Abstract
Translation initiation in eukaryotes is a highly regulated and rate-limiting process. It results in the assembly and disassembly of numerous transient and intermediate complexes involving over a dozen eukaryotic initiation factors (eIFs). This process culminates in the accommodation of a start codon marking the beginning of an open reading frame at the appropriate ribosomal site. Although this process has been extensively studied by hundreds of groups for nearly half a century, it has been only recently, especially during the last decade, that we have gained deeper insight into the mechanics of the eukaryotic translation initiation process. This advance in knowledge is due in part to the contributions of structural biology, which have shed light on the molecular mechanics underlying the different functions of various eukaryotic initiation factors. In this review, we focus exclusively on the contribution of structural biology to the understanding of the eukaryotic initiation process, a long-standing jigsaw puzzle that is just starting to yield the bigger picture.
INTRODUCTION
Translation of messenger RNA (mRNA) into protein is a highly conserved process in all living cells. It mobilizes nearly half of the energy production of the cell and abides to numerous and intricate pathways of regulation. In eukaryotes, in contrast to eubacteria, translation regulation is by far more sophisticated, owing in part to the increase in complexity of the eukaryotic 80S ribosome, mainly characterized by larger ribosomal RNA (rRNA) chains, additional ribosomal proteins (~20 more proteins), and a larger number of translation regulation factors.
Eukaryotic translation initiation (reviewed in 63) starts with the assembly of a ternary complex (TC) consisting of methionine initiator transfer RNA (Met-tRNAi), eukaryotic initiation factor 2 (eIF2), and guanosine triphosphate (GTP), which is then recruited to the 40S subunit (thoroughly reviewed in 56 and 57), aided by eIF1, eIF1A, and eIF3 (subunits a–m) to form a 43S preinitiation complex (PIC) (8, 49, 87). The binding of these factors to the 40S subunit produces an open state to which the TC readily binds (reviewed in 2). The eIF2-GTPase-activating protein eIF5 binds the 43S PIC along with the TC. The 43S PIC is then recruited to the capped 5′ end of an mRNA bound to the cap-binding complex formed by eIF4F (in turn composed of eIFs 4E, 4A, and 4G), eIF4A, and eIF4B. The resulting 48S initiation complex (IC), in the open POUT conformation in which Met-tRNAi is not fully accommodated in the P site, scans the 5′ region until it encounters the AUG start codon in a suitable initiation context (60, 81). The attachment of mRNA to the 43S PIC triggers the relocation of eIF3b along with its closely associated eIF3i and eIF3g subunits, from the solvent side to the intersubunit face of the 40S subunit (131) (also reviewed in 57) to bind below the TC in the proximity of helix 44 (h44). This relocation of the eIF3b-i-g module may represent the molecular origin of the scanning stimulation role of this trimeric eIF3 module (29, 101). Recognition of AUG triggers the scanning arrest of the 48S IC and induces a positional change of the 40S head relative to its body, back to the closed state in which Met-tRNAi is fully engaged in the P site (the so-called PIN state) (58, 81). As a result of start-codon recognition, eIF1 is then released or at least relocated from the P site along with the C-terminal tail of eIF1A (23, 98, 99, 158). This eIF1 movement is believed to allow an irreversible release of free inorganic phosphate (Pi) from GTP hydrolyzed on eIF2 upon AUG recognition (3), resulting in dissociation of eIF2 and eIF5 from the late-stage IC, which is composed of the 40S subunit, mRNA, eIF1A, eIF3, and fully accommodated Met-tRNAi in the P site (reviewed in 143). The dissociation of eIF2 exposes one of the binding sites for eIF5B [which engages the CCA end of the Met-tRNAi (75, 153)], and the interplay between eIF5B and Met-tRNAi allows both to acquire their 60S-binding-competent conformations (37, 75, 153). Finally, the interaction of eIF5B with the ribosome promotes 60S joining and, after GTP hydrolysis on eIF5B, this factor together with eIF1A leaves the 80S IC, rendering the ribosome elongation competent (42, 130).
The mRNA translation process is cyclic, and thus the default pathway for a new round of protein synthesis after termination is the reuse of posttermination ribosomes (62). After termination, the posttermination ribosomal complexes [i.e., mRNA and deacylated tRNA-bound ribosomes] are split into free 60S subunits and deacylated tRNA- and mRNA-bound 40S subunits by a process called ribosome recycling (109, 110).
The purpose of this review is not to revisit the field of eukaryotic translation initiation, as recent excellent and exhaustive reviews already exist in the literature (57), but rather to highlight the quintessential contribution of the field of structural biology to the understanding of this very complex process. Therefore, we must apologize in advance for not citing numerous pivotal studies unrelated to structural biology, even if they have contributed tremendously to shaping the field of translation initiation in eukaryotes as we know it today. Instead, we focus exclusively on the structural studies, mainly by X-ray crystallography and cryo-electron microscopy (cryo-EM), which yielded the pieces of the giant jigsaw puzzle that translation initiation presents, allowing us to start assembling the bigger picture. In the following, we discuss each piece of the initiation puzzle separately from the structural viewpoint and explain how these different structures have contributed to better understanding of the molecular mechanics underlying the various functions of different initiation factors and regulators.
THE CENTERPIECES: THE EUKARYOTIC RIBOSOME STRUCTURES AND EUKARYOTIC INITIATION FACTOR 3
Two main multi-component complexes, the ribosome complex and eukaryotic initiation factor 3, compose the beating heart of mRNA translation initiation.
The Eukaryotic Ribosome Complex
Unlike any other jigsaw puzzle, composition of the picture of translation initiation in eukaryotes must start with the centerpieces: the structures of the ribosomal 40S, 60S, and 80S complexes that will bear the factors forming various initiation complexes at different stages of the process. The ribosome is a hybrid complex composed of a core of rRNA surrounded and enmeshed by dozens of ribosomal proteins. In its core, the ribosome is universally conserved among all orders of life. It is composed of two subunits—a small subunit (SSU) and a large subunit (LSU)—with both exhibiting the same type of hybrid composition of protein and rRNA. The space between the small and large subunits contains three major binding sites called A (aminoacyl), P (peptidyl), and E (exit) sites, where the tRNAs carrying the amino acids and various translation factors interact during the different stages of translation. While the ribosome is generally conserved, its structure and regulation differ substantially between bacteria and eukaryotes (thoroughly reviewed in 95 and 151).
In bacteria, the ribosome (also called 70S for its sedimentation coefficient) has a total molecular weight of ~2.5 MDa, and its mature rRNA is subdivided into three parts designated as 16S, 23S, and 5S. The 16S rRNA chain binds some 20 ribosomal proteins and forms the SSU (30S). The 23S and the 5S rRNA, jointly, bind some 33 ribosomal proteins and form the LSU (50S).
In eukaryotes, the ribosome (called 80S) is much larger than its prokaryotic counterpart and has a larger and more variable molecular weight; in yeast, for example, its molecular weight is approximately 4 MDa (see review in Reference 72 for a comprehensive description of the 80S structure). Its mature rRNA is divided generally into 18S, 28S, 5.8S, and 5S. The 18S rRNA chain binds some 32 ribosomal proteins as it forms the SSU (40S). Together, the 28S, 5.8S, and 5S rRNAs bind some 47 ribosomal proteins and form the LSU (60S). In addition to possessing more numerous ribosomal proteins, the eukaryotic rRNA is substantially larger (Figure 1a).
Figure 1.
Structure of the 80S eukaryotic ribosome and the rRNA expansion segments (ESs) of the small ribosomal subunit. (a) The eukaryote-specific ribosomal features (in red) compared to the bacterial ribosome. Small subunit (SSU) shown from (b) the solvent side and (c) the intersubunit face. Ribosomal proteins are shown as gray surface, and the 18S rRNA as yellow ribbons. (d,e) Involvement of ES6 and ES7 in the binding of eukaryotic initiation factor 3 (eIF3) as deduced by the cryo-electron microscopy structure of the 43S mammalian preinitiation complex (30). Additional abbreviation: LSU, large subunit.
Indeed, the eukaryotic ribosome contains RNA insertions that are called expansion segments (ESs) (44, 33, 157). These ESs can be of substantial size, up to several hundreds of nucleotides, and can be accompanied by extensions of ribosomal proteins, as demonstrated by the pioneering 40S, 60S, and 80S structures obtained by X-ray crystallography and cryo-EM, to which we owe the exquisite and detailed structural knowledge of the eukaryotic ribosomes (4, 6, 7, 11, 14, 15, 51, 67, 71, 80, 115, 147).
Although the SSU is now a known giant piece of the puzzle, the functions of some of its features such as the RNA ESs are yet to be elucidated. Indeed, it is commonly accepted that ESs have evolved to ensure certain eukaryote-specific functions. The structure of several of these ESs could not be determined accurately because of their high level of flexibility, as demonstrated by cryo-EM studies. In addition, the role of most of these ESs has not been identified. Nevertheless, one could reasonably speculate on their structural role, as they accompany the introduction of additional ribosomal proteins compared to the bacterial ribosome precursor. Several of these ESs are located in regions devoid of any proteins and seem to be simply “hanging off” from both ribosomal subunits, far from any eukaryote-specific ribosomal proteins and extensions. Indeed, on the SSU, for example, ESs 3, 6, 7, 9, and 12 point toward the solvent, far from any other ribosomal components (Figure 1b,c). In this context, the structures of complexes formed by the ribosome with different protein factors at different stages of the translation process can shed light on the role of some of these ESs. For instance, the structure of the mammalian 43S PIC (30) clearly displays the interactions of ESs 6 and 7 with the eukaryotic initiation factor 3 (eIF3) (a key factor that is discussed further in the review). In fact, helix A of ES6 interacts with the peripheral module b-i-g of eIF3 (Figure 1d), and ES7 interacts with subunits a and c of the eIF3 octamer core (Figure 1e). We anticipate that other ESs could similarly be involved in certain stages of translation regulation.
The Eukaryotic Initiation Factor 3 Complex
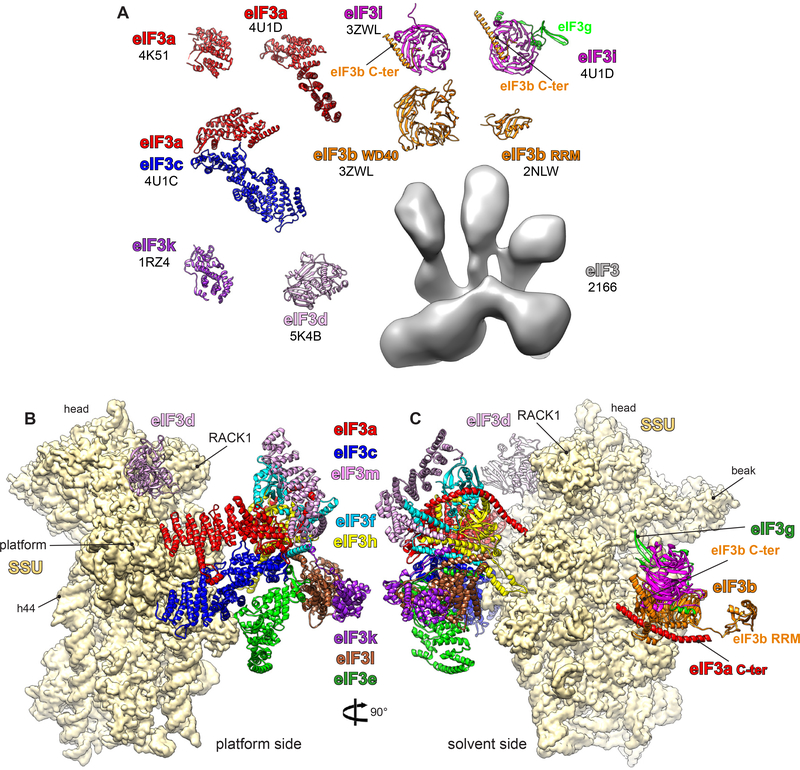
Although recent studies show the involvement of eIF3 in other processes along the translation regulation pathway, such as its role in the stop-codon read-through (18), eIF3 is the initiation factor par excellence that accompanies the initiation process all the way from the beginning to the end. As the largest and most complex initiation factor, eIF3 is involved in virtually all steps of initiation, including the attachment of 43S PICs to mRNA (through the interaction with eIF4G), the ribosomal recruitment of the TC, and ensuring the fidelity of the scanning process for the correct start codon (thoroughly reviewed in 55). Although eIF3 is an essential protein deeply involved in the regulation of the initiation process, its size and composition can vary dramatically among organisms. Indeed, in mammals, eIF3 is an ~800-kDa multimeric complex that comprises 13 subunits (a–m) (Figure 2). Six subunits (a, c, e, k, l, and m) contain PCI domains (proteasome component), which consist of N-terminal helical repeats followed by a winged helix domain that mediates PCI polymerization, and two subunits (f and h) contain MPN (Mpr1/Pad1 N-terminal) domains, which consist of a β-barrel surrounded by α-helices and β-strands that function to promote assembly of multi-protein complexes. The PCI/MPN subunits form the so-called conserved octameric structural core of eIF3.
Figure 2.
Structures of eIF3 fragments and subunits by X-ray crystallography, nuclear magnetic resonance, and cryo-electron microscopy (cryo-EM). (a) Different fragments of eIF3 subunits are colored variably, and the accession codes are written in black below the name of each subunit. (b,c) Small subunit (SSU) shown in yellow surface, and eIF3 in colored ribbons. Different eIF3 subunits are labeled and colored variably. The displayed eIF3 structure is a model derived from an ~6-Å cryo-EM map of the mammalian 43S preinitiation complex (30), which was in part interpreted thanks to the high-resolution structures of eIF3 fragments. Additional abbreviations: C-ter, C-terminus; eIF, eukaryotic initiation factor; h44, helix 44; RRM, RNA recognition motif.
The remaining subunits (j, b, d, g, and i) are flexibly linked to the PCI/MPN core. Domains in these subunits include RNA recognition motif (RRM) domains (eIF3b and eIF3g) and WD40 b-propeller domains (eIF3b and eIF3i). eIF3b, eIF3i, and eIF3g form a separate module that binds to the PCI/MPN core through its interaction with the C-terminal domain (CTD) of eIF3a. The last subunit, eIF3j, is substoichiometric and loosely attached to eIF3. Most eukaryotes encode the complete set of eIF3 subunits probably arranged similarly to the abovementioned mammalian factor. However, Saccharomyces cerevisiae and related yeasts possess only six subunits—only two PCI (a and c) and four noncore (b, i, g, and j) subunits. Other exceptions can be found in more exotic species such as kinetoplastids (pathogenic protozoa), where subunit m is lacking (94, 118).
The first three-dimensional structure of eIF3 was derived by EM through the random conical tilt (RCT) technique in the context of the native (pre)initiation complex purified directly from cell extracts (137). RCT used to yield deformed structures at very modest resolutions, especially in the early days of the technique after the factor bound to the 40S subunit was first visualized in 1992. Despite these shortcomings, the study already provided the binding position to the 40S subunit and clues about the function of this factor as an anti-LSU-association factor.
Since then, several pioneering cryo-EM studies revealed the organization of the five-lobed PCI/MPN core of mammalian eIF3 and confirmed the similarity of its topology with those of the proteasome lid and the COP9 signalosome (114, 132, 133, 140). In parallel, several crystallography and nuclear magnetic resonance (NMR) studies were successful in determining the structures of various fragments and several subunits of eIF3 (34, 36, 53, 70, 77, 79, 149) (Figure 2). However, the low resolution of eIF3 in these pioneering cryo-EM studies (12 to 20 Å) was insufficient to reveal the architecture of the PCI/MPN core. In addition, the high-resolution structures of eIF3 fragments in these X-ray crystallography and NMR studies were far too sparse to allow their exploitation for the interpretation of the low-resolution cryo-EM maps and reveal the fine molecular assembly of eIF3.
More recently, and thanks to the tremendous technological advances in the cryo-EM field mainly brought about by the advent of the direct electron detector cameras, we were successful in solving the structure of the mammalian 43S PIC [previously solved at ~12 Å (49)] at nearly 6 Å (30), showing the fine architecture of eIF3 in this specific context. According to this study, the PCI/MPN core resides on the back of the 40S subunit, making two contact points via its left arm and head with ribosomal proteins (rp’s) eS1/eS26 and uS15/eS27, respectively. Two additional densities, on the solvent side underneath the rRNA helix 16 (h16) and on the head behind RACK1, were attributed to the peripheral domains of eIF3. The ~6-Å cryo-EM structure of eIF3 was interpreted, finally yielding a fairly reliable atomic model totaling more than 70% of the mass of mammalian eIF3. This interpretation was made possible thanks to the high-resolution structures of eIF3 fragments, to homologous subunits from the proteasome lid and COP9 signalosome structures (9, 78), and to the assignment of eIF3 octamer core subunits using immunolabeled cryo-EM (114).
The factor eIF3 plays many essential roles in the initiation process. In fact, it is required in nearly all types of translation initiation, canonical and noncanonical. The first proposed role for eIF3 was to promote the dissociation of the 80S ribosome into SSU and LSU and to act as an antiassociation factor for these ribosomal subunits. This role was finally explained by the first structure of the mammalian 43S PIC (49), as the N-terminal end of eIF3c was observed to interfere with the intersubunit bridge B4, situated between rpL30e and rpS13e. In addition, eIF3 was shown to stabilize the binding of the eIF2 TC to the 40S subunit, a role that was in part explained by the structure of the mammalian 43S PIC (30), where a contact was observed between eIF3d and the eIF2α-D1 domain.
Together with eIF4G, eIF3 also plays a key role in the promotion of the recruitment of the mRNA to the SSU (13, 142). However, the interaction between eIF3 and eIF4G was never observed in structural terms, and to date, the molecular mechanics underlying this function remain elusive. Last but not least, eIF3 was observed to play a role in start-codon recognition (100, 107, 144), a role that was explained only recently by two cryo-EM studies (81, 131), which are discussed in more detail further below in the Discussion section.
THE BULK PIECES: eIF2 TERNARY COMPLEX, eIF1, AND eIF1A
Following the jigsaw puzzle analogy, these pieces can be compared to the bulk pieces that stitch together the different parts of the image.
The eIF2 Ternary Complex
The eIF2 TC is composed of eIF2 bound to a molecule of GTP, and it recruits the initiator methionylated Met-tRNA (tRNAiMet). eIF2 in turn is composed of three subunits: eIF2α, eIF2β, and eIF2γ. The eIF2γ subunit, a GTPase, was first crystallized nearly 15 years ago (123) (Figure 3a), revealing for the first time the strong structural homology existing between eIF2γ and elongation factor EF-Tu (eEF1A in eukaryotes). Further structural studies established the architecture of eIF2γ in both eukaryotes and archaea, either nucleotide-free or in interaction with GDP (guanosine diphosphate) or GTP (102, 119, 134, 154, 155) (Figure 3a), thus providing a valuable insight into the GTP hydrolysis mechanism and the initiator tRNA binding to the latter eIF2 subunit (reviewed in 124). At the same time, several NMR and X-ray studies undertook the structure determination of other eIF2 subunits, α and β (24, 31, 61, 64, 145, 156), which already provided clues about the structure of the full eIF2 factor (Figure 3a). Shortly after, the first structures of full archaeal eIF2 factors confirmed the predicted topology and demonstrated the flexibility of eIF2 (138, 155) (Figure 3a). However, the interaction of the initiator tRNA with the eIF2 complex remained mysterious until the first structure of The archaeal eIF2 TC appeared (125) (Figure 3b). Very briefly, the initiator tRNA interacts from its CCA end with eIF2γ domain II and from its elbow with domains D1 and D2 of eIF2α. The GTP interacts with the G domain of eIF2γ. Finally, after nearly a decade of structural studies, the eIF2 TC has yielded its structure, demonstrating the profound differences between EF-Tu and eIF2γ in the way they interact with the tRNA despite their sequence homology. Interestingly, another structure of the TC shows a very different tRNA binding constellation and conformation of eIF2, but these features have never been observed in the context of an IC (139).
Figure 3.
Structures of the eIF2-TC fragments and subunits. (a) Different fragments of eIF2 subunits are colored variably, and the accession codes are written in black below the name of each subunit. (b) Structure of the TC seen from two different orientations. (c) The TC in the context of the SSU is shown in yellow surface, and eIF3 in colored ribbons. Different eIF3 subunits are labeled and colored variably. The conformation of the TC is a model derived from a ~12Å cryo-EM map, then confirmed by the ~6-Å cryo-EM map of the mammalian 43S preinitiation complex (30, 49). (d,e) Blowup on the TC seen from two different orientations. Additional abbreviations: eIF, eukaryotic initiation factor; EM, electron microscopy; GDP, guanosine diphosphate; GTP, guanosine triphosphate; SSU, small subunit; TC, ternary complex; tRNA, transfer RNA.
In spite of this tremendous progress in the elucidation of the TC structure, the interaction pattern of the TC on the 40S has remained unknown. It was at first predicted by hydroxyl radical cleavage (HRC) to involve domain III of eIF2γ and h44 on the SSU (129). Only after the first cryo-EM structure of the mammalian 43S PIC was solved that this piece of the puzzle could be clipped in and the actual interaction of the TC with the 40S be unveiled (49) (Figure 3c–e). However, cryo-EM showed some differences in the predicted binding, as the TC interacted mostly with the 40S through domain D1 of eIF2α and the initiator tRNA anticodon stem-loop that is inserted in the P site. In fact, domain III of eIF2γ does not directly contact h44; rather, it comes in proximity of h44 at the level of an internal loop predicted by HRC. In addition, domain D2 of eIF2α interacts in a different manner with the initiator tRNA elbow in the context of the 40S compared to the structure of the TC alone.
Comparison of the structure of the TC alone versus the structure in the context of the 40S subunit allows us to explain the mechanics of the initiator tRNA recruitment to the (pre)initiation complex, the epicenter of canonical initiation in eukaryotes.
eIF1 and eIF1A
As for eIF1 and eIF1A, from the viewpoint of structural biology, these factors were among the first to be studied because of their relatively small size and good solubility. In spite of their small size, their importance in translation initiation is pivotal, as they play a crucial role in the start-codon selection. The structure of eIF1 was first solved by NMR in solution (41), showing a small globular domain accompanied by two unstructured N- and C-terminal tails. eIF1A is somehow similar in topology. It has roughly the same size as eIF1 (~17 kDa) and includes one central globular domain, accompanied by two unstructured tails. eIF1A is a homolog of initiation factor 1 (IF1) in bacteria and was assumed to bind similarly to the latter on the SSU (i.e., near the A site) (22) (Figure 5). As for eIF1, although its binding site was deduced from HRC experiments (83), it has not been visualized.
Figure 5.
Model of the structure of the mammalian 43S preinitiation complex. (a) The 43S complex seen from the solvent side. Thick red dashed line reflects the continuity of eIF3a structure that remains unsolved at this region. (b) The 43S complex seen from the beak side. Transparent green surface reflects the shape of the N-terminal part of DHX29 that was not modeled. Black dashed circle indicates the binding site of eIF3j, and dashed red circle indicates the position of the dsRBD of DHX29. Notice that eIF3j and DHX29 dsRBD overlap on the same ribosomal position. Abbreviations: C-ter, C-terminus; dsRBD, DHX, DExH-box helicase; double-stranded RNA binding domain; eIF, eukaryotic initiation factor; h44, helix 44; RRM, RNA recognition motif; SSU, small subunit; tRNA, transfer RNA.
An NMR study revealed the existence of two distinct binding sites for eIF5, another important initiation factor (117). More than a decade after the first NMR structure of eIF1, the X-ray structure of eIF1 in the context of the yeast SSU was solved (115), confirming the predicted binding of this factor near the P site on the SSU (Figure 5). More recent X-ray crystallography studies revealed the structure of the 40S subunit in interaction with factors eIF1 and 1A (84, 150). eIF1 and eIF1A were observed to play an important role in start-codon selection and in ensuring the fidelity of the scanning mechanism (106). In structural terms, the presence of these factors appears to produce certain conformational changes of the SSU, mainly focused on the A site region and, more generally, the upper portion of h44. In fact, this region of h44 is squeezed between eIF1A and eIF1. This conformational modulation can have other important implications, as it also concerns the portion of h44 facing the eIF2 TC (eIF2γ domain III) and may play a role in the recruitment of the latter (150). The structure of the eIF1-eIF1A-40S complex also explained the binding synergy between these factors since they both act directly on h44 as a common binding site. It is worth mentioning that the presence of eIF1 and eIF1A was observed to play a role in modulating the opening of the so-called mRNA latch, marking the mRNA channel exit (103). However, because of the different SSU head conformations in the four molecules forming the crystallographic asymmetric unit in the eIF1-eIF1A-40S structure, it was not possible to corroborate this conclusion. One should bear in mind that eIF1 and eIF1A are initiation factors and that only their structure in the context of the (pre)initiation complex can provide the mechanistic interpretation for their observed roles, which are discussed further in this review.
THE MISSING PIECES: eIF3j, eIF3g, THE CAP-BINDING COMPLEX, AND eIF5
Just as in assembling a jigsaw puzzle, once the main image is starting to form, one is left with a bunch of pieces that appear not to go anywhere in the image and with a number of pieces that are nowhere to be found.
eIF3j
For a long time, the factor eIF3j (j/HCR1 in yeast) had been considered part of the eIF3 complex. However, because eIF3j is substoichiometric compared to other eIF3 subunits and also because it appears to perform its functions independently from the context of the full eIF3, it came to be considered as an eIF3-associated factor. To date, there are no available structures for the full-length eIF3j; only one structure containing a fraction of the N-terminal tail of this factor in interaction with eIF3b-RRM was solved by NMR [PDBID 2KRB (35)]. More recently, a cryo-EM structure of the 40S-eIF1-eIF1A-eIF3j complex from yeast was solved (8), which showed that eIF3j’s globular body resides at the intersubunit face of the 40S subunit, in contact with eIF1A, near the A site (Figure 5), where it could be projecting its N-terminal tail toward h16 and eIF3b-RRM, elements with which it was also found to interact. The full extent of the molecular roles of eIF3j is still unclear, but it appears to also be implicated in the stringent selection of the start-codon (35), stop-codon read-through, and termination/recycling processes (17), roles that might be even more important than its role in the initiation process. Interestingly, eIF3j was also shown to be involved in selective mRNA translation (88).
eIF3g
As for eIF3g, it is a piece that is without a doubt part of eIF3. It binds solidly to the eIF3i WD40 domain (36, 81) and is part of the so-called eIF3b-i-g module, which was found positioned at the mRNA channel entrance on the 40S solvent side (Figure 2). eIF3g encompasses an RRM (PDBID 2CQ0, unpublished) of a thus far unknown position. The exact function of eIF3g in the initiation process is still poorly understood; however, the eIF3g-RRM moiety appears to play a role in the stimulation of scanning (29), a role that is discussed later in this review.
Cap-Binding Complex
The cap-binding complex (CBC) is still one of the components of the eukaryotic initiation system most resistant to structural investigations. In spite of the recent advances in the structural biology, the structure of the entire CBC has not yet been determined. The CBC is composed of eIF4F, eIF4A, and eIF4B. eIF4F in turn is composed of the cap-binding protein eIF4E, the scaffold protein eIF4G, and the DEAD-box RNA helicase eIF4A. eIF4A exists in two forms within the CBC, alone and as part of eIF4F. The CBC is pivotal for the recruitment of the capped mRNA to the 43S PIC and the unwinding of its 5′ structured untranslated region (UTR). The eIF4G scaffold interacts with the cap-eIF4E complex and the eIF3–40S complex, thus ensuring the recruitment of the mRNA. The cap consists of an N7-methylguanosine linked to the first 5′ nucleotide. In mammals and higher eukaryotes more generally, the two riboses immediately following the cap are 2′-O methylated. Importantly, the helicase activity of eIF4A is stimulated by both eIF4G and eIF4B (further references can be found in 76).
From the structural viewpoint, of all the factors composing the CBC, eIF4E is certainly the best characterized. Early on, the X-ray structure of eIF4E showed the interaction mechanism between the modified guanine base and eIF4E (90) (Figure 4a). Accordingly, eIF4E traps the cap by stacking the modified guanine base between two tryptophans (Figure 4b). Shortly after, a second structure was published, which showed the interaction mechanism between eIF4E and eIF4G, through the conserved Tyr-X-X-X-X-Leu-ϕ peptide segment (91). Later work corroborated these early studies and revealed further details about the guanine base–binding site (21) and the existence of conformational coupling between eIF4E and eIF4G (47). Further structural studies revealed the molecular mechanics of regulation related to the inhibition of the eIF4E-eIF4G interaction by small peptides called eIF4E-binding peptides (5, 108, 128). Later structures from different species highlighted the existence of large species-specific variability in the eIF4E-eIF4G interface. In the yeast complex, for instance, the eIF4E-eIF4G interface is largely extended compared to other species such as Drosophila melanogaster and human (48).
Figure 4.
Structures of fragments and factors composing the cap-binding complex (CBC). (a) Different structures of fragments and factors of the CBC are shown, colored variably, and the accession codes are written in black below the name of each fragment and factor. (b) Blowup on the N7-methyl-guanosine-binding pocket of eIF4E. (c) A model of the CBC based on the different available partial structures. The model illustrates the scaffold role of eIF4G carrying functionally different factors to the (pre)initiation complex, which is in a way comparable to eIF3 and its octamer core. Additional abbreviations: eIF, eukaryotic initiation factor; mRNA, messenger RNA; IRES, internal ribosomal entry site; PABP, polyadenylate-binding protein.
As for the DEAD-box RNA helicase eIF4A, the crystal structure of its ATPase was obtained almost at the same time as the structure of eIF4E (16) (Figure 4a). Nearly a decade after that, the full crystal structure eIF4A from yeast was obtained in interaction with a portion from eIF4G, thus revealing the mechanism of interaction between these two essential factors (127). Other later structures detailed the mechanism of translation inhibition by the tumor suppressor Pdcd4 through the inhibition of eIF4A activity after their binding (82, 148).
Last, we come to eIF4G, the most elusive among all three eIF4F factors. It is considered a scaffold factor that structures the CBC. To date, only fractions of eIF4G have been solved along with, and in interaction with, eIF4E and eIF4A (Figure 4a). eIF4G is an ~1600-residue-long protein that encompasses several domains known to bind a number of initiation-related factors [polyadenylate-binding protein (PABP), eIF4E, eIF4A/internal ribosomal entry site (IRES), eIF4A, and Mnk-1, from the N- to C-terminus. The first X-ray structure revealed the structure of a conserved HEAT domain from eIF4GII, corresponding to the eIF4A/IRES-binding domain (92). Then the structure of the C-terminal region of eIF4G was determined, revealing the existence of two atypical HEAT domains representing the second binding site eIF4A and the Mnk1 binding site, the kinase responsible for the phosphorylation of eIF4E (12). A few years after that, a very interesting structure was solved, representing a complex composed of the PABP, a fraction of mRNA, and the eIF4G PABP fragment (122). This structure served to unravel the molecular basis for the crucial interaction between the CBC and PABP. Finally, the structure of a fraction of death-associated protein 5 (DAP5/p97), which is a homolog of eIF4G, was solved (146), corroborating the structure of the early eIF4A/IRES HEAT domain but additionally suggesting an alternative eIF4G initiation pathway.
The last of the CBC proteins is eIF4B, a multi-domain protein that appears to promote the association of the mRNA to the 40S subunit and stimulates the RNA helicase activity of eIF4A (105, 121). The structure of eIF4B is still unknown; however, an NMR study revealed the structure of its RRM (RNA recognition motif) but interestingly revealed that, on its own, the core RRM domain has a relatively weak interaction with RNA and probably requires the N- and C-terminus extensions for high-affinity binding with its RNA targets (40).
In spite of the lack of a structure of the full CBC or even of the full eIF4F, these fragmentary structures have furnished valuable structural insight into the function of the different fragments/subunits of the CBC and have already allowed the demonstration of some part of the molecular basis for the way the different factors of this complex are assembled. However, it remains absolutely crucial to solve the structure of the full CBC in the context of functional (pre)initiation complexes that can reveal the molecular mechanics underlying the mRNA recruitment and 5′ UTR unwinding processes.
eIF5
Like other initiation factors, eIF5 has been studied extensively by molecular biology. eIF5 plays several important roles during the eukaryotic initiation process. Its N-terminal domain (NTD) is known to be a GTPase-activator protein (GAP) for eIF2 bound in the context of its TC along with GTP. The C-terminal region (CTD) of eIF5 stimulates the interactions between several translation factors, including eIF1 and the β subunit of eIF2. However, only a few partial structures of this factor have been published thus far. The NMR solution structure of eIF5 revealed fold similarities with eIF1 and the eIF2β N-terminus (α/β fold) (27), probably indicative of its capacity to bind structured RNA targets. The CTD of eIF5, by contrast, is structurally quite different, as it is formed by a dozen bundled helices of a HEAT domain, as revealed by its X-ray structure (19). eIF5’s CTD is known to play a key role in the formation of a complex by eIF1, eIF2β, and the N-terminal domain (NTD) tail of eIF3c. While these early structures failed to reveal how eIF5 can conduct its various functions, a recent study detailed its interactions with the IC (103). According to this study, during scanning, eIF5 along with eIF3c-NTD help in anchoring eIF1 at the P site, where eIF1 maintains the PIC in the open conformation. eIF1A-NTT also binds eIF5 (86) and prevents it from binding to eIF2β-NTT. Next, the start-codon recognition causes the arrest of scanning, enhanced by eIF1A-NTT binding to the codon-anticodon duplex and eIF1 dissociates, also assisted by the NTD eIF3c. eIF5 is now available for binding with eIF2β-NTT. The release of eIF1 promotes ejection of eIF2 that is bound to eIF5. In the future, it is hoped that structures of eIF5 in the context of functional (pre)initiation complexes will corroborate these findings and reveal the molecular mechanics of this sophisticated pattern of interactions.
TWO BRAND-NEW PIECES: DHX29 AND ABCE1
Two pieces were found lately, despite the fact that they were never thought to be missing: DHX29, a DExH-box helicase, and ATP binding cassette E1 (ABCE1), a highly conserved and essential ABC-type NTPase known to be a recycling factor. Indeed, neither of two factors were previously thought to have a clear involvement in the initiation process.
The DHX29 Helicase
DHX29 is required for scanning on mRNAs with highly structured 5′ UTSs that occur in higher eukaryotes (111). The silencing of DHX29 results in accumulation of free mRNA and the disassembly of polysome, symptomatic of initiation defects. Although the function of DHX29 has now been extensively studied for several years, an understanding of its molecular mechanism is still lacking. Indeed, even though its global binding site on the SSU near the mRNA channel entrance was predicted, it remains unknown whether it acts as a typical helicase unwinding RNA. A partial homology model of DHX29 was proposed, and the functions of its different domains were analyzed (32). Globally, DHX29 is a protein composed of ~1,400 residues that can hydrolyze any triphosphate nucleotide (NTPase) and contains a helicase core made of RecA1 and RecA2 domains, followed by a winged helix, a ratchet domain, and an OB domain. DHX29 has a long NTD (~550 residues) preceding the RecA1 domain and encloses a double-stranded RNA binding domain (dsRBD). Shortly after this first model appeared, the structure of the mammalian 43S PIC was solved at intermediate resolution (~12 Å) by cryo-EM (49), revealing the general shape of DHX29 bound to the SSU on h16, where it was predicted to bind (Figure 5). In addition, the 43S structure revealed the extension of DHX29 to the intersubunit side of the SSU through what appeared to be a helical linker, reaching a small triangular domain near the A site. Very interestingly, a later study revealed that eIF3j and this DHX29 domain overlap on the same ribosomal side, and therefore DHX29 and eIF3j are mutually exclusive (8) (Figure 5). Later on, the structure of the same mammalian PIC was solved at ~6 Å (30) and suggested this intersubunit domain to be the dsRBD. Also, the RecA1 domain appears to interact directly with the eIF3b-i-g module through, mainly, eIF3i. However, in spite of the knowledge of these structures, the molecular mechanism of action of DHX29 is still elusive, and future studies of DHX29 in complex with the (pre)initiation complex will be required to address this long-standing question.
ABC-type NTPase ABCE1
As for ABCE1, it was for a long time regarded exclusively as a ribosomal recycling factor. After termination, the posttermination ribosomal complexes made of mRNA and deacylated tRNA-bound ribosomes are split, yielding LSU 60S subunits and deacylated tRNA- and mRNA-bound SSU 40S subunits by a process called ribosome recycling (109, 110). It was shown that ABCE1 is able to perform recycling over a wide range of Mg2+ concentrations. ABC-like proteins constitute a vast family of proteins, and they are present in organisms from all kingdoms of life. Most ABC-family members contain membrane domains that enable them to transport solutes across membranes (93). Typically, ABC-type enzymes act as either homo- or heterodimers, and during its catalytic cycle, their nucleotide-binding domains (NBDs) close when bound to ATP. After ATP hydrolysis, the NBDs switch to an open state, acting like a pair of tweezers. It has been proposed that binding and hydrolysis of ATP as well as release of Pi promote conformational structural changes in the NBDs that are transmitted to other internal domains of the ABC enzyme or to other associated partners (116).
Furthermore, it has been demonstrated under physiological conditions that ABC-like enzymes (including ABCE1) can bind and hydrolyze any natural nucleotide triphosphate and not only ATP molecules (110). The structure of ABCE1 was solved by cryo-EM in the context of the ribosomal recycling complex (10), and we do not dwell on its function in this context. However, recent structural studies suggested its involvement in the initiation process as well and demonstrated its role as an antiribosomal association factor (54, 69). Interestingly, these studies show ABCE1 in complex with the SSU in a very different conformation compared to that observed in context of the recycling complex, where the NBDs are fully closed and the iron-sulfur cluster (FeS) domain is rotated by ~180° so as to acquire the proper orientation for direct binding to h44 on the SSU. This same orientation was also observed in the context of the mammalian 48S late-stage IC (131), along with eIF3, the eIF3 TC, and mRNA (89) (Figure 5). This newly discovered role of ABCE1 as an antiribosomal association factor and its possible function in the formation of the IC is consistent with the observation that ribosomal subunits’ reassociation is an intrinsic molecular behavior in nature (52), established very early on, and strongly indicates the need for antiassociation factors to allow translation initiation to proceed.
Future studies must be conducted to reveal the exact effect of ABCE1 in the recruitment and the formation of different initiation complexes, but already the existing structures tend to confirm its role in the process and explain the antiribosomal association role as a result of disrupting the B3 intersubunit bridge, implicating h44 on the SSU.
THE REMAINING PIECES: eIF2B, eIF6, AND eIF5B
These last pieces of the puzzle are treated separately here because they occur at different stages of the initiation process, off the (pre)initiation complexes, which does not, however, diminish their importance in any way.
eIF2B
Protein eIF2B is the guanine nucleotide exchange factor that exchanges GDP for GTP on the γ subunit of eIF2. Its activity is inhibited by stress-induced phosphorylation of eIF2α. eIF2B is a heterodecameric complex of two copies each of α, β, γ, δ, and ε subunits. The crystal structure of eIF2Bα was the first subunit to be obtained (59, 65), then were the structures of the β and δ subunits (74), and soon after, the structure of the entire eIF2B was solved (66). These structures explained the molecular mechanism of the nucleotide exchange and finally explained the effect of the eIF2α phosphorylation, which yields what is referred to as a nonproductive eIF2-eIF2B complex that blocks the GDP exchange on the eIF2γ subunit.
eIF6
Next, eIF6 is a factor that binds to the 60S subunit and plays a role as an antiribosomal association factor. However, as it inhibits translation initiation, it was first considered an initiation factor. The structures of archaeal aIF6 and its eukaryotic homolog from yeast eIF6 were solved by X-ray crystallography (46), revealing their quasi-identical architecture composed of a pentagram of α/β domains. Nearly a decade later, the cryo-EM structure of the 60S subunit-bound eIF6 was solved at low resolution (43)—sufficient, nevertheless, to allow the general molecular mechanism of the intersubunit association inhibition to be deciphered. Indeed, the cryo-EM structure of the 60S-eIF6 revealed the binding site of eIF6 to be on the sarcin-ricin loop (SRL), posed to disrupt the B6 intersubunit bridge, thus preventing the SSU from associating with the LSU. Shortly after this cryo-EM structure came out, a similar complex (60S-eIF6) from Tetrahymena thermophila was solved by X-ray crystallography at high resolution, corroborating the proposed mechanism of action (71). Finally, the archaeal 50S-aIF6 complex was solved by cryo-EM at intermediate resolution, confirming a mechanism of action of aIF6 in ribosomal antiassociation, similarly to its eukaryotic counterpart (45).
eIF5B
Finally, eIF5B is a late-acting initiation factor that marks the last stage of eukaryotic translation initiation. It stimulates the association of the 40S SSU (carrying a P-site initiator tRNA) to the 60S LSU. eIF5B is the eukaryotic homolog of the bacterial IF2, also a GTPase, responsible for the proper accommodation of the initiator tRNA and the stimulation of the ribosomal subunit association. The X-ray structure of IF2 was solved both nucleotide-free and in complex with GDP and GTP (120), thus demonstrating the changes in the shape of IF2/eIF5B upon hydrolysis of GTP, characterized by a rotation of domain IV, which already suggested a mechanical action to be at the origin of its function. Importantly, this same domain IV interacts with eIF1A in the context of the ribosome (IF1 in bacteria) at a late stage of translation initiation (25). It was only after the determination of the structure of 80S-Met-tRNAiMet-mRNA-eIF5B that the molecular mechanics of eIF5B action were unraveled (37). This structure suggests that the specific interactions between eIF5B domain IV and the initiator tRNA stabilize large-scale conformation changes mainly involving domain IV. The simultaneous recognition of the initiator tRNA by domain IV and the recognition of the SRL by the G-domain through the binding of a fully maturated 60S LSU stimulate the GTPase activity of eIF5B. The stimulatory activity of eIF5B in the association of both ribosomal subunits can in part be explained by the numerous contacts of eIF5B with both subunits owing to its large surface.
DISCUSSION
The Other (Pre)initiation Complexes
Since the first cryo-EM structure of the mammalian PIC was reconstructed (49), other eukaryotic (pre)initiation complexes have been solved, both from mammals and from yeast. These structures captured different stages of translation initiation, starting from the formation of the 43S PIC (8, 30, 36), through the assembly of the 48S complex during scanning for the start codon and its recognition (60, 81), all the way to formation of the 48S late-stage IC after completion of the scanning process (131), and, finally, after the completion of the process, the ribosomal subunit joining (37). The determination of these structures greatly expanded our understanding of the initiation process in eukaryotes and provided successive frames representing the progress of initiation in time, thus shedding light on the molecular mechanisms underlying the regulation of this process.
Accordingly, initiation begins right after the completion of ribosomal recycling where the 40S subunit is dissociated from the 60S subunit but still carries deacetylated tRNA and mRNA. The tRNA and the mRNA are released with the aid of eIFs 1, 1A, and 3 (Figure 6, step ❶). Thus free postrecycled 40S complexes are then ready for formation of the 43S PIC. At this stage, ABCE1 is able to bind to the 40S complex, preventing premature 60S joining very shortly after subunit splitting. ABCE1 and the 43S PIC factors can coexist at this early stage (Figure 6, step ❷). The attachment of a new mRNA marks the start of the 48S IC assembly and triggers the relocation of the eIF3b-i-g module to the intersubunit face of the SSU, thereby chasing away ABCE1 and setting the eIF2 TC in the scanning-competent conformation, as previously described (131) (Figure 6, steps ❸ and ❹). Importantly, ABCE1 in the late-stage 48S IC was initially misattributed to eIF3 i and g subunits (131), an error that was subsequently corrected (89); the correction was corroborated by other studies presenting structures of ABCE1 bound to the 40S SSU (54, 69) (see 89 for more details).
Figure 6.
Simplified model of eukaryotic translation initiation. The model is based on different structures of eukaryotic (pre)initiation complexes. Different stages of the process are indicated by the circled numbers. The thick, curved green arrow indicates the eIF3b-i-g module relocation from the solvent to the intersubunit side of the 40S subunit, and the red curved arrow indicates its relocation away from the intersubunit face, probably back to the solvent side, until the release of eIF3. Dark gray arrows indicate the conformational change of the eIF2 ternary complex upon eIF3b-i-g relocation and departure to and from the intersubunit face of the 40S, which may reflect its previously described PIN and POUT conformations. Abbreviations: ABCE1, ATP binding cassette E1; C-ter; C-terminus; eIF, eukaryotic initiation factor; GDP, guanosine diphosphate; mRNA, messenger RNA; Pi, inorganic phosphorous; RRM, RNA recognition motif; tRNA, transfer RNA.
Once the start codon is recognized, the eIF3b-i-g module moves away from the 40S intersubunit face, probably because of the release of eIF1 and the conformational rearrangement of the eIF2 TC, leading to the PIN conformation in the closed state of the PIC (Figure 6, step ❺). The departure of eIF3b-i-g from the intersubunit face of the 40S clears up the binding site for ABCE1 to come back again to playing the antiassociation role (Figure 6, steps ❺ and ❻). Meanwhile, eIF5-stimulated GTP hydrolysis on eIF2 produces irreversible release of the Pi, and these factors leave the 48S IC (Figure 3, step ❻). It is not known at which exact moment eIF3 leaves the late-stage initiation complex, and it was proposed that eIF3 actually remains bound to the 40S subunit even past subunit joining during a few elongation cycles (112, 141).
At this stage, ABCE1 is likely to be the only factor preventing premature ribosomal subunit joining (this antiassociation activity was previously misassigned to eIF3i and eIF3g in the mammalian 48S late-stage initiation complex; see 131). The ejection of eIF2 allows eIF5B to bind to the initiator tRNA-CCA end and to be subsequently accommodated at the GTPase binding site (Figure 6, step ❼) once ABCE1 is ejected. The binding of eIF5B stimulates ribosomal subunit joining and marks the end of the initiation process. eIF5B then departs (along with eIF1A that remained attached to the A site) upon GTP hydrolysis (Figure 6, step ❽). The ribosome is now elongation competent.
The Other Initiation Puzzles
We cannot conclude this review on the contribution of the structures of the initiation factors and complexes without mentioning that translation initiation in eukaryotes can occur noncanonically as well (i.e., in a cap-independent manner). The biggest representative of noncanonical initiation is the IRES-dependent initiation. The way IRES-dependent initiation is generally regulated is quite different from cap-dependent initiation.
IRESs are special mRNA molecules with a structured 5′ UTR. They are capable of recruiting the ribosomal complexes through a noncanonical mechanism, circumventing the scanning process for the start codon. Most known IRESs are of viral origin and can be roughly classified into four types, from type 1 to 4 (68). Each type may require a different number of initiation and transacting factors. For instance, types 1 and 2 require nearly a full set of initiation factors, whereas type-4 IRESs require none. Over the past 15 years, many structural studies have attempted to elucidate the molecular basis for IRES-dependent initiation in its various types. We do not discuss the molecular mechanism of noncanonical translation initiation here, but it is useful to mention the major structural studies that have helped elucidate many of its aspects. These studies have been more successful for IRESs of types 3 and 4, which are highly structured and compact compared to IRESs of types 1 and 2, which are less structured and substantially larger in size. The contributions by cryo-EM to this field have been incomparably more significant than those by X-ray crystallography and NMR, which were confined to determination of small, highly structured fragments of IRESs, mainly from hepatitis C virus and other closely related IRESs (HCV-like IRESs, all from type 3; most of these studies are reviewed in 85) and to some extent the cricket paralysis virus (CrPV) IRES (26) (CrPV, type 4, is reviewed in 28). Indeed, cryo-EM has yielded the very first structures of IRES initiation complexes nearly 15 years ago (136), providing a first glance of the interaction between the HCV IRES (type 3) and the SSU. During the following few years, other structures of IRESs where solved at increasingly higher resolutions in different research groups, providing additional insight on the interactions between type-3 and −4 IRESs (mainly HCV and CrPV) and the ribosome (20, 39, 126, 135). However, the resolutions of these studies were not sufficient to allow reliable atomic modeling of the studied IRESs and consequently were incapable of unraveling the intricate mechanism of IRES-dependent initiation. A breakthrough came in 2013, after the advent of the direct detector devices. The number of cryo-EM studies at near-atomic resolutions virtually exploded, providing very detailed snapshots of the dynamic processes underlying type-3 and −4 IRES initiation (1, 38, 50, 73, 96, 97, 113, 152, 153).
In contrast to this large amount of detailed structural results for type-3 and −4 IRESs, IRESs of types 1 and 2 have remained elusive to structural studies despite their tremendous relevance to animal and human health.
CONCLUDING REMARKS
The function of biological molecules has always been investigated and understood thanks to molecular biology by directly observing the impact of their alterations on the formation of supramolecular complexes in vitro and the phenotype of the studied model organisms in vivo. A few decades ago, structural biology bloomed out and offered a new complementary approach to gain further access to the function of biological molecules. Since then, the idea that structure of a molecule is tightly linked to its function became a universally accepted concept.
Translation initiation is one of the many areas of study in biology strongly affected by the capabilities and recent advances of single-particle cryo-EM, the technique of structural investigation that has just been recognized by the 2017 Nobel Prize in Chemistry. Given the complexity of the multistep process of initiation and its regulation, with wide-ranging implications in cancer and other human diseases, the new ability to solve structures of large multi-factor complexes at high resolution can be expected to have a profound impact in the battle against human disease.
ACKNOWLEDGEMENTS
This work was supported by HHMI, the NIH grant R01 GM29169 (to J.F.), and the ANR @RAction program grant for CryoEM80S, ANR-14-ACHN-0024, and this work benefits from funding from the state, managed by the French National Research Agency as part of the Investments for the Future program (to Y.H.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Abeyrathne PD, Koh CS, Grant T, Grigorieff N, Korostelev AA. 2016. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. eLife 5:e14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitken CE, Lorsch JR. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol 19:568–76 [DOI] [PubMed] [Google Scholar]
- 3.Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 20:251–62 [DOI] [PubMed] [Google Scholar]
- 4.Anger AM, Armache J-P, Beninghausen O, Habeck M, Subklewe M., et al. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497:80–85 [DOI] [PubMed] [Google Scholar]
- 5.Apadopoulos E, Jenni S, Kabha E, Takrouri KJ, Yi T, et al. 2014. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. PNAS 111:E3187–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, et al. 2010. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. PNAS 107:19748–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, et al. 2010. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. PNAS 107:19754–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aylett CHS, Boehringer D, Erzberger JP, Schaefer T, Ban N. 2015. Structure of a yeast 40S–eIF1–eIF1A–eIF3–eIF3j initiation complex. Nat. Struct. Mol. Biol 22:269–71 [DOI] [PubMed] [Google Scholar]
- 9.Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, et al. 2012. Near-atomic resolution structural model of the yeast 26S proteasome. PNAS 109:14870–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482:501–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrmann E, et al. 2015. Structural snapshots of actively translating human ribosomes. Cell 161:845–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. 2006. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 14:913–23 [DOI] [PubMed] [Google Scholar]
- 13.Benne R, Hershey JWB. 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Bio. Chem 253:3078–87 [PubMed] [Google Scholar]
- 14.Ben-Shem A, Garreau de Loubresse N, Molnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334:1524–29 [DOI] [PubMed] [Google Scholar]
- 15.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. 2010. Crystal structure of the eukaryotic ribosome. Science 330:1203–9 [DOI] [PubMed] [Google Scholar]
- 16.Benz J, Trachsel H, Baumann U. 1999. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae—the prototype of the DEAD box protein family. Structure 7:671–79 [DOI] [PubMed] [Google Scholar]
- 17.Beznosková P, Cuchalová L, Wagner S, Shoemaker CJ, Gunišová S, et al. 2013. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLOS Genet. 9:e1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beznosková P, Wagner S, Jansen ME, von der Haar T, Valášek LS. 2015. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 43:5099–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bieniossek C, Schütz P, Bumann M, Limacher A, Uson I, Baumann U. 2006. The crystal structure of the carboxy-terminal domain of human translation initiation factor eIF5. J. Mol. Biol 360:457–65 [DOI] [PubMed] [Google Scholar]
- 20.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. 2005. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13:1695–706 [DOI] [PubMed] [Google Scholar]
- 21.Brown CJ, Verma CS, Walkinshaw MD, Lane DP. 2009. Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site. Cell Cycle 8:1905–11 [DOI] [PubMed] [Google Scholar]
- 22.Carter AP, Clemons WM Jr., Brodersen DE, Morgan-Warren RJ, Hartsch T et al. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498–501 [DOI] [PubMed] [Google Scholar]
- 23.Cheung Y-N, Maag D, Mitchell SF, Fekete CA, Algire MA, et al. 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21:1217–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho S, Hoffman DW. 2002. Structure of the β subunit of translation initiation factor 2 from the archaeon Methanococcus jannaschii: a representative of the eIF2β/eIF5 family of proteins. Biochemistry 41:5730–42 [DOI] [PubMed] [Google Scholar]
- 25.Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, et al. 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol 20:7183–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colussi TM, Costantino DA, Zhu J, Donohue JP, Korostelev AA, et al. 2015. Initiation of translation in bacteria by a structured eukaryotic IRES RNA. Nature 519:110–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, et al. 2006. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry 45:4550–58 [DOI] [PubMed] [Google Scholar]
- 28.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. 2008. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol 15:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cuchalová L, Kouba T, Herrmannová A, Dányi I, Chiu W, Valášek L. 2010. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol 30:4671–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, et al. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525:491–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhaliwal S, Hoffman DW. 2003. The crystal structure of the N-terminal region of the alpha subunit of translation initiation factor 2 (eIF2α) from Saccharomyces cerevisiae provides a view of the loop containing serine 51, the target of the eIF2α-specific kinases. J. Mol. Biol 334:187–95 [DOI] [PubMed] [Google Scholar]
- 32.Dhote V, Sweeney TR, Kim N, Hellen CU, Pestova TV. 2012. Roles of individual domains in the function of DHX29, an essential factor required for translation of structured mammalian mRNAs. PNAS 109:E3150–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dube P, Bacher G, Stark H, Mueller F, Zemlin F, et al. 1998. Correlation of the expansion segments in mammalian rRNA with the fine structure of the 80S ribosome; a cryoelectron microscopic reconstruction of the rabbit reticulocyte ribosome at 21 Å resolution. J. Mol. Biol 279:403–21 [DOI] [PubMed] [Google Scholar]
- 34.ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. 2007. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J. Biol. Chem 282:8165–74 [DOI] [PubMed] [Google Scholar]
- 35.ElAntak L, Wagner S, Herrmannová A, Karásková M, Rutkai E, et al. 2010. The indispensable N-terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1-RRM and with eIF1A in stringent AUG selection. J. Mol. Biol 396:1097–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, et al. 2014. Molecular architecture of the 40S-eIF1-eIF3 translation initiation complex. Cell 158:1123–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández IS, Bai X-C, Hussain T, Kelley AC, Lorsch JR, et al. 2013. Molecular architecture of a eukaryotic translational initiation complex. Science 342:1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández IS, Bai X-C, Murshudov G, Scheres SHW, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157:823–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. 2013. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol 20:150–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleming K, Ghuman J, Yuan X, Simpson P, Szendröi A, et al. 2003. Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B. Biochemistry 42:8966–75 [DOI] [PubMed] [Google Scholar]
- 41.Fletcher CM, Pestova TV, Hellen CU, Wagner G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18:2631–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol 27:2384–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R. 2010. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. Biol. Chem. 285:14848–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerbi SA. 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA In Ribosomal RNA—Structure, Evolution, Processing, and Function in Protein Synthesis, ed. Zimmermann RA Dahlberg AE, pp. 71–87. Boca Raton, FL: CRC Press [Google Scholar]
- 45.Greber BJ, Boehringer D, Godinic-Mikulcic V, Crnkovic A, Ibba M, et al. 2012. Cryo-EM structure of the archaeal 50S ribosomal subunit in complex with initiation factor 6 and implications for ribosome evolution. J. Mol. Biol 418:145–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groft CM, Beckmann R, Sali A, Burley SK. 2000. Crystal structures of ribosome anti-association factor IF6. Nat. Struct. Biol 12:1156–64 [DOI] [PubMed] [Google Scholar]
- 47.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, et al. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115:739–50 [DOI] [PubMed] [Google Scholar]
- 48.Grüner S, Peter D, Weber R, Wohlbold L, Chung MY, et al. 2016. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell 64:467–79 [DOI] [PubMed] [Google Scholar]
- 49.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, et al. 2013a. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153:1108–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, et al. 2013. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503:539–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, et al. 2013. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 494:385–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henshaw EC, Guiney DG, Hirsch CA. 1973. The ribosome cycle in mammalian protein synthesis. I. The place of monomeric ribosomes and ribosomal subunits in the cycle. J. Biol. Chem 248:4367–76 [PubMed] [Google Scholar]
- 53.Herrmannová A, Daujotyte D, Yang JC, Cuchalová L, Gorrec F, et al. 2012. Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre-initiation complex assembly. Nucleic Acids Res. 40:2294–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, et al. 2017. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat. Struct. Mol. Biol 24:453–60 [DOI] [PubMed] [Google Scholar]
- 55.Hinnebusch AG. 2006. eIF3: a versatile scaffold for translation initiation complexes. TIBS 31:553–62 [DOI] [PubMed] [Google Scholar]
- 56.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem 83:779–812 [DOI] [PubMed] [Google Scholar]
- 57.Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci 42:589–611 [DOI] [PubMed] [Google Scholar]
- 58.Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4:a011544 Deleted in proof [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiyama TB, Ito T, Imataka H, Yokoyama S. 2009. Crystal structure of the α subunit of human translation initiation factor 2B. J. Mol. Biol 392:937–51 [DOI] [PubMed] [Google Scholar]
- 60.Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, et al. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159:597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito T, Marintchev A, Wagner G. 2004. Solution structure of human initiation factor eIF2α reveals homology to the elongation factor eEF1B. Structure 12:1693–704 [DOI] [PubMed] [Google Scholar]
- 62.Jackson RJ. 2007. The missing link in the eukaryotic ribosome cycle. Mol. Cell 28:356–58 [DOI] [PubMed] [Google Scholar]
- 63.Jackson RJ, Hellen CUT, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol 11:113–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia MZ, Horita S, Nagata K, Tanokura M. 2010. An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2α and the 3′ end fragment of 16S rRNA. J. Mol. Biol 398:774–85 [DOI] [PubMed] [Google Scholar]
- 65.Kakuta Y, Tahara M, Maetani S, Yao M, Tanaka I, Kimura M. 2004. Crystal structure of the regulatory subunit of archaeal initiation factor 2B (aIF2B) from hyperthermophilic archaeon Pyrococcus horikoshii OT3: a proposed structure of the regulatory subcomplex of eukaryotic IF2B. Biochem. Biophys. Res. Commun 319:725–32 [DOI] [PubMed] [Google Scholar]
- 66.Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, et al. 2016. Crystal structure of eukaryotic translation initiation factor 2B. Nature 531:122–25 [DOI] [PubMed] [Google Scholar]
- 67.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. 2015. Structure of the human 80S ribosome. Nature 520:640–45 [DOI] [PubMed] [Google Scholar]
- 68.Kieft JS. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci 33:274–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kiosze-Becker K, Ori A, Gerovac M, Heuer A, Nürenberg-Goloub E, et al. 2016. Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry. Nat. Commun 7:13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khoshnevis S, Gunišová S, Vlčková V, Kouba T, Neumann P, et al. 2014. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res. 42:4123–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334:941–48 [DOI] [PubMed] [Google Scholar]
- 72.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. 2012. Atomic structures of the eukaryotic ribosome. Trends Biochem. Sci 37:189–98 [DOI] [PubMed] [Google Scholar]
- 73.Koh CS, Brilot AF, Grigorieff N, Korostelev AA. 2014. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. PNAS 111:9139–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuhle B, Eulig NK, Ficner R. 2015. Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res. 43:9994–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuhle B, Ficner R. 2014. Structural insight into the recognition of amino-acylated initiator tRNA by eIF5B in the 80S initiation complex. BMC Struct. Biol 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar P, Hellen CU, Pestova TV. 2016. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 30:1573–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee AS, Kranzusch PJ, Doudna JA, Cate JH. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536:96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, et al. 2014. Crystal structure of the human COP9 signalosome. Nature 512:161–65 [DOI] [PubMed] [Google Scholar]
- 79.Liu Y, Neumann P, Kuhle B, Monecke T, Schell S, et al. 2014. Translation initiation factor eIF3b contains a nine-bladed b-propeller and interacts with the 40S ribosomal subunit. Structure 22:923–30 [DOI] [PubMed] [Google Scholar]
- 80.Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, et al. 2016. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. PNAS 113:12174–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, et al. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59:399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loh PG, Yang H-S, Walsh MA, Wang Q, Wang X, et al. 2009. Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J. 28:274–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 17:2786–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lomakin IB, Steitz TA. 2013. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500:307–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res. 139:166–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luna RE, Arthanari H, Hiraishi H, Akabayov B, Tang L, et al. 2013. The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemistry 52:9510–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Majumdar R, Bandyopadhyay A, Maitra U. 2003. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J. Biol. Chem 278:6580–87 [DOI] [PubMed] [Google Scholar]
- 88.Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, et al. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159:1086–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mancera-Martínez E, Brito Querido J, Valasek LS, Simonetti A, Hashem Y. 2017. ABCE1: a special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol. 12:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951–61 [DOI] [PubMed] [Google Scholar]
- 91.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3:707–16 [DOI] [PubMed] [Google Scholar]
- 92.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7:193–203 [DOI] [PubMed] [Google Scholar]
- 93.McKeegan KS, Borges-Walmsley MI, Walmsley AR. 2003. The structure and function of drug pumps: an update. Trends Microbiol. 11:21–29 [DOI] [PubMed] [Google Scholar]
- 94.Meleppattu S, Kamus-Elimeleh D, Zinoviev A, Cohen-Mor S, Orr I, Shapira M. 2015. The eIF3 complex of Leishmania-subunit composition and mode of recruitment to different cap-binding complexes. Nucleic Acids Res. 43:6222–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melinkov S, et al. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol 19:560–67 [DOI] [PubMed] [Google Scholar]
- 96.Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, et al. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell 57:422–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernández IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5:e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nag N, Lin KY, Edmonds KA, Yu J, Nadkarni D, et al. 2016. eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res. 44:7441–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nanda JS, Saini AK, Muñoz AM, Hinnebusch AG, Lorsch JR. 2013. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem 288:5316–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nielsen KH, Szamecz B, Valásek L, Jivotovskaya A, Shin BS, Hinnebusch AG. 2004. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 23:1166–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nielsen KH, Valášek L, Sykes C, Jivotovskaya A, Hinnebusch AG. 2006. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol. Cell. Biol 26:2984–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nikonov O, Stolboushkina E, Nikulin A, Hasenöhrl D, Bläsi U, et al. 2007. New insights into the interactions of the translation initiation factor 2 from archaea with guanine nucleotides and initiator tRNA. J. Mol. Biol 373:328–36 [DOI] [PubMed] [Google Scholar]
- 103.Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, et al. 2017. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep. 18:2651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, et al. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26:41–50 [DOI] [PubMed] [Google Scholar]
- 105.Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854–59 [DOI] [PubMed] [Google Scholar]
- 107.Pestova TV, Kolupaeva VG. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peter D, Igreja C, Weber R, Wohlbold L, Weiler C, et al. 2015. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell 57:1074–87 [DOI] [PubMed] [Google Scholar]
- 109.Pisarev AV, Hellen CUT, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131:286–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, et al. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37:196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135:1237–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pöyry TAA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ. 2007. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev. 21:3149–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. 2015. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun 6:7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Querol-Audi J, et al. 2013. Architecture of human translation initiation factor 3. Structure 21:920–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331:730–36 [DOI] [PubMed] [Google Scholar]
- 116.Rees DC, Johnson E, Lewinson O. 2009. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol 10:218–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, et al. 2008. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J. Biol. Chem 283:1094–103 [DOI] [PubMed] [Google Scholar]
- 118.Rezende AM, Assis LA, Nunes EC, da Costa Lima TD, Marchini FK, et al. 2014. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates—identification of conserved and divergent features based on orthologue analysis. BMC Genom. 15:1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roll-Mecak A, Alone P, Cao C, Dever TE, Burley SK. 2004. X-ray structure of translation initiation factor eIF2γ: implications for tRNA and eIF2α binding. J. Biol. Chem 279:10634–42 [DOI] [PubMed] [Google Scholar]
- 120.Roll-Mecak A, Cao C, Dever TE, Burley SK. 2000. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103:781–92 [DOI] [PubMed] [Google Scholar]
- 121.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol 10:1134–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. 2012. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48:375–86 [DOI] [PubMed] [Google Scholar]
- 123.Schmitt E, Blanquet S, Mechulam Y. 2002. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 21:1821–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schmitt E, Naveau M, Mechulam Y. 2010. Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett. 584:405–12 [DOI] [PubMed] [Google Scholar]
- 125.Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux PD, Perez J, et al. 2012. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat. Struct. Mol. Biol 19:450–54 [DOI] [PubMed] [Google Scholar]
- 126.Schüler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, et al. 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol 13:1092–96 [DOI] [PubMed] [Google Scholar]
- 127.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, et al. 2008. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein–protein interactions. PNAS 105:9564–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sekiyama N, Arthanari H, Papadopoulos E, Rodriguez-Mias RA, Wagner G, Léger-Abraham M. 2015. Molecular mechanism of the dual activity of 4EGI-1: dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. PNAS 112:E4036–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shin B-S, Kim J-R, Walker SE, Dong J, Lorsch JR, Dever TE. 2011. Initiation factor eIF2γ promotes eIF2–GTP–Met–tRNAiMet ternary complex binding to the 40S ribosome. Nat. Struct. Mol. Biol 18:1227–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shin B-S, Maag D, Roll-Mecak A, Arefin MS, Burley SK, et al. 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111:1015–25 [DOI] [PubMed] [Google Scholar]
- 131.Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martinez E, Renaud A, et al. 2016. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell 63:206–17 [DOI] [PubMed] [Google Scholar]
- 132.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310:1513–15 [DOI] [PubMed] [Google Scholar]
- 133.Smith MD, Arake-Tacca L, Nitido A, Montabana E, Park A, Cate JH. 2016. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure 24:886–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sokabe M, Yao M, Sakai N, Toya S, Tanaka I. 2006. Structure of archaeal translational initiation factor 2 βγ-GDP reveals significant conformational change of the β-subunit and switch 1 region. PNAS 103:13016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118:465–75 [DOI] [PubMed] [Google Scholar]
- 136.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, et al. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–62136. [DOI] [PubMed] [Google Scholar]; Srivastava S,Verschoor A Frank J 1992. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol 226:301–4 [DOI] [PubMed] [Google Scholar]
- 137.Srivastava S, Verschoor A, Frank J. 1992. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol 226:301–4 [DOI] [PubMed] [Google Scholar]
- 138.Stolboushkina E, Nikonov S, Nikulin A, Bläsi U, Manstein DJ, et al. 2008. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the α- and β-subunits. J. Mol. Biol 382:680–91 [DOI] [PubMed] [Google Scholar]
- 139.Stolboushkina E, Nikonov S, Zelinskaya N, Arkhipova V, Nikulin A, et al. 2013. Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAfMet. J. Mol. Biol 425:989–98 [DOI] [PubMed] [Google Scholar]
- 140.Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, et al. 2011. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). PNAS 108:20473–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Szamecz B, Rutkai E, Cuchalová L, Munzarová V, Herrmannová A, et al. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 22:2414–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Trachsel H, Staehelin T. 1979. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim. Biophys. Acta 565:305–14 [DOI] [PubMed] [Google Scholar]
- 143.Valášek LS. 2012. ‘Ribozoomin’—translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci 13:305–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Valášek LS, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol 24:9437–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vasile F, Pechkova E, Nicolini C. 2008. Solution structure of the β-subunit of the translation initiation factor aIF2 from archaebacteria Sulfolobus solfataricus. Proteins 70:1112–15 [DOI] [PubMed] [Google Scholar]
- 146.Virgili G, Frank F, Feoktistova K, Sawicki M, Sonenberg N, et al. 2013. Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 21:517–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Voorhees RM, Fernández IS, Scheres SHW, Hegde RS. 2014. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157:1632–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Waters LC, Strong SL, Ferlemann E, Oka O, Muskett FW, et al. 2011. Structure of the tandem MA-3 region of Pdcd4 protein and characterization of its interactions with eIF4A and eIF4G: molecular mechanisms of a tumor suppressor. J. Biol. Chem 286:17270–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei Z, et al. 2004. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J. Biol. Chem 279:34983–90 [DOI] [PubMed] [Google Scholar]
- 150.Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. 2013. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat. Struct. Mol. Biol 20:1015–17 [DOI] [PubMed] [Google Scholar]
- 151.Wilson DN, Doudna Cate JH. 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol 4:a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, et al. 2014. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 34:3042–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, et al. 2014. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol 21:721–27 [DOI] [PubMed] [Google Scholar]
- 154.Yatime L, Mechulam Y, Blanquet S, Schmitt E. 2006. Structural switch of the γ subunit in an archaeal aIF2αγ heterodimer. Structure 14:119–28 [DOI] [PubMed] [Google Scholar]
- 155.Yatime L, Mechulam Y, Blanquet S, Schmitt E. 2007. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. PNAS 104:18445–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yatime L, Schmitt E, Blanquet S, Mechulam Y. 2005. Structure-function relationships of the intact aIF2α subunit from the archaeon Pyrococcus abyssi. Biochemistry 44:8749–56 [DOI] [PubMed] [Google Scholar]
- 157.Yokoyama T, Suzuki T. 2008. Ribosomal RNAs are tolerant toward genetic insertions: evo- lutionary origin of the expansion segments. Nucleic Acids Res. 36:3539–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang F, Saini AK, Shin B-S, Nanda J, Hinnebusch AG. 2015. Conformational changes in the P site and mRNA entry channel evoked by AUG recognition in yeast translation preinitiation complexes. Nucleic Acids Res. 43:2293–312 [DOI] [PMC free article] [PubMed] [Google Scholar]