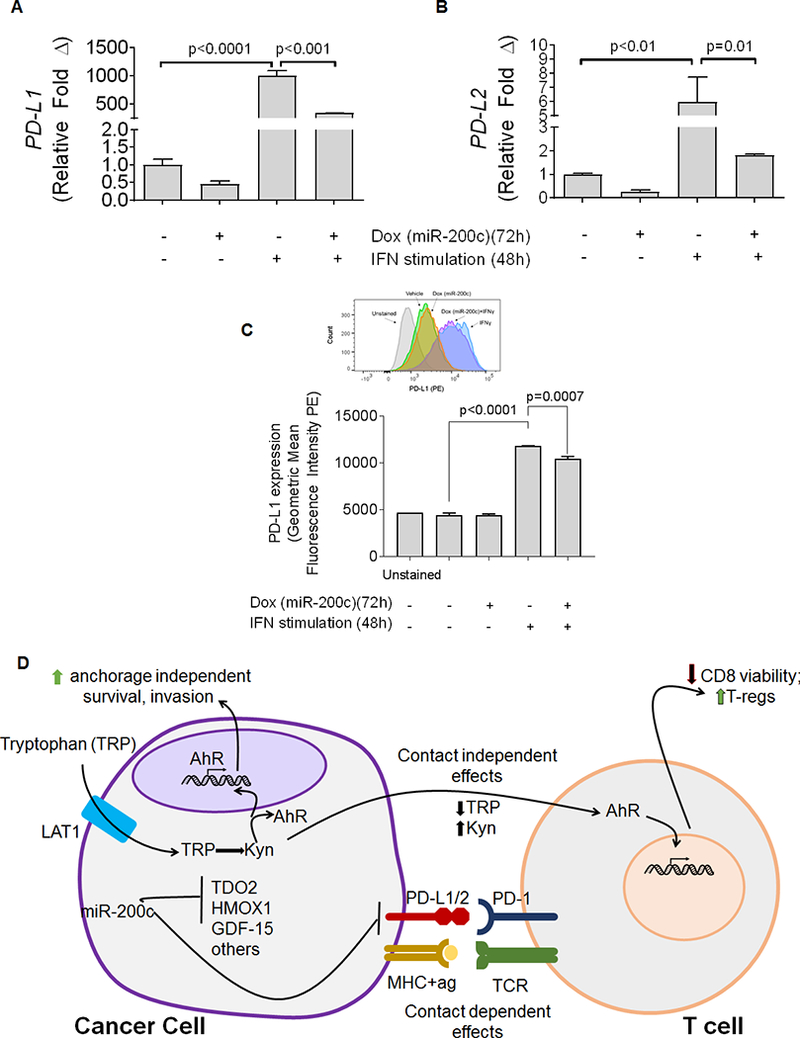
BT549-TripZ-200c cells were treated with DOX to induce expression of miR-200c, or vehicle control. (A) Relative PD-L1 mRNA levels were determined by qRT-PCR in 24 hours after DOX treatment began, cells were treated with interferon γ (IFNγ, 10 ng/mL) or vehicle. Cells were harvested 72 hours after the commencement of DOX treatment. ****P<0.0001, as determined by one-way ANOVA. (B) Relative PD-L2 mRNA levels were determined by qRT-PCR in BT549 cells expressing a stably transduced, DOX-inducible miR-200c vector were treated with DOX to induce expression of miR-200c, or vehicle control. 24 hours after DOX treatment began, cells were treated with IFNγ (10 ng/mL) or vehicle. Cells were harvested 72 hrs after DOX treatment. ****P<0.0001, as determined by one-way ANOVA. (C) PD-L1 expression was measured by flow cytometry. BT549 cells expressing a stably transduced, DOX-inducible miR-200c vector were treated with DOX to induce expression of miR-200c, or vehicle control. 24 hours after DOX treatment began, cells were treated with IFNγ (10 ng/mL) or vehicle. Cells were harvested 72 hours after the commencement of DOX treatment and stained for PD-L1. n=3 samples, data shown is representative of two independent experiments. *** p<0.001, **** p<0.0001, as determined by unpaired t-test. (D) Model of factors repressed by restoration of miR-200c to TNBC and their published effects on T cells.
Discussion
Here we report a novel mechanism of regulation of tumor tryptophan catabolism, involving post-transcriptional regulation by miR-200c. The miR-200 family is well-described as a critical determinant of the epithelial phenotype that directly targets and suppresses transcriptional repressors of E-cadherin such as ZEB1 and ZEB2, Twist, and Snail, to mediate normal EMT that facilitates rearrangement of epithelial cells during development, as well as oncogenic EMT in carcinomas, which can facilitate metastasis (19,20). Many mesenchymal and neuronal genes involved in motility, invasiveness, anoikis resistance, chemo-resistance and stem cell-like properties such as Tropomyosin-Related Kinase B (TrkB), Neurotrophin 3 (NTF3), Moesin, Tubulin Beta 3 Class III (TUBB3), and Suppressor Of Zeste 12 Protein Homolog (SUZ12) are targeted by miR-200c (21,30). In many de-differentiated, aggressive carcinomas, including TNBC, miR-200 family members are often lost or repressed by micro-deletions or silenced via methylation (31,32). Here, restoration of miR-200c to TNBC cells repressed ZEB1, restored E-cadherin, profoundly reversed a patient-derived, pan-cancer EMT signature (26), and significantly repressed genes encoding immune suppressive factors. GSEA analysis of genes reduced by induction of miR-200c in TNBC revealed a change in a number of pathways involved in immune modulation including allograft rejection, complement and coagulation pathways, and tryptophan metabolism.
Analysis of genes increased by induction of miR-200c demonstrated the expected gain of epithelial characteristics, but also indications of increased glycolysis and oxidative phosphorylation. While this alteration was unexpected, a recent study of the metabolic properties of an immortalized breast epithelial cell line and its EMT counterpart also found glycolysis and oxidative phosphorylation to be associated with the epithelial phenotype, while amino acid anaplerosis and fatty acid oxidation drove the mesenchymal phenotype (33). Interestingly, the tryptophan transporter LAT1 was deemed “essential for mesenchymal cell survival” (33).
Multiple clinical trials evaluating inhibitors of IDO activity such as Indoximod (D-1-methyl-tryptophan, NewLink Genetics) and INCB024360 (Incyte) either have already been conducted or are underway. However, these compounds do not inhibit TDO2, which evolved separately to metabolize tryptophan (34,35). Data presented here, in our previous report, and by others, suggests that TDO2 may be relevant in TNBC and other types of cancer (11–13). Thus, new drugs that inhibit both IDO and TDO2 may be more effective in some cancer types including TNBC. TDO2-mediated production of Kyn supports tumor cell anchorage independent growth and invasion in an autocrine manner by binding to and activating AhR (11–13). Indeed AhR was recently linked to inflammation and tryptophan metabolism in breast cancer (36). Kyn can also act in a paracrine fashion by binding AhR in T cells to suppress T-cell mediated tumor rejection (12,37) via expansion of regulatory T-cells (14) and direct effects on cytotoxic T cells (38). In addition to increased TDO2 upon NFκB activation in TNBC (11), we now report post-transcriptional control of TDO2 by miR-200c, explaining why this enzyme is expressed in TNBC, but not ER+ lines, which have high levels of miR-200c (16). TDO2 is decreased as a consequence of direct targeting of the TDO2 3’UTR by miR-200c and Kyn production is consequently reduced. Conversely, transfection of TDO2 lacking the 3’UTR containing miR-200c binding site into ER+ cells increased expression of Kyn and growth on soft agar.
GSEA analysis of genes altered by miR-200c showed a reduction in pathways involved in immune-suppression and tolerance. Interestingly, normal EMT enables extravillar cytotrophoblasts to invade into the maternal decidual stroma and produce immune-suppressive substances that ensure embryo maintenance/fetal tolerance during pregnancy (23). We demonstrate that restoration of miR-200c suppresses the ability of TNBC to make similar immune-suppressive factors. The miR-200 family typically serves as the “brakes” by which EMT is kept at bay in normal epithelium or ER+ breast cancer. We propose that loss or silencing of the miR-200 family permits TNBC to undergo EMT and co-opt this invasive/immune suppressive program. In support of this theory, low levels of the miR-200 family member, miR-141, permit successful trophoblast uterine invasion and immune suppression to facilitate successful tolerance of the fetus, whereas miR-141 is abnormally high in pre-eclamptic placentae (when these processes fail) (39).
Mellor and Munn described the role of tryptophan catabolism in the prevention of T-cell-driven complement activation and inflammation during pregnancy (40). Inhibition of IDO or TDO2-mediated tryptophan catabolism in pregnant mice induced fetal loss, supporting its role in maternal fetal tolerance (40). The immune-suppressive effects of both tryptophan depletion and increased tryptophan metabolites such as Kyn are well established (12,14,41). TDO2 also plays a role in endometrial decidualization where it suppresses the activation of decidual T-cells to maintain pregnancy (42).
HO-1 is an anti-inflammatory, immune-suppressive, pro-angiogenic enzyme involved in suppression of CD8+ T-cells through expansion of T-regulatory cells necessary for fetal tolerance during pregnancy (43). Gestational stress in mice led to lower HO-1, increased cytotoxic CD8+ T-cells and intrauterine growth restriction (44). Here we show that restoration of miR-200c significantly decreased levels of HMOX-1 (encoding HO-1) and HO-1 protein is overexpressed in breast cancer and predicts shorter overall survival (45). In clear-cell renal cell carcinoma cells, miR-200c directly targets HMOX-1 (46). In a mouse mammary carcinoma model, HO-1 inhibition reduced tumor growth to a degree comparable to PD-1 blockade (47). GDF-15 is highly expressed in placenta and decidual stromal cells in the uterus and has been found to promote tolerogenic dendritic cells during pregnancy (48). In malignant gliomas, depletion of GDF-15 enhanced natural killer cell-mediated clearance of tumors (49) and is also though to help tumor cells evade immune surveillance by macrophages (50). In the normal prostate, GDF-15 is inversely correlated with inflammation (51); however, circulating GDF-15 levels are elevated in prostate cancer patients compared to men without prostate cancer. GDF-15 was significantly decreased by restoration of miR-200c to TNBC even in the face of NFκB stimulation.
Our data support recent reports that PD-L1 (CD274) is targeted by miR-200c (28,29), and we confirmed this finding at the protein level by flow cytometry. Restoration of miR-200c also reduced PD-L2 (CD273) induction by IFNγ. Like PD-L1, PD-L2 is expressed on both antigen-presenting cells and tumor cells and interacts with PD-1 on T-cells to inhibit anti-tumor function (52). High tumor PD-L2 indicated a better clinical response in patients treated with pembrolizumab (Keytruda), suggesting that PD-1 blockade may function through inhibition of both ligands (53).
Restoration of miR-200c decreased metastasis in an immune competent genetically engineered mouse model of claudin-low TNBC (27) and although changes in the immune response were not examined in that study, comparison of the data with the human TNBC cells studied here revealed overlap in the genes reduced upon induction of miR-200c, including TDO2, HMOX-1, IKBKB, PLCG1 and PD-L1 (Supplemental Figure S1B). We observed that NFκB stimulating cytokines upregulate TDO2, HO-1, and GDF-15 protein in TNBC, but this induction by inflammatory stimuli is inhibited in the face of miR-200c as are baseline levels of these proteins. While the ability of miR-200c to repress NFκB function may also be due to miR-200c affecting upstream regulators of this pathway such as IKKB, data presented here and by others indicate that TDO2, HMOX-1 (46), IL-6 (54), and PD-L1 (28,29) are directly targeted by miR-200c. This insight into both the inflammation-mediated transcriptional and miRNA-mediated post-transcriptional regulation of specific immune-suppressive factors produced by TNBC cells may lead to new therapeutic strategies. During pregnancy, infection can trigger pre-term birth by activating the immune system, so dampening the immune response to infection and non-self antigens is advantageous in that setting. However, chronic inflammation may contribute to cancer by prompting epithelial cells to make immune suppressive factors such as those described here, which would impede the anti-tumor immune response. Furthermore, we previously documented that NFκB activity increased in TNBC cells under anchorage independent conditions (7,11) and indeed TDO2 and HMOX-1 increased in TNBC under these conditions as well (8,11). Thus, circulating tumor cells, which are able to resist the stress of anchorage independent survival during dissemination, may also be particularly able to generate immune suppressive metabolites that contribute to immune evasion. Interestingly, HMOX-1 emerged in a recent synthetic lethal screen for genes supporting metastasis in a mouse mammary carcinoma model (55).
In a recent comparison of two isogenic mammary carcinoma cell lines, the more mesenchymal line expressed higher PD-L1, lower MHC-1, and contained increased T regulatory cells and M2 macrophages in the tumor stroma (56); however, specific effectors or regulatory mechanisms or were not identified. The current study reveals multiple genes/proteins made by TNBC reminiscent of those used by trophoblasts to achieve fetal tolerance including TDO2, PD-L1/2, HMOX-1 and GDF-15. While future studies are necessary to determine how to utilize this information to reverse tumor-induced tolerance and re-activate the immune system, some of these enzymes (TDO2 and HO-1) and ligands (GDF-15) are targetable. The miR-200c family regulates the epigenetic modifier PcG protein Enhancer of Zeste Homolog 2 (EZH2), which affects the adaptive response to immunotherapies in melanoma (57). While other lines of evidence have linked EMT and immune suppression in cancer (26,56,58), our study functionally links miR-200c to an early embryonic program of immune suppressive factors that ensure fetal tolerance. Therapeutic strategies targeting these tumor-derived non-contact dependent immune suppressive factors may increase the efficacy of checkpoint inhibitors in TNBC.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the use of the University of Colorado Cancer Center/National Institute of Health/National Cancer Institute Cancer Core Support Grant P30 CA046934, specifically the Tissue Culture Core and the DNA Sequencing Core at the University of Colorado-Anschutz Medical Campus. We thank Lei Zhao, University of Iowa for technical expertise and Nicole Spoelstra and Jill Slansky, PhD, for valuable editing.
FINANCIAL SUPPORT
DOD BCRP Breakthrough Level 2 award W81XWH-15–1-0039 to JKR; CU Cancer Center’s Women’s Event/The Salah Foundation to JKR; F99 CA212230–01 NCI Pre to Postdoctoral Fellow Transition Award to TJR; NRSA T32 CA190216–01A1 to JLC; NRSA F31 CA203486–01A1 to LIG.
Footnotes
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest.
References
- 1.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008;26(8):1275–81 doi 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13(15 Pt 1):4429–34 doi 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the National Comprehensive Cancer Network. Cancer 2012;118(22):5463–72 doi 10.1002/cncr.27581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun 2017;8:15773 doi 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339(6119):580–4 doi 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol 1994;124(4):619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe EN, Cochrane DR, Cittelly DM, Richer JK. miR-200c targets a NF-kappaB up-regulated TrkB/NTF3 autocrine signaling loop to enhance anoikis sensitivity in triple negative breast cancer. PLoS One 2012;7(11):e49987 doi 10.1371/journal.pone.0049987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton VN, Christenson JL, Gordon MA, Greene LI, Rogers TJ, Butterfield K, et al. Androgen receptor supports an anchorage-independent, cancer stem cell-like population in triple-negative breast cancer. Cancer Res 2017. doi 10.1158/0008-5472.CAN-16-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009;461(7260):109–13 doi 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farris JC, Pifer PM, Zheng L, Gottlieb E, Denvir J, Frisch SM. Grainyhead-like 2 Reverses the Metabolic Changes Induced by the Oncogenic Epithelial-Mesenchymal Transition: Effects on Anoikis. Mol Cancer Res 2016;14(6):528–38 doi 10.1158/1541-7786.MCR-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Amato NC, Rogers TJ, Gordon MA, Greene LI, Cochrane DR, Spoelstra NS, et al. A TDO2-AhR Signaling Axis Facilitates Anoikis Resistance and Metastasis in Triple-Negative Breast Cancer. Cancer Res 2015;75(21):4651–64 doi 10.1158/0008-5472.CAN-15-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011;478(7368):197–203 doi 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 13.Novikov O, Wang Z, Stanford EA, Parks AJ, Ramirez-Cardenas A, Landesman E, et al. An Aryl Hydrocarbon Receptor-Mediated Amplification Loop That Enforces Cell Migration in ER-/PR-/Her2- Human Breast Cancer Cells. Mol Pharmacol 2016;90(5):674–88 doi 10.1124/mol.116.105361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol 2010;185(6):3190–8 doi 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ 2002;9(10):1069–77 doi 10.1038/sj.cdd.4401073. [DOI] [PubMed] [Google Scholar]
- 16.Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer 2010;1(6):306–19 doi 10.1007/s12672-010-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bockmeyer CL, Christgen M, Muller M, Fischer S, Ahrens P, Langer F, et al. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat 2011;130(3):735–45 doi 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 18.Castilla MA, Diaz-Martin J, Sarrio D, Romero-Perez L, Lopez-Garcia MA, Vieites B, et al. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS One 2012;7(10):e47709 doi 10.1371/journal.pone.0047709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10(5):593–601 doi 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 20.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22(7):894–907 doi 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia 2012;17(1):65–77 doi 10.1007/s10911-012-9244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries B, Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015;6(9):6472–98 doi 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piechowski J Trophoblastic-like transdifferentiation: A key to oncogenesis. Crit Rev Oncol Hematol 2016;101:1–11 doi 10.1016/j.critrevonc.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 24.Costanzo V, Bardelli A, Siena S, Abrignani S. Exploring the links between cancer and placenta development. Open Biol 2018;8(6) doi 10.1098/rsob.180081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holtan SG, Creedon DJ, Haluska P, Markovic SN. Cancer and pregnancy: parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin Proc 2009;84(11):985–1000 doi 10.1016/S0025-6196(11)60669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res 2016;22(3):609–20 doi 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knezevic J, Pfefferle AD, Petrovic I, Greene SB, Perou CM, Rosen JM. Expression of miR-200c in claudin-low breast cancer alters stem cell functionality, enhances chemosensitivity and reduces metastatic potential. Oncogene 2015;34(49):5997–6006 doi 10.1038/onc.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014;5:5241 doi 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 2017;6(1):e1263412 doi 10.1080/2162402X.2016.1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell 2010;39(5):761–72 doi 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, et al. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One 2010;5(1):e8697 doi 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, et al. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol 2009;114(3):457–64 doi 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Halldorsson S, Rohatgi N, Magnusdottir M, Choudhary KS, Gudjonsson T, Knutsen E, et al. Metabolic re-wiring of isogenic breast epithelial cell lines following epithelial to mesenchymal transition. Cancer Lett 2017;396:117–29 doi 10.1016/j.canlet.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki S, Tone S, Takikawa O, Kubo T, Kohno I, Minatogawa Y. Expression of indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase in early concepti. Biochem J 2001;355(Pt 2):425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Shin N, Koblish HK, Yang G, Wang Q, Wang K, et al. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood 2010;115(17):3520–30 doi 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- 36.Vacher S, Castagnet P, Chemlali W, Lallemand F, Meseure D, Pocard M, et al. High AHR expression in breast tumors correlates with expression of genes from several signaling pathways namely inflammation and endogenous tryptophan metabolism. PLoS One 2018;13(1):e0190619 doi 10.1371/journal.pone.0190619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilotte L, Larrieu P, Stroobant V, Colau D, Dolusic E, Frederick R, et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci U S A 2012;109(7):2497–502 doi 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene LI, Bruno T, Christenson JL, D’Alessandro A, Culp-Hill R, Torkko K, et al. A Role for Tryptophan-2,3-dioxygenase in CD8 T Cell Suppression and Evidence of Tryptophan Catabolism in Breast Cancer Patient Plasma. Mol Cancer Res (Accepted 2018); 10.1158/1541-7786.MCR-18-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ospina-Prieto S, Chaiwangyen W, Herrmann J, Groten T, Schleussner E, Markert UR, et al. MicroRNA-141 is upregulated in preeclamptic placentae and regulates trophoblast invasion and intercellular communication. Transl Res 2016;172:61–72 doi 10.1016/j.trsl.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Mellor AL, Sivakumar J, Chandler P, Smith K, Molina H, Mao D, et al. Prevention of T cell-driven complement activation and inflammation by tryptophan catabolism during pregnancy. Nat Immunol 2001;2(1):64–8 doi 10.1038/83183. [DOI] [PubMed] [Google Scholar]
- 41.Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol 2006;176(11):6752–61. [DOI] [PubMed] [Google Scholar]
- 42.Li DD, Gao YJ, Tian XC, Yang ZQ, Cao H, Zhang QL, et al. Differential expression and regulation of Tdo2 during mouse decidualization. J Endocrinol 2014;220(1):73–83 doi 10.1530/JOE-13-0429. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher A, Wafula PO, Teles A, El-Mousleh T, Linzke N, Zenclussen ML, et al. Blockage of heme oxygenase-1 abrogates the protective effect of regulatory T cells on murine pregnancy and promotes the maturation of dendritic cells. PLoS One 2012;7(8):e42301 doi 10.1371/journal.pone.0042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solano ME, Kowal MK, O’Rourke GE, Horst AK, Modest K, Plosch T, et al. Progesterone and HMOX-1 promote fetal growth by CD8+ T cell modulation. J Clin Invest 2015;125(4):1726–38 doi 10.1172/JCI68140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noh SJ, Bae JS, Jamiyandorj U, Park HS, Kwon KS, Jung SH, et al. Expression of nerve growth factor and heme oxygenase-1 predict poor survival of breast carcinoma patients. BMC Cancer 2013;13:516 doi 10.1186/1471-2407-13-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao C, Peng FH, Peng LK. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma 2014;61(6):680–9 doi 10.4149/neo_2014_083. [DOI] [PubMed] [Google Scholar]
- 47.Muliaditan T, Opzoomer JW, Caron J, Okesola M, Kosti P, Lall S, et al. Repurposing Tin Mesoporphyrin as an Immune Checkpoint Inhibitor Shows Therapeutic Efficacy in Preclinical Models of Cancer. Clin Cancer Res 2018;24(7):1617–28 doi 10.1158/1078-0432.CCR-17-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segerer SE, Rieger L, Kapp M, Dombrowski Y, Muller N, Dietl J, et al. MIC-1 (a multifunctional modulator of dendritic cell phenotype and function) is produced by decidual stromal cells and trophoblasts. Hum Reprod 2012;27(1):200–9 doi 10.1093/humrep/der358. [DOI] [PubMed] [Google Scholar]
- 49.Roth P, Junker M, Tritschler I, Mittelbronn M, Dombrowski Y, Breit SN, et al. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin Cancer Res 2010;16(15):3851–9 doi 10.1158/1078-0432.CCR-10-0705. [DOI] [PubMed] [Google Scholar]
- 50.Ratnam NM, Peterson JM, Talbert EE, Ladner KJ, Rajasekera PV, Schmidt CR, et al. NF-kappaB regulates GDF-15 to suppress macrophage surveillance during early tumor development. J Clin Invest 2017;127(10):3796–809 doi 10.1172/JCI91561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambert JR, Whitson RJ, Iczkowski KA, La Rosa FG, Smith ML, Wilson RS, et al. Reduced expression of GDF-15 is associated with atrophic inflammatory lesions of the prostate. Prostate 2015;75(3):255–65 doi 10.1002/pros.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2(3):261–8 doi 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 53.Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, et al. PD-L2 Expression in Human Tumors: Relevance to Anti-PD-1 Therapy in Cancer. Clin Cancer Res 2017;23(12):3158–67 doi 10.1158/1078-0432.CCR-16-1761. [DOI] [PubMed] [Google Scholar]
- 54.Hong L, Sharp T, Khorsand B, Fischer C, Eliason S, Salem A, et al. MicroRNA-200c Represses IL-6, IL-8, and CCL-5 Expression and Enhances Osteogenic Differentiation. PLoS One 2016;11(8):e0160915 doi 10.1371/journal.pone.0160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knott SRV, Wagenblast E, Khan S, Kim SY, Soto M, Wagner M, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 2018;554(7692):378–81 doi 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res 2017;77(15):3982–9 doi 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zingg D, Arenas-Ramirez N, Sahin D, Rosalia RA, Antunes AT, Haeusel J, et al. The Histone Methyltransferase Ezh2 Controls Mechanisms of Adaptive Resistance to Tumor Immunotherapy. Cell Rep 2017;20(4):854–67 doi 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Akalay I, Janji B, Hasmim M, Noman MZ, Andre F, De Cremoux P, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res 2013;73(8):2418–27 doi 10.1158/0008-5472.CAN-12-2432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


