Abstract
The androgen receptor (AR) is a key driver and therapeutic target in androgen-sensitive prostate cancer, castration resistant prostate cancer (CRPC), and CRPC resistant to abiraterone and enzalutamide, two second generation inhibitors of AR signaling. Since current AR inhibitors target a functioning C-terminal ligand binding domain (LBD), the identification and characterization of co-factors interacting with the N-terminal domain (NTD) of AR may lead to new approaches to target AR signaling in CRPC. Using a pulldown approach coupled with proteomics, we have identified Hsp70 as a co-factor for the NTD of AR in prostate cancer cells. Hsp70 inhibition using siRNA or small molecules indicated that Hsp70 played an important role in the expression and transactivation of endogenous AR. Prostate specific antigen (PSA) promoter/enhancer-driven luciferase assays showed that Hsp70 was also required for transactivation of AR mutant lacking LBD. Furthermore, clonogenic assays showed that an Hsp70 inhibitor, either alone or in synergy with enzalutamide, can inhibit the proliferation of 22Rv1, a widely-used enzalutamide-resistant CRPC prostate cancer cell line. These findings suggest that Hsp70 is a potential therapeutic target for the treatment of enzalutamide-resistant CRPC.
Keywords: Hsp70, AR, prostate cancer, Hsp70 inhibitor, castration resistant prostate cancer
INTRODUCTION
The androgen receptor (AR) is a member of the steroid receptor family of nuclear transcription factors and plays a key role in prostate cancer (PCa), from PCa initiation to progression to castration resistant prostate cancer (CRPC) (1). High AR expression levels in primary PCa have been correlated with increased proliferation, aggressiveness and worse prognosis (2,3). Thus, the AR signaling axis provides an excellent therapeutic target for PCa and androgen deprivation therapy (ADT) remains an effective treatment, either individually or in combination with other therapies, not only for patients with metastatic or recurrent PCa (4,5), but also for locally advanced cases (6). ADT is not curative and virtually all treated patients relapse and develop CRPC. In CRPC, AR is constitutively active, transactivates many AR-target genes, and drives cancer cell proliferation (7). Abiraterone and enzalutamide, two second generation AR signaling targeting agents, were developed for the treatment of CRPC (8–10) and these two new drugs can prolong survival of CRPC patients from 4–6 months (11–14). The development of resistance to abiraterone or enzalutamide is associated with elevated serum prostate specific antigen (PSA) in the majority of treated patients, suggesting that AR is activated again in CRPC tumors resistant to abiraterone or enzalutamide (15). New strategies of targeting AR signaling may lead to novel therapies for CRPC patients relapsed after treatment with abiraterone or enzalutamide.
AR contains a ligand binding domain (LBD) at the C-terminus, a DNA binding domain (DBD) linked to the LBD by a hinge region and an N-terminal domain (NTD) which is the least conserved of the four domains (16). The expression of AR splice variants (AR-Vs) such as AR-V7 is thought to contribute to the re-activation of AR signaling following abiraterone or enzalutamide treatment. The expression of AR-Vs is up-regulated in CRPC tissue samples as compared to hormone-dependent tumors (17–19). Also, androgen ablation up-regulates AR3/AR-V7 expression in prostate cancer cell lines (20). Since these AR variants lack the ligand binding domain (LBDAR), they are independent of androgens and insensitive to all agents targeting LBD directly or indirectly, including abiraterone and enzalutamide. One potential approach to inhibit ARvs is to target the N-terminal domain of AR (NTDAR). NTD can recruit co-factors that are essential for AR transactivation. Around 70 NTDAR co-factors have been reported (21). A more complete identification of NTDAR interacting proteins may lead to new approaches to target NTDAR.
Hsp70 family members are important chaperones that are required for protein folding, transport and degradation (22). The Hsp70-HSP90 chaperone machinery regulates about 20% of the eukaryotic proteins (23). In humans, eight Hsp70 isoforms share 52–99% amino acid sequence homology. They have some similar functions but also their own characteristics. Hsp70–1a and Hsp70–1b are almost identical (99% amino acid homology), encoded by genes HSPA1A and HSPA1B (Hsp70 refers to Hsp70–1a/b in this paper). They are mainly expressed in the cytosol, but stress can induce their nuclear localization (24). HSPA8 codes Hsc70 (86% homology to HSPA1A), which is constitutively expressed at a high level in both the cytosol and nucleus (25). Hsp70–6 (HSPA6) is the only other stress-induced isoform, which can be induced only under severe stress (26). The Hsp70–6 was reported to express in blood, dendritic cells and monocytes (25,27). Grp78 (HSPA5) is confined to endoplasmic reticulum (ER) (28). Hsp70–9 (HSPA9) only expresses in mitochondria (29). All Hsp70s contain an N-terminal nucleotide binding domain (NBD) and a C-terminal substrate binding domain (SBD), which is separated by a linker (30). NBD acts as an ATPase, while SBD binds to client protein. SBD possess a β-sheet sandwich to provide a hydrophobic pocket and a α-helices lid (31). When substrate is absent, ATP-bound NBD has a low ATPase activity. After substrate loosely binds to the SBD, the interaction between NBD and SBD is disrupted. J-proteins will then bind to the NBD to stimulate ATP hydrolysis (32,33). The ADP-bound NBD will cause the closing of the lid at the SBD and result in tight binding between SBD and substrate (34).
In this study, we stably transfected C4–2 prostate cancer cell line with GFP-tagged NTDAR expression vector, performed GFP pulldown and mass spectrometry analysis of co-factors associated with transfected GFP-NTDAR, and identified Hsp70 as a co-factor bound to NTDAR. Subsequently, we characterized the role of Hsp70 in AR action in prostate cancer cells.
MATERIALS AND METHODS
Plasmid construction
Human AR and its deletion mutants, amino acid 1–560 (N-terminal domain, NTD), 1–665 (NAR), 558–920 (DNA Binding domain, Hinge region and ligand-binding domain, DBDH-LBD), 668–920 (ligand-binding domain, LBD) coding sequence, were inserted separately into pEGFP-C1 vector, generating GFP-AR, GFP-NTDAR, GFP-NAR, GFP-DBDH-LBDAR, and GFP-LBDAR, GFP-ARa.a.1−293 and GFP-ARa.a.294−556 plasmids (35). Hsc70, Hsp70 and its Nucleotide Binding domain (NBD a.a. 1–386), and Substrate Binding domain (SBD a.a. 386–641) coding sequence were inserted into pCMV-3Tag-1A vector to make Flag-Hsc70, Flag-Hsp70, Flag-NBD and Flag-SBD plasmids. Hsp70 coding sequence was inserted into pGEX-2T plasmid, generating pGEX-Hsp70 plasmid.
Cell Culture and transfection
PC3, 22Rv1, LNCaP cell lines were purchased from ATCC (Manassas, VA, USA) and C4–2 cell line was a gift from Dr. Leland WK Chung. If not specially described, all cell lines were cultured at 37°C, 5% CO2 in RPMI 1640 media with 10% Fetal Bovine Serum (FBS), L-glutamine and penicillin-streptomycin. Cell lines 22Rv1, LNCaP and C4–2 were authenticated in 2016 using DNA fingerprinting by examining microsatellite loci in a multiplex PCR reaction (AmpFlSTR® Identifiler® PCR Amplification Kit, Applied Biosystems, Foster City, CA) by the University of Pittsburgh Cell Culture and Cytogenetics Facility. Cell line PC3 was obtained from ATCC in 2016. ATCC performed authentication for PC3 cell line using short tandem repeat profiling. Phenol-red free RPMI 1640 with 5% charcoal stripped FBS (CSS) was used as androgen free medium. PolyJet™ In Vitro DNA Transfection Reagent (SignaGen) was used for plasmids transfection according to manufacturer’s instruction.
RNA interference
Individual siRNAs specific to Hsp70 were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). The most effective sequence used was 5’-AGAUCAGCGAGGCGGACAAGAAGAA-3’ (identified as siHSPA1A). Hsp70 was knocked down in C4–2 cells plated in 6-well plates using the DharmaFECT™ Transfection Reagents (Formulation 3). All procedures followed the manufacturer’s instruction, with 50 nM (final concentration) siRNA and 10 μL reagent for each well. Cells were lysed after 72 hours of transfection.
Stable Cell Lines Generation
C4–2 cells were transfected with 2.0 μg GFP-NTD or pEGFP-C1 in 6-well plates at 70–80% confluency. After 24 hours of transfection, half of the cells from each well were reseeded in 10 cm dishes containing 10 ml regular RPMI1640 medium. After another 24 hours, original medium was replaced by RPMI1640 containing 1000 μg/ml G418 for selection of stably transfected cells. The selection medium was changed every week until the colonies formed were large enough to be picked up. One single colony was transferred to one well of a 96-well plate. As the colony expanded, it was reseeded to a 48-well, followed by a 12-well, then a 10 cm dish. The C4–2 GFP-NTD or GFP stable cell lines were monitored under fluorescence microscopy (Nikon Eclipse TS100, Prior Scientific Lumen 200 Fluorescence Illumination System) and confirmed by Western blotting.
Immunoblotting and Antibodies
Cells were lysed in RIPA buffer (150mM NaCl, 50mM Tris-HCl, 1% NP-40, 0.1%SDS, 0.5% Sodium Deoxycholate, pH8.0). Antibodies used included: Hsp70 (sc-1060, Santa Cruz Biotechnology, Dallas, TX, USA), AR (sc-816, Santa Cruz Biotechnology), γ-tubulin (sc-17787, Santa Cruz Biotechnology), GAPDH (FL-335, sc-25778, Santa Cruz Biotechnology), and secondary antibody IgG-HRP (sc-2004 sc-2005, Santa Cruz Biotechnology). Hsp90 (SPA-835-D), Hsp40 (SPA-400-D), HOP (SRA-1500-D), and Hsp27 (SPA-800D) antibodies were from Stressgen Bioreagents (Victoria, BC, CA); Flag antibody (F1804, St. Louis, MO, USA) was from SIGMA-ALDRICH and GFP antibody (TP401, Houston, TX, USA) was from Torrey Pines Biolabs. Most of the Western blotting membranes were developed by films. Some were observed by VersaDoc imaging system (4000 MP, Bio-Rad, Hercules, CA, USA) following the manufacture’s instruction. Densitometry was performed using Image J software (36). Band intensities were normalized to GAPDH or γ-tubulin individually.
Immunoprecipitation and mass spectrometry (MS) based label free quantitation
Immunoprecipitation was performed using lysates from the three C4–2 GFP-NTD cell lines and three GFP cells individually. For each sub-cell line, two confluent 15 cm dishes of cells were lysed in the lysis buffer (175 mM NaCl, 20 mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% Glycerol, 2 mM EDTA, pH7.5). Lysate was pre-cleaned by adding 50 μL Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) and rotated for 1 hour at 4 °C. A total volume of 20ul Anti-GFP mAb-Agarose (Medical & Biological Laboratories CO., LTD., Nagoya, Japan) was pre-blocked by 2.5% Albumin bovine/ fraction V, pH 7.0 for 1 hour before adding to the lysate. After 2 hours rotation, the anti-GFP agarose was separated from the lysate and washed by 1 ml lysis buffer containing 175 mM NaCl on the rotator for 10 min and repeated three more times with lysis buffer containing 250 mM NaCl. The agarose was then eluted by adding 20 μL Glycine-HCl, pH 2.5 and vortexing for 5 min. The process was repeated to get a total 40 μL elution, and 5% of the total elution was used for silver staining to verify the quality of immunoprecipitation. The silver staining was performed using the Pierce Silver Stain Kit (Cat # 24612, Thermo Scientific) following the instructions.
A total of six samples were analyzed by high accuracy mass spectrometer (LTQ/Orbitrap Velos) online coupled with reverse phase liquid chromatography as described (37) after the immunoprecipitation and isolation of the resultant tryptic peptides. Peptide identification and label free quantitation of identified proteins were carried out using MaxQuant/Andromeda software (38). The iBAQ value generated by MaxQuant, which is the summed precursor peptide intensity of a protein divided by the number of theoretically detectable, was used as label free quantitation metric (39). Selected proteins with increased abundance in NTD samples were verified using Skyline (40), an alternative software for examining full MS intensity of peptide ions for label free quantitation.
Co-Immunoprecipitation (co-IP)
PC3 or C4–2 prostate cancer cells were transfected with various plasmids for 36 hours. The cells were then lysed in the lysis buffer consisting of 250 mM NaCl, 20 mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% Glycerol, and 2 mM EDTA, pH7.5. Lysate was pre-cleaned by adding 50 μL Protein A/G PLUS-Agarose and 1-hour rotation at 4°C. Then, 8 μl anti-GFP mAb-Agarose (Medical & Biological Laboratories CO., LTD.) or 10 μL anti-Flag M2 affinity gel (SIGMA-ALDRICH) was pre-blocked using 2.5% albumin bovine/fraction V, pH 7.0 for 1 hour before adding to the lysate. After rotation for 2 hours, the anti-GFP agarose was separated from the lysate and washed 3 times with 1 ml lysis buffer on the rotator for 10 min. For endogenous co-IP, two confluent 150 mm dishes of 22RV1 cells were lysed in 1 mL lysis buffer (175 mM NaCl, 20 mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% Glycerol, 2 mM EDTA, pH 7.5), 3 μg Hsp70 antibody or normal mouse IgG (Santa Cruz Biotechnology, Inc.) was added to lysate for overnight incubation at 4 °C. Next, 70 μL of Protein A/G PLUS-Agarose was added to the lysate for an additional 5 hours of incubation at 4 °C. Then the Protein A/G PLUS-Agarose was separated from the lysate and washed by 1 mL 175 mM lysis buffer on the rotator for 10 min and repeated three more times. For fractionation, NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) were used to extract cytoplasmic protein. A total of 1 ml Cytoplasmic Extraction Reagent I (CER I) was used for one 15 cm dish according to manufacturer’s instructions. The nuclear extraction was obtained by adding 800 μl lysis buffer (175 mM NaCl, 20 mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% Glycerol, 2 mM EDTA, pH7.5) to the nuclear pellet. The same amount of total protein was used as input for both cytoplasmic and nuclear extract.
Glutathione S-Transferase (GST) fused protein purification and in vitro protein binding assay
GST tagged Hsp70 was expressed and purified as previously described (41) with some adjustment. BL21 competent cells were infected by pGEX2B or pGEX-Hsp70 and selected by ampicillin LB plate. One colony was picked up and incubated in 5 mL LB medium for overnight at 37 °C on a 250-rpm shaker. A 2 mL aliquot of the 5 ml total cell volume was diluted into 100 mL LB medium and cultured at 37 °C on a 250-rpm shaker until OD600 reached 0.6. IPTG was then added (final concentration 1 mM) and the solution was incubated at 29 °C on a 250-rpm rotator for 3 hours. Cells were lysed in lysis buffer (175 mM NaCl, 20mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% Glycerol, 2 mM EDTA, pH7.5) and sonicated at 15% power for 10 sounds for 3 times. Pierce Glutathione Spin Columns (0.2 mL, Thermo Scientific) were used to purify GST and GST-Hsp70 followed manufacturer’s instruction. Recombinant human androgen receptor protein was purchased from SIGMA-ALDRICH. 30 ng AR and 30 ng GST or GST-Hsp70 were used for in vitro binding assay. Binding buffer (1.5 mM Tris-HCl, 1 mM Sodium Molybdate, 10% Glycerol, 1mM EDTA, pH 7.5) was added with Protease Inhibitor Cocktails and Albumin bovine/ fraction V, pH 7.0 (final 2.5%). The proteins were incubated at 4 °C for overnight on rotator. Pierce Glutathione Agarose was used to pull down GST or GST-HSp70 following the manufacturer’s instruction.
Inhibitors and Antagonist
Hsp90 inhibitor 17-AAG (Tanespimycin, see chemical structure in (42)) was purchased from LC Laboratories (Woburn, MA, USA), AR antagonist MDV3100 (Enzalutamide) from Selleck Chemicals. Hsp70 inhibitor VER-155008 (see chemical structure in (43)) was purchased from SIGMA-ALDRICH.. The novel Hsp70 inhibitor UPCMLD18BBQU015254 (18BBQU) was synthesized as previously reported (44–46).
PSA luciferase assay
The PSA luciferase vector containing 6.1kb PSA promoter region was a generous gift from Dr. Marianne Sadar. TK promoter driven green Renilla luciferase vector was purchased from Thermo Scientific (Rockford, IL). In some experiments in this study, PSA luciferase plasmid and Renilla plasmid were transiently transfected into cells. We also generated the C4–2 cell line stably transfected with both the PSA luciferase vector and Renilla luciferase vector (47). Luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega). All procedures followed the manufacturer’s instructions.
Quantitative Real-time PCR
Total RNA was extracted using the TRIzol RNA isolation system (Invitrogen, Carlsbad, CA). To synthesize first strand cDNA, reverse transcription PCR was performed using 2 μg of RNA in 20 μL of reaction buffer using GoScriptTM Reverse Transcription System (Promega, Madison, WI) by following the suggested protocol. The quantitative real-time PCR was performed using ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) with 2x SYBR Green PCR Master Mix (Thermo Scientific). The mRNA expression levels of various genes were calculated by normalizing to β-actin expression level. The primer sequences were as follows: AR, TGGATGGATAGCTACTCCGG and CCCAGAAGCTTCATCTCCAC; PSA, AGGCCTTCCCTGTACACCAA and GTCTTGGCCTGGTCATTTCC; β-actin, AGGCATCCTCACCCTGAAGTA and CACACGCAGCTCATTGTAGA, hK2, CTGTCAGAGCCTGCCAAGAT and GCAAGAACTCCTCTGGTTCG; TMPRSS2, CTGCCAAGGTGCTTCTCATT and CTGTCACCCTGGCAAGAATC.
Clonogenic assay
Prostate cancer cell line 22Rv1 was plated at a density of ~1500 cells per 10 cm dish. Cells were treated with DMSO or Hsp70 inhibitor, either VER-155008 or 18BBQU, at indicated concentrations (0 – 3 μM VER-155008, or 0 – 50 μM 18BBQU), in the presence of 0, 5 μM, or 10 μM of MDV3100 (Enzalutamide). The media was changed every five days for 14 days. The colonies were rinsed with PBS before fixation using acetic acid/methanol 1:7 (v/v). Crystal violet solution (0.5%) was used for staining. ImageJ (36) was used to count colony number followed suggested protocol.
RESULTS
Proteomics identification of Hsp70 family proteins as NTDAR-interacting factors
To identify proteins associated with NTDAR, we stably transfected C4–2 prostate cancer cells with GFP-NTDAR or GFP expression vector and established three C4–2-GFP-NTDAR sublines and three C4–2-GFP sublines (Figure 1A and 1B). The expression of GFP-NTDAR and GFP proteins in the stable sublines were analyzed and confirmed using fluorescent microscopy and Western blot. The proteins bound to GFP-NTDAR were pulled down individually from total cell lysates of three different C4–2-GFP-NTDAR sublines using anti-GFP beads. The three C4–2-GFP sublines were used as controls in the parallel pull-down experiments. SDS-PAGE followed by silver staining revealed proteins bands in all three independent GFP-NTDAR pull-down samples, with some of the strong bands having molecular weight around 50–70 kDa (Figure 1C). Mass spectrometry analysis identified Hsp70 as the most abundant protein pulled down by GFP-NTDAR and not by GFP in the C4–2 lysates.
Figure 1.
Identification of Hsp70 and Hsc70 as co-factors bound to GFP-NTDAR in C4–2 cells. A. Representative C4–2-GFP-NTDAR and C4–2-GFP stable sublines were imaged by fluorescence microscopy. B. Western blot analysis of AR, GFP-NTDAR, and GFP expression in 3 C4–2-GFP and 3 C4–2-GFP-NTDAR sublines. AR antibody detects both endogenous AR and transfected GFP-NTDAR. C. Silver staining of proteins precipitated from total cell lysates of 3 C4–2-GFP and 3 C4–2-GFP-NTDAR sublines using anti-GFP beads. D. Skyline analysis for Hsp70 and Hsc70 in data from mass spectrometry. Hsp70 (HSPA1A) and Hsc70 (HSPA8) showed an increase of peptide intensity greater than 50-fold. Each peptide’s intensity (peak area) in each of the 6 samples was shown as the bar.
Database searching of all six raw files against human uniprot database using the Andromeda search engine (38) led to the identification of a total of 183 proteins (including GFP) after filtering out common contaminating proteins such as keratins. We then applied label-free quantitation to compare protein abundance between GFP-NTDAR and GFP control using MaxQuant software. Among these 182 proteins (excluding GFP), a total of 34 proteins (Supplemental Table S1) matched the following criteria: (1) abundance in all three GFP-NTDAR samples was higher than in all three GFP samples; (2) fold change greater than 3 comparing mean iBAQ values of GFP-NTDAR vs. GFP; (3) at least two peptides were identified. Among these 34 proteins, five were Hsp70 isoforms. Tubulin, a previously reported NTDAR co-factor, was also found (48). Compared to the known co-factor or other co-factors among the 34 proteins, the Hsp70 family had a higher number of peptides recognized and much higher fold change in relative abundance. There were also many SWI/SNF complex subunits and related proteins in the 34 proteins.
The relative abundance of some selected proteins, including the five Hsp70s, was further verified using Skyline (40). Extraction of full MS intensities using Skyline required more manual examination and thus the results could be considered more accurate. We started our analysis on Hsp70 (HSPA1A) and Hsc70 (HSPA8) because these two members of the Hsp70 family are among the most characterized. The relative abundance of peptides belonging to Hsp70 and Hsc70 is shown in bar graphs in Figure 1D. For every peptide, the abundance was much higher in each of the triplicate NTD samples than in the GFP samples. Although these two proteins share 86% homology, the peptides selected were unique and non-overlapping to each protein (Supplemental Figure 1). Table 1 shows the mean abundance of each peptide and enrichment fold in the NTD samples as compared to the GFP controls. The other Hsp70 family members were also included in Table 1.
Table 1.
Mean full MS intensity of peptide ions for label free quantitation of Hsp70 family
| Gene | Peptide | GFP | NTD | FoldChange |
|---|---|---|---|---|
| HSPA1A | FGDPVVQSDMK | 1.56E+06 | 1.82E+08 | 116.94 |
| LVNHFVEEFK | 1.95E+06 | 1.18E+08 | 60.22 | |
| NQVALNPQNTVFDAK | 6.85E+06 | 4.60E+08 | 67.17 | |
| HSPA8 | NSLESYAFNMK | 1.78E+07 | 5.84E+08 | 32.73 |
| SQIHDIVLVGGSTR | 4.98E+06 | 3.26E+08 | 65.47 | |
| VC[+57]NPIITK | 1.92E+07 | 1.05E+09 | 54.57 | |
| HSPA5 | ELEEIVQPIISK | 2.84E+07 | 2.66E+08 | 9.39 |
| ITPSYVAFTPEGER | 3.83E+07 | 2.68E+08 | 6.99 | |
| NQLTSNPENTVFDAK | 6.20E+07 | 5.25E+08 | 8.46 | |
| SQIFSTASDNQPTVTIK | 3.21E+07 | 2.59E+08 | 8.08 | |
| HSPA9 | DAGQISGLNVLR | 8.75E+06 | 1.84E+08 | 21.03 |
| QAVTNPNNTFYATK | 4.90E+06 | 2.58E+08 | 52.65 | |
| TTPSVVAFTADGER | 1.05E+07 | 2.89E+08 | 27.48 | |
| VQQTVQDLFGR | 8.40E+06 | 1.91E+08 | 22.77 | |
| HSPA6 | ATAGDTHLGGEDFDNR | 2.74E+06 | 2.37E+08 | 86.52 |
| IINEPTAAAIAYGLDR | 7.65E+05 | 1.71E+08 | 223.60 |
Co-IP of AR with Hsp70 is more efficient than with Hsc70
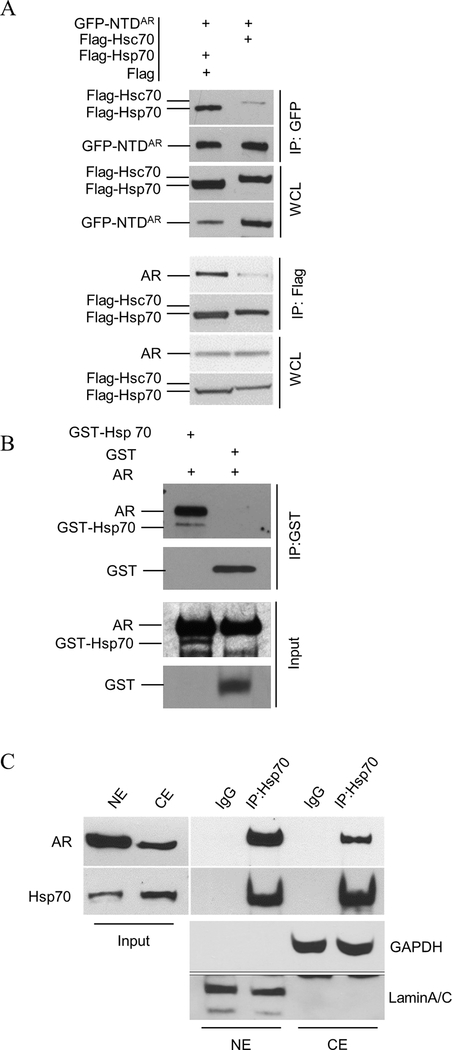
The proteomics analysis identified both Hsp70 and Hsc70 as co-factors associated with GFP-NTDAR. To compare Hsp70 and Hsc70 in their interactions with AR, we performed a co-IP analysis of AR and Hsp70 or Hsc70, separately. As expected, both Hsp70 and Hsc70 were co-immunoprecipitated with GFP-NTDAR or AR (Figure 2A). Hsp70 was pulled down more efficiently than Hsc70 by the anti-GFP antibody in the presence of GFP-NTDAR. In addition, AR was pulled down more efficiently by Flag-Hsp70 than by Flag-Hsc70. These observations suggest that Hsp70 was more efficient and/or stable than Hsc70 in the interaction with AR.
Figure 2.
Verification of the interaction between the Hsp70 family and AR. A. Flag-Hsp70 or Flag-Hsc70 was transfected with GFP-NTDAR into PC3 cells. In the lower panel, C4–2 cells were transfected with Flag-Hsp70 or Flag-Hsc70. Whole cell lysates (WCL) were prepared for co-IP using anti-GFP or Flag agarose beads as indicated. Flag vector was added to the transfection of Flag-Hsp70 group but not to the Flag-Hsc70 group. Less flag-Hsp70 vector was used due to the robust Flag-Hsp70 expression; flag vector was added such that equal amount of plasmids was added in each transfection reaction. B. GST and GST-Hsp70 purified from E. coli were used in the GST-pulldown of recombinant human androgen receptor. GST or GST-Hsp70 was mixed with human AR protein and then subjected to pulldown using glutathione-conjugated agarose. C. C4–2 cells were treated with 1 nM R1881 for 24 hours before fractionation. A total of 4 dishes were lysed. The nuclear extraction was obtained by adding 800 μL lysis buffer (175mM NaCl, 20 mM Tris-HCl, 1.5 mM MgCl2, 0.5% NP-40, 15% glycerol, 2mM EDTA, pH7.5) to nuclear pellet of 4 dishes. Equal total protein amounts were used as input for both cytoplasmic (CE) and nuclear extract (NE).
We next performed an in vitro binding assay to test if Hsp70 could directly bind to AR. Using commercially available AR protein and GST-Hsp70 fusion protein purified from E. coli, Figure 2B shows that AR was pulled down with GST-Hsp70, indicating direct binding of Hsp70 to AR.
We also investigated whether Hsp70 binding to AR could occur in the nucleus and/or cytoplasm using the C4–2 prostate cancer cell line as a model. Cytoplasmic and nuclear extractions were used separately in co-immunoprecipitations. Figure 2C showed that AR and Hsp70 could co-immunoprecipitate in both the nucleus and cytoplasm.
The SBD of Hsp70 interacts with the NTD of AR
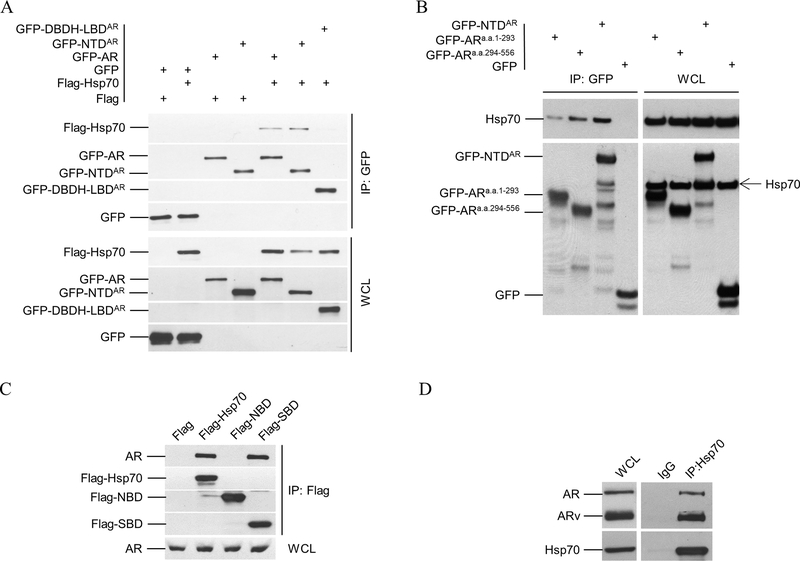
To define AR domain(s) interacting with Hsp70, we co-transfected Flag- Hsp70 with GFP-tagged AR or truncated AR vectors into PC-3 or C4–2 prostate cancer cells for co-IP. Figure 3A shows that GFP-AR and GFP-NTDAR, but not GFP or GFP-DBDH-LBDAR, could be immunoprecipitated by Flag-Hsp70 in PC-3 cells. Figure 3B shows that GFP-NTDAR co-immunoprecipitates with endogenous Hsp70 in C4–2 cells. Furthermore, Figure 3B indicates that both the N-terminal portion (a.a. 1–293) and the C-terminal portion (a.a. 294–556) of NTDAR could co-immunoprecipitate with Hsp70, with the a.a. 294–556 region being more effective than the a.a. 1–293 sequence in pulling down Hsp70. Both the a.a. 294–556 and a.a. 1–293 regions are less effective than the intact NTDAR in the co-IP experiment. These observations suggested that Hsp70 binds to AR through NTDAR, with the a.a. 294–556 being the most important region for the binding. We have previously shown that region a.a. 294–556 is required for androgen-independent AR nuclear localization in CRPC cells (35). The binding of Hsp70 to this region of AR may suggest a role for Hsp70 in promoting androgen-independent AR nuclear localization in CRPC cells, however further studies will be required to fully elucidate the significance of Hsp70 binding to the C-terminal portion (a.a. 294–556) of NTDAR.
Figure 3.
Co-immunoprecipitation (co-IP) analysis of Hsp70 and AR interaction. A. Flag-tagged Hsp70 was co-transfected with GFP-tagged AR or its deletion mutants into PC3 cells. Whole cell lysates (WCL) were prepared for co-IP using anti-GFP beads. Both WCL and immnoprecipitates were analyzed by immunoblotting using anti-Flag antibody and then anti-GFP antibody. B. C4–2 cells were transfected with expression vectors for GFP-tagged NTD, AR(1–293), and AR(294–556), along with GFP control. Co-IP were performed similarly as in A. The WCL and immnoprecipitates were analyzed by immunoblotting using anti-Hsp 70 antibody and then anti-GFP antibody. C. Expression vectors for GFP-tagged Nucleotide Binding domain (NBD a.a. 1–386) and Substrate Binding Domain (SBD a.a. 386–641) of Hsp70 were transfected into C4–2 cells separately. Co-IP and immunoblotting were performed similarly as in A using anti-Flag and anti-AR antibodies. D. 22Rv1 cell lysates were prepare for co-IP using anti-Hsp70 antibody or IgG. 22Rv1 cells express full-length AR and its splice variants, including AR-V7 that lacks LBD. The WCL and immnoprecipitates were analyzed by immunoblotting using anti-AR antibody and then anti-Hsp70 antibody. All experiments were reproduced at least twice.
To determine the Hsp70 domain(s) responsible for binding to AR, we transfected Hsp70 and its two domains NBD and SBD into C4–2 cells. Figure 3C shows that Flag-Hsp70 and Flag-SBD, but not Flag-NBD, co-immunoprecipitated endogenous AR. We also tested the co-immunoprecipitation of SBD and AR truncated mutations (Supplemental Figure 2). As expected, NTD and NAR constructs, but not DBDH-LBD and LBD, could bind to the SBD of Hsp70, which further suggested that the interaction between Hsp70 and AR was mediated through the SBD of Hsp70 and the NTD of AR, and was independent of LBD.
To further demonstrate that binding of Hsp70 to AR in vivo did not require the LBD, we performed immunoprecipitation with antibody against Hsp70 in 22Rv1 cells, which express both full-length AR and AR splice variants (49–51). The predominant AR splice variant in 22Rv1 is AR-V7 that lacks the LBD. In Figure 3D, the anti-Hsp70 antibody also precipitated both full-length AR and AR-Vs, indicating that the LBD was not required for AR binding to Hsp70.
Hsp70 inhibition down-regulated endogenous, but not exogenous, AR expression
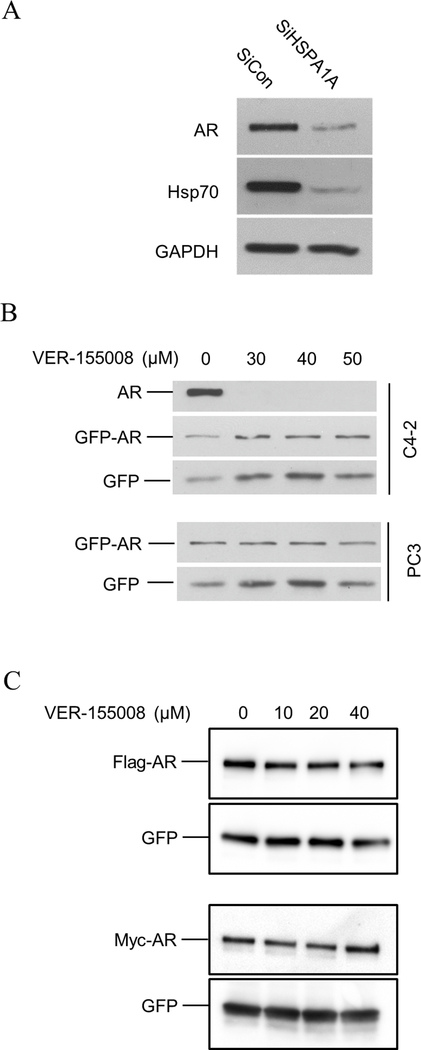
To investigate the role of Hsp70 in regulating AR function, we first performed a Hsp70 knockdown in C4–2 cells. AR protein levels were decreased when Hsp70 was reduced by siRNA (Figure 4A). Similarly, in LNCaP cells, the AR protein was down-regulated when Hsp70 was down-regulated in the presence of an increasing dosage of siRNA targeting Hsp70 (Supplemental Figure 3).
Figure 4.
Effect of Hsp70 knockdown or inhibition on endogenous and transfected AR expression. A. Hsp70 was knocked down by HSPA1A siRNA in C4–2 cells. B. AR-positive C4–2 and AR-negative PC3 cells were transiently transfected with GFP-AR along with GFP as a control. Forty-eight hours after the transfection, the cells were treated with different dose of VER-155008 for 24 hours prior to harvest for Western blot analysis using anti-AR or anti-GFP antibody. C. C4–2 were transfected with Flag-AR or Myc-AR along with GFP expression vector as a control. Twenty-four hours after the transfection, the cells were treated with different dose of VER-155008 for 24 hours prior to harvest for Western blot analysis using anti-Flag, anti-Myc, or anti-GFP antibody.
We transfected GFP-AR to C4–2 and PC3 cells to test the impact of Hsp70 inhibition on transfected GFP-AR using the commercially available Hsp70 inhibitor VER-155008 (Sigma-Aldrich). VER-155008 is a small molecule, ATP-competitive inhibitor of Hsp70 (IC50 = 500 nM) (52). To our surprise, GFP-AR did not decrease as the endogenous AR did when C4–2 cells were treated with 30–50 μM VER-155008 for 24 hours (Figure 4B). Similarly, the GFP-AR level was not reduced by VER-155008 treatment in transfected PC3 cells (Figure 4B). To rule out the potential impact of the GFP tag on AR stability upon VER-155008 treatment, we also tested the effect of VER-155008 on transfected Flag-AR and Myc-AR in C4–2 cells. VER-155008 did not affect Flag-AR or Myc-AR level in C4–2 cells (Figure 4C).
Hsp70 inhibition reduces transcriptional activity of AR and truncated AR lacking LBD
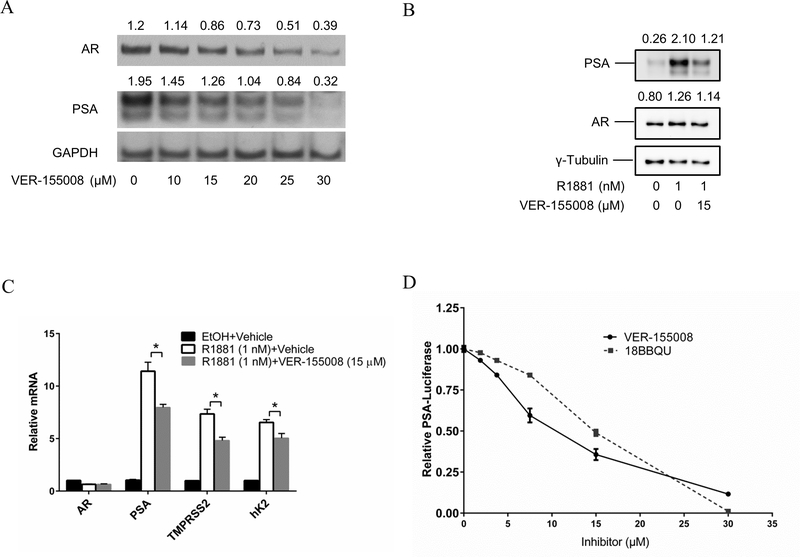
To evaluate the effect of Hsp70 inhibition on AR activity, we tested whether the Hsp70 inhibitor VER-155008 could decrease the AR-target gene PSA expression in C4–2 cells. As expected, a Western blot showed that VER-155008 induced a dose-dependent down-regulation of AR (Figure 5A). However, VER-155008 induced a more profound down-regulation of PSA (Figure 5A), suggesting that VER-155008 not only inhibits AR expression but could also inhibit AR transcriptional activity.
Figure 5.
Effect of Hsp70 inhibition on AR transcriptional activity. A. C4–2 cells cultured in RPMI1640 with 5% CSS were treated with Hsp70 inhibitor VER-155008 at the indicated concentrations, in the presence of 1 nM R1881. Cell lysates were prepared 24 hours after the treatment and then analyzed by Western blotting using anti-AR, anti-PSA, and anti-GAPDH antibodies. B. Protein expression in C4–2 cells cultured in 5 % CSS RPMI 1640 media were treated with vehicle, 1 nM R1881, or 1 nM R1881 plus 15 μM VER-155008 for 72 hours, then analyzed by western blotting using anti-AR, anti-PSA, and anti-γ-Tubulin antibodies. C. Relative mRNA expression in C4–2 cells treated as in B, then harvested for quantitative real-time PCR analysis. D. C4–2 cells stably transfected with both the PSA-luciferase plasmid and Renilla plasmid (47) were treated with either VER-155008 or another Hsp70 inhibitor, 18BBQU (UPCMLD18BBQU015254), in the presence of 1 nM R1881 for 24 hours. Luciferase assays were performed as described previously (66). To verify 18BBQU impact on AR protein level, C4–2 cells treated with 18BBQU in parallel were tested by immunoblotting using anti-AR antibody (Figure S4).
To further test whether Hsp70 could inhibit AR transcription activity, we investigated whether PSA expression at both protein and mRNA levels could be inhibited in C4–2 cells by 15 μM VER-155008, a dosage that had no significant impact on the AR protein level (see Figure 5A). Figure 5B shows that the R1881 induction of PSA protein could be inhibited by 15 μM VER-155008, a dose that did not reduce the AR protein level. Figure 5C shows that the R1881 induction of PSA mRNA was also inhibited by 15 μM VER-155008, along with the inhibition of two additional AR-target genes, TMPRSS2 and hK2, at the mRNA level. Further experiments using a PSA promoter-based luciferase assay also showed a dose-dependent VER-155008 inhibition of the AR transactivation of the PSA promoter (Figure 5D). We then tested the effect of another Hsp70 inhibitor, UPCMLD18BBQU015254 (18BBQU), in the PSA promoter-based luciferase assay. Different from VER-155008, which is an active site ATP-competitive inhibitor, 18BBQU inhibits Hsp70 by competing for the binding of Hsp40 to Hsp70 (53). Figure 5D shows that 18BBQU also inhibited the PSA promoter-driven luciferase activity with a similar dose response. As a control, we showed that 15 μM 18BBQU did not inhibit the AR protein level in C4–2 cells (Supplemental Figure 4).
VER-155008 inhibits the NAR transactivation of the PSA promoter-driven luciferase activity
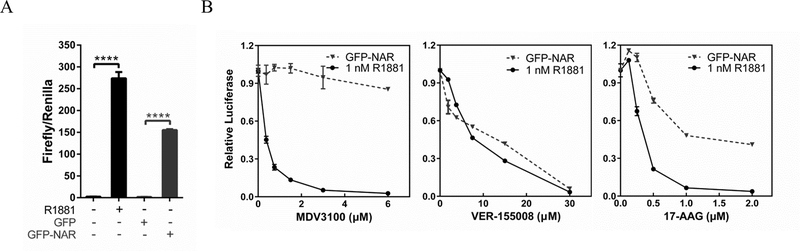
The Hsp70 binding to NTDAR suggests that Hsp70 inhibition of AR transcriptional activity is likely mediated through the NTDAR, instead of the LBD. Since AR splice variants contain NTDAR while lacking a LBD, targeting Hsp70 may also inhibit AR splice variants. GFP-NAR (AR without LBD) behaves the same as the AR splice variants in transactivation of AR-target genes (54), providing a good model for AR splice variants lacking the LBD. Untransfected C4–2 cells exhibited very low PSA promoter-driven luciferase activity under androgen-free conditions, and a high activity upon addition of 1 nM R1881 (Figure 6A). In the absence of androgens, C4–2 cells transfected with GFP also exhibited very low PSA-luciferase activity whereas the cells transfected with GFP-NAR displayed robust luciferase activity (Figure 6A), indicating that GFP-NAR could transactivate the PSA promoter in the absence of androgens. As expected, MDV3100 inhibited R1881-induced but not GFP-NAR-driven PSA-luciferase activity in C4–2 cells (Figure 6B). In contrast, VER-155008 inhibited R1881-induced and GFP-NAR-driven PSA-luciferase activities similarly and effectively (Figure 6B). We also tested if inhibition of another heat shock protein, Hsp90, could influence GFP-NAR transcriptional activity. Figure 6B shows that the Hsp90 inhibitor 17-AAG at 1 μM or higher doses effectively inhibited R1881-induced, but only partially inhibited GFP-NAR-driven, PSA-luciferase activities. These observations suggest that Hsp70 modulation of AR activity is distinctly different from Hsp90 regulation of AR.
Figure 6.
VER-155008 inhibition of the transcriptional activity of GFP-NAR (mutant AR lacking LBD) in PSA promoter-driven luciferase assay. A. To determine GFP-NAR transcriptional activities, C4–2 cells were transfected with PSA 6.0 luciferase and Renilla plasmids, along with GFP or GFP-NAR, in RPMI1640 with 5% CSS for 48 hours and then the luciferase activities were measured. As controls, C4–2 cells were transfected with PSA 6.0 luciferase plasmid and Renilla plasmid in RPMI1640 with 5% CSS for 24 hours and then treated with 1 nM R1881 or vehicle for another 24 hours prior to luciferase assay. B. The luciferase assays in A were used to test the effects of VER-155008, 17-AAG, or MDV3100 on the transcriptional activity of androgen-induced endogenous AR or the transfected GFP-NAR. Twenty-four hours after the transfection, C4–2 cells were cultured for another 24 hours in the presence of 1 nM R1881 or ethanol vehicle plus indicated doses of VER-155008, 17-AAG or MDV3100 prior to harvest.
An Hsp70 inhibitor, either alone or in combination with enzalutamide, could inhibit 22Rv1 cell growth
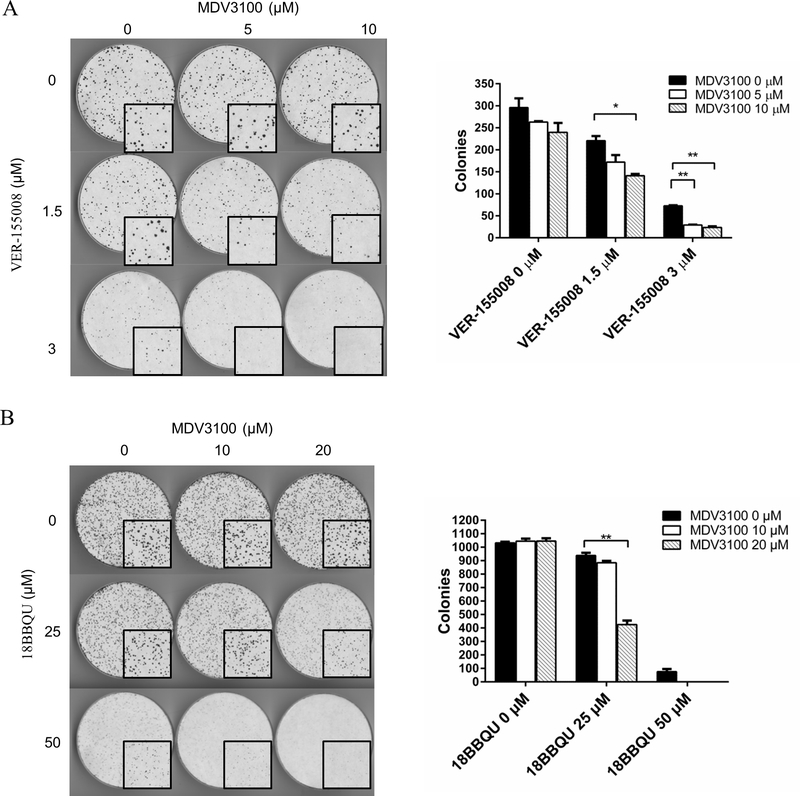
The prostate cancer cell line 22Rv1 is resistant to MDV3100, and this resistance is thought to be due to its expression of AR splice variants in addition to the full-length AR (55). Since Hsp70 can bind to the NTDAR (see Figure 3) and a Hsp70 inhibitor inhibited GFP-NARAR (AR lacking LBD) transcriptional activity (see Figure 5), targeting the NTDAR via Hsp70 inhibition may synergize with LBDAR targeting AR-antagonists such as enzalutamide. Enzalutamide alone had no significant inhibition of 22Rv1 colony formation, whereas it inhibited the colony formation in the presence of VER-155008 (Figure 7A). The inhibition of 22Rv1 colony formation by Hsp70 inhibitor combined with enzalutamide was greater than the sum of the inhibition of 22Rv1 colony formation by Hsp70 inhibitor and enzalutamide individually, suggesting an enhanced inhibition of growth by the combination. Similar results were obtained when a more selective Hsp70 inhibitor, 18BBQU, was used instead of VER-155008 in the 22Rv1 colony formation assay (Figure 7B). These observations suggested that Hsp70 inhibition can potentiate enzalutamide to inhibit 22Rv1 cell growth.
Figure 7.
Hsp70 inhibitor inhibits the growth and enhances the sensitivity to MDV3100 of 22Rv1 cells. Cells were plated at equal density (1500 cells/10 cm dish) and treated with vehicle, VER-155008 (A) or 18BBQU (B), in the absence or presence of MDV3100 at the indicated concentrations. The media was changed every 5 days for 14 days. Image was scanned at 300 dpi in black and white for colony quantification using ImageJ. The required colony size was set as 20 pixel^2. The quantitative data are shown at the right side of the images.
DISCUSSION
The studies presented herein identified Hsp70 as an important AR co-factor that could bind to the NTD of AR through its substrate-binding domain. Hsp70 inhibition inhibited transactivation of both full-length AR and mutant AR lacking the LBD. Furthermore, Hsp70 inhibition synergized with MDV3100 in the inhibition of 22Rv1 cell growth in the colony formation assay. These findings suggest that Hsp70 is an important regulator of AR in prostate cancer cells.
Chaperones are required for a proper functioning of steroid receptors, including AR. Most previous studies have addressed the interactions between chaperones and the ligand-binding domain (LBD) of steroid receptors. However, consistent with our findings that Hsp70 can interact with the AR NTD directly, Hsp70 inhibitor was recently shown to inhibit the level and transcriptional activity of both AR and ARv7 (56) supporting a ligand-independent role for Hsp40/Hsp70 chaperones in the regulation of AR. Hsp70 was shown to enhance the degradation of expanded polyglutamine repeat AR in murine motor neuron hybrid cells (57), also suggesting Hsp70 interaction with the N-terminal domain of AR where polyglutamine repeat is localized. Others have observed that a conserved motif in the NTD could interact with the COOH terminus of the Hsp70-interacting protein (CHIP), which promotes AR degradation, thereby negatively regulating AR transcriptional activity (58). The NTD structure is characterized as “regions of intrinsic disorder” with limited stable secondary structure that conforms to a “collapsed disordered conformation” (59). Hsp70 may act as a chaperone to facilitate the proper folding of the NTD, such that it can interact with other AR co-factors more efficiently. Hsp70 was reported to also influence the LBD (60); however, we did not detect their interactions via co-IP in this study. More studies will be needed to define the mechanisms of Hsp70 modulation of AR function.
In addition to direct binding to AR, Hsp70 also can regulate expression of endogenous AR in prostate cancer cells (see Figure 4). Since Hsp70 inhibition did not inhibit exogenous AR expression under the control of the CMV promoter, Hsp70 is unlikely to regulate AR at the protein level. Another possibility for Hsp70 regulation of AR expression is that Hsp70 inhibition can result in the inhibition of endogenous AR mRNA expression. A recent study reported the effect of Hsp70 regulation of the AR promoter through the activation and nuclear translocation of YB-1 (61). This finding is consistent with our observation that Hsp70 inhibitor inhibits endogenous AR but not the transfected AR under CMV promoter control.
Hsp70 appears to be an excellent target for inhibiting AR-positive prostate cancer cells. Hsp70 inhibition could cause a reduction of endogenous AR expression as well as inhibition of AR transcriptional activity (see Figures 4, 5 and 6). Hsp70 inhibition could inhibit both full-length AR and AR lacking the LBD, since Hsp70 interacts with the NTD of AR. Current prostate cancer therapies such as enzalutamide inhibit AR function through the AR ligand-binding domain (LBD) (62). The expression of AR variants, which lake the LBD, are also likely to be one major mechanism in the development of enzalutamide resistance in prostate cancer (63). Our results suggest that Hsp70 inhibitors can confer enzalutamide inhibition of 22Rv1, a cell line that expresses AR splice variants and is resistant to enzalutamide (see Figure 7). Therefore, the development of Hsp70 small molecule inhibitors which could inhibit both full-length AR and its splice variants may lead to new therapies for prostate cancer resistant to enzalutamide, which is at present a major clinical challenge.
Hsp70 inhibition may also inhibit AR-independent pathways in prostate cancer cells, because Hsp70 acts as a chaperone for many proteins. For instance, galeterone, which is an AR depleting agent has also been shown to activate the unfolded protein response and induce apoptosis in AR-negative prostate cancer cell lines (64). Recently galeterone analogs were shown to interact with Hsp70 and could induce apoptosis even in AR-negative PC3 cells, potentially through the induction of sustained endoplasmic reticulum stress and deregulated calcium homeostasis (65). The inhibition of AR-independent signaling may enhance the inhibitory effect of Hsp70 inhibitors in prostate cancer cells. However, strong inhibition of this chaperone may also cause potential toxicity to prostate cancer patients. Hsp70 inhibitor alone significantly inhibited colony formation in 22Rv1 cells, suggesting that Hsp70 inhibition was cytotoxic (see Figure 7). It will be important to determine if a therapeutic window can be achieved for Hsp70 inhibitors in the treatment of prostate cancer. Ideally, small molecules that can specifically block Hsp70 binding to AR, if they can be developed, will be less toxic to patients, but more research will be needed to explore the interactions between Hsp70 and AR and the possibilities to block these interactions.
In summary, our study identified Hsp70 as an important co-factor that binds to the NTD of AR and suggests that targeting Hsp70 can inhibit AR signaling in prostate cancer cells, including those resistant to enzalutamide. The development of novel Hsp70 inhibitors may lead to new treatments for prostate cancer patients resistant to the current portfolio of anti-androgen therapies.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yifeng Jing, Erica Parrinello, Katherine Willett and Aiyuan Zhang for technical support, and members of the Wang Lab for discussion. This work was funded in part by NIH grants NIH R01 CA186780 (Z. Wang), NIH 1P50 CA180995, and NIH R50 CA211242 (L.E. Pascal). The proteomics analysis was performed in the Cancer Proteomics Facility (CPF) at the University of Pittsburgh and was supported in part by award P30 CA047904.
Author’s financial support:
Zhou Wang, National Cancer Institute, R01 CA186780 wangz2@upmc.edu
Zhou Wang, National Cancer Institute, P50 CA180995 wangz2@upmc.edu
Laura E. Pascal, National Cancer Institute, R50 CA211242 pascalle@upmc.edu
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell 2010;18(1):11–22 doi 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan MJ, Osman I, Khan FM, Vengrenyuk Y, Capodieci P, Koscuiszka M, et al. Androgen receptor expression is associated with prostate cancer-specific survival in castrate patients with metastatic disease. BJU international 2010;105(4):462–7 doi 10.1111/j.1464-410X.2009.08747.x. [DOI] [PubMed] [Google Scholar]
- 3.Li R, Wheeler T, Dai H, Frolov A, Thompson T, Ayala G. High level of androgen receptor is associated with aggressive clinicopathologic features and decreased biochemical recurrence-free survival in prostate: cancer patients treated with radical prostatectomy. The American journal of surgical pathology 2004;28(7):928–34. [DOI] [PubMed] [Google Scholar]
- 4.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. The Journal of urology 2002;167(2 Pt 2):948–51; discussion 52. [PubMed] [Google Scholar]
- 5.Pagliarulo V, Bracarda S, Eisenberger MA, Mottet N, Schroder FH, Sternberg CN, et al. Contemporary role of androgen deprivation therapy for prostate cancer. European urology 2012;61(1):11–25 doi 10.1016/j.eururo.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messing EM, Manola J, Yao J, Kiernan M, Crawford D, Wilding G, et al. Immediate versus deferred androgen deprivation treatment in patients with node-positive prostate cancer after radical prostatectomy and pelvic lymphadenectomy. The lancet oncology 2006;7(6):472–9 doi 10.1016/S1470-2045(06)70700-8. [DOI] [PubMed] [Google Scholar]
- 7.Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. The American journal of pathology 2004;164(1):217–27 doi 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324(5928):787–90 doi 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danila DC, Morris MJ, de Bono JS, Ryan CJ, Denmeade SR, Smith MR, et al. Phase II multicenter study of abiraterone acetate plus prednisone therapy in patients with docetaxel-treated castration-resistant prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010;28(9):1496–501 doi 10.1200/JCO.2009.25.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet 2010;375(9724):1437–46 doi 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364(21):1995–2005 doi 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013;368(2):138–48 doi 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371(5):424–33 doi 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367(13):1187–97 doi 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 15.Boudadi K, Antonarakis ES. Resistance to Novel Antiandrogen Therapies in Metastatic Castration-Resistant Prostate Cancer. Clin Med Insights Oncol 2016;10(Suppl 1):1–9 doi 10.4137/CMO.S34534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrine reviews 1997;18(3):306–60. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer research 2009;69(6):2305–13 doi 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer research 2009;69(1):16–22 doi 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PloS one 2011;6(4):e19059 doi 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proceedings of the National Academy of Sciences of the United States of America 2010;107(39):16759–65 doi 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M. The androgen receptor gene mutations database: 2012 update. Hum Mutat 2012;33(5):887–94 doi 10.1002/humu.22046. [DOI] [PubMed] [Google Scholar]
- 22.Manos-Turvey A, Brodsky JL, Wipf P. The Effect of Structure and Mechanism of the Hsp70 Chaperone on the Ability to Identify Chemical Modulators and Therapeutics. Top Med Chem 2016;19:81–129. [Google Scholar]
- 23.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature 2011;475(7356):324–32 doi 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 24.Hageman J, Kampinga HH. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell stress & chaperones 2009;14(1):1–21 doi 10.1007/s12192-008-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. Febs Lett 2007;581(19):3702–10 doi DOI 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 26.Leung TK, Rajendran MY, Monfries C, Hall C, Lim L. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B’) and isolation of its cDNA and genomic DNA. The Biochemical journal 1990;267(1):125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America 2004;101(16):6062–7 doi 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 1986;46(2):291–300. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharyya T, Karnezis AN, Murphy SP, Hoang T, Freeman BC, Phillips B, et al. Cloning and subcellular localization of human mitochondrial hsp70. The Journal of biological chemistry 1995;270(4):1705–10. [DOI] [PubMed] [Google Scholar]
- 30.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell 2006;125(3):443–51 doi 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996;272(5268):1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proceedings of the National Academy of Sciences of the United States of America 1991;88(7):2874–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proceedings of the National Academy of Sciences of the United States of America 1999;96(10):5452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Molecular cell 2012;48(6):863–74 doi 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Dar JA, Masoodi KZ, Eisermann K, Isharwal S, Ai J, Pascal LE, et al. The N-terminal domain of the androgen receptor drives its nuclear localization in castration-resistant prostate cancer cells. J Steroid Biochem Mol Biol 2014;143:473–80 doi 10.1016/j.jsbmb.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9(7):671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang FT, Zeng XM, Kim W, Balasubramani M, Fortian A, Gygi SP, et al. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proceedings of the National Academy of Sciences of the United States of America 2013;110(39):15722–7 doi DOI 10.1073/pnas.1308014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011;10(4):1794–805 doi 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 39.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature 2011;473(7347):337–42 doi Doi 10.1038/Nature10098. [DOI] [PubMed] [Google Scholar]
- 40.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26(7):966–8 doi DOI 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harper S, Speicher DW. Purification of proteins fused to glutathione S-transferase Protein Chromatography: Springer; 2011. p 259–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimopoulos MA, Mitsiades CS, Anderson KC, Richardson PG. Tanespimycin as antitumor therapy. Clin Lymphoma Myeloma Leuk 2011;11(1):17–22 doi 10.3816/CLML.2011.n.002, S2152–2650(11)70122–7 [pii]. [DOI] [PubMed] [Google Scholar]
- 43.Schlecht R, Scholz SR, Dahmen H, Wegener A, Sirrenberg C, Musil D, et al. Functional analysis of Hsp70 inhibitors. PloS one 2013;8(11):e78443 doi 10.1371/journal.pone.0078443, PONE-D-13–33675 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manos-Turvey A, Al-Ashtal HA, Needham PG, Hartline CB, Prichard MN, Wipf P, et al. Dihydropyrimidinones and -thiones with improved activity against human polyomavirus family members. Bioorg Med Chem Lett 2016;26(20):5087–91 doi S0960–894X(16)30912-X [pii], 10.1016/j.bmcl.2016.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ireland AW, Gobillot TA, Gupta T, Seguin SP, Liang M, Resnick L, et al. Synthesis and structure-activity relationships of small molecule inhibitors of the simian virus 40 T antigen oncoprotein, an anti-polyomaviral target. Bioorg Med Chem 2014;22(22):6490–502 doi 10.1016/j.bmc.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huryn DM, Brodsky JL, Brummond KM, Chambers PG, Eyer B, Ireland AW, et al. Chemical methodology as a source of small-molecule checkpoint inhibitors and heat shock protein 70 (Hsp70) modulators. Proceedings of the National Academy of Sciences of the United States of America 2011;108(17):6757–62 doi 10.1073/pnas.1015251108, 1015251108 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JK, Skoda EM, Zhou J, Parrinello E, Wang D, O’Malley K, et al. Small Molecule Antagonists of the Nuclear Androgen Receptor for the Treatment of Castration-Resistant Prostate Cancer. ACS Med Chem Lett 2016;7(8):785–90 doi 10.1021/acsmedchemlett.6b00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu ML, Horbinski CM, Garzotto M, Qian DZ, Beer TM, Kyprianou N. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer research 2010;70(20):7992–8002 doi 10.1158/0008-5472.can-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sramkoski RM, Pretlow TG 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, et al. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim 1999;35(7):403–9 doi 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 50.Tepper CG, Boucher DL, Ryan PE, Ma AH, Xia L, Lee LF, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res 2002;62(22):6606–14. [PubMed] [Google Scholar]
- 51.Libertini SJ, Tepper CG, Rodriguez V, Asmuth DM, Kung HJ, Mudryj M. Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res 2007;67(19):9001–5 doi 67/19/9001 [pii], 10.1158/0008-5472.CAN-07-1072. [DOI] [PubMed] [Google Scholar]
- 52.Massey AJ, Williamson DS, Browne H, Murray JB, Dokurno P, Shaw T, et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother Pharmacol 2010;66(3):535–45 doi 10.1007/s00280-009-1194-3. [DOI] [PubMed] [Google Scholar]
- 53.Wisen S, Bertelsen EB, Thompson AD, Patury S, Ung P, Chang L, et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol 2010;5(6):611–22 doi 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol 2014;54:49–59 doi 10.1016/j.biocel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer research 2013;73(2):483–9 doi 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moses MA, Kim YS, Rivera-Marquez GM, Oshima N, Watson MJ, Beebe KE, et al. Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer research 2018;78(14):4022–35 doi 10.1158/0008-5472.CAN-17-3728, 0008–5472.CAN-17–3728 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet 2002;11(5):515–23. [DOI] [PubMed] [Google Scholar]
- 58.He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, et al. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). The Journal of biological chemistry 2004;279(29):30643–53 doi 10.1074/jbc.M403117200, M403117200 [pii]. [DOI] [PubMed] [Google Scholar]
- 59.McEwan IJ. Intrinsic disorder in the androgen receptor: identification, characterisation and drugability. Mol Biosyst 2012;8(1):82–90 doi 10.1039/c1mb05249g. [DOI] [PubMed] [Google Scholar]
- 60.Knapp RT, Wong MJ, Kollmannsberger LK, Gassen NC, Kretzschmar A, Zschocke J, et al. Hsp70 cochaperones HspBP1 and BAG-1M differentially regulate steroid hormone receptor function. PloS one 2014;9(1):e85415 doi 10.1371/journal.pone.0085415, PONE-D-13–33245 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kita K, Shiota M, Tanaka M, Otsuka A, Matsumoto M, Kato M, et al. Heat shock protein 70 inhibitors suppress androgen receptor expression in LNCaP95 prostate cancer cells. Cancer Sci 2017;108(9):1820–7 doi 10.1111/cas.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jung ME, Ouk S, Yoo D, Sawyers CL, Chen C, Tran C, et al. Structure-activity relationship for thiohydantoin androgen receptor antagonists for castration-resistant prostate cancer (CRPC). J Med Chem 2010;53(7):2779–96 doi 10.1021/jm901488g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence MG, Obinata D, Sandhu S, Selth LA, Wong SQ, Porter LH, et al. Patient-derived Models of Abiraterone- and Enzalutamide-resistant Prostate Cancer Reveal Sensitivity to Ribosome-directed Therapy. European urology 2018. doi S0302–2838(18)30445–7 [pii], 10.1016/j.eururo.2018.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruno RD, Gover TD, Burger AM, Brodie AM, Njar VC. 17alpha-Hydroxylase/17,20 lyase inhibitor VN/124–1 inhibits growth of androgen-independent prostate cancer cells via induction of the endoplasmic reticulum stress response. Mol Cancer Ther 2008;7(9):2828–36 doi 10.1158/1535-7163.MCT-08-0336, 7/9/2828 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCarty DJ, Huang W, Kane MA, Purushottamachar P, Gediya LK, Njar VCO. Novel galeterone analogs act independently of AR and AR-V7 for the activation of the unfolded protein response and induction of apoptosis in the CWR22Rv1 prostate cancer cell model. Oncotarget 2017;8(51):88501–16 doi 10.18632/oncotarget.19762, 19762 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo W, Keener AL, Jing Y, Cai L, Ai J, Zhang J, et al. FOXA1 modulates EAF2 regulation of AR transcriptional activity, cell proliferation, and migration in prostate cancer cells. Prostate 2015;75(9):976–87 doi 10.1002/pros.22982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.