Dear Editor,
Dynamic and reversible N6-methyladenosine (m6A) RNA methylation has been found to greatly impact gene expression, leading to the field of epitranscriptomics.1 Unlike m6A that is an internal modification, a terminal modification at mRNA cap in higher eukaryotes exists, termed as N6,2′-O-dimethyladenosine (m6Am) (Fig. 1a). The first and sometimes the second nucleotide after the N7-methylguanosine (m7G) cap can be methylated at the 2′-hydroxyl group; and when the first nucleotide is 2′-O-methyladenosine (Am), it can be further methylated at the N6 position to become m6Am. m6Am was first identified in animal cells and virus mRNA in 19752; several years later the methyltransferase was partially purified and was proposed to be a species whose molecular weight is ~65 KD.3 Only very recently, m6Am was found to be reversible as well: the first m6A demethylase FTO also catalyzed the demethylation of m6Am, depending on its sub-cellular localizations.4,5 By changing FTO levels, m6Am at mRNA cap was also suggested to impair DCP2-mediated mRNA decapping.4 However, the methyltransferase of m6Am is not unambiguously identified, significantly hindering the functional and mechanistic study of m6Am.
Fig. 1.
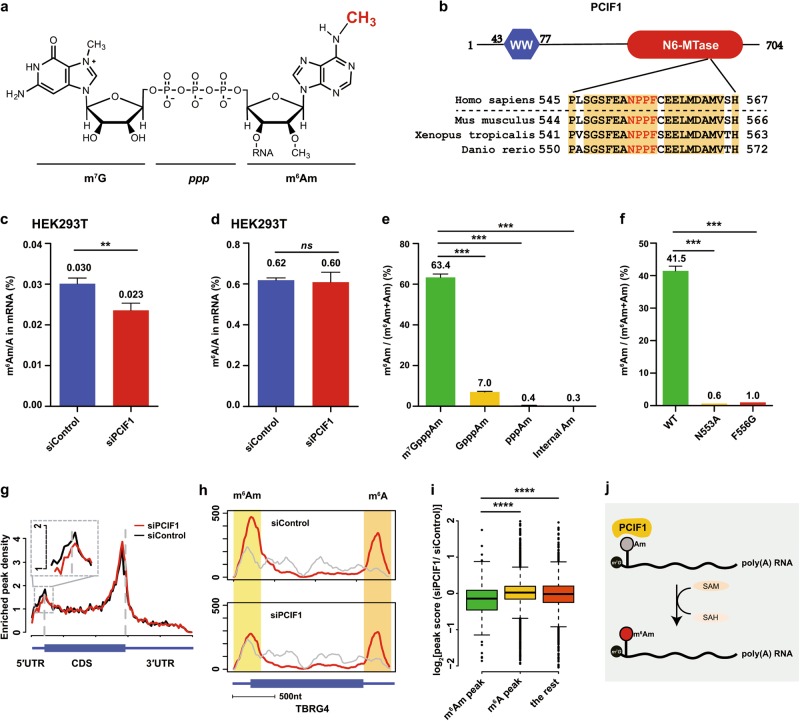
Identification of N6,2′-O-dimethyladenosine methyltransferase. a Chemical structure of m6Am, which is adjacent to the m7G mRNA cap. b Cartoon view of the predicted domain structure of PCIF1, with the conserved “NPPF” motif in the zoom-in view. A sequence alignment is shown below to highlight the high conservation of the key residues for PCIF1 orthologues. Residue 43–77 (blue segment) represents the WW domain, and the red segment denotes the putative catalytic methyltransferase domain. c LC-MS/MS quantification of the m6Am/A ratios of HEK293T polyA + RNA treated with control or PCIF1 siRNA (n = 3). d LC-MS/MS quantification of the m6A/A ratios of HEK293T polyA+ RNA treated with control or PCIF1 siRNA (n = 3). e Quantification of the m6Am/(Am + m6Am) ratios in RNA probes starting with different cap structure (n = 3). f Quantification of the methylation activity of WT and mutant PCIF1 proteins (n = 3). g Distribution of enriched m6A/m6Am peak density across mRNA segments of control and PCIF1 knockdown samples using an m6A-seq protocol with random priming. Each segment was normalized according to its average length in Ref-seq annotation. h One representative transcript harboring m6Am and m6A peaks. The m6Am peak at the 5′-terminal is significantly decreased upon PCIF1 knockdown, while the m6A peak at the 3′-UTR stays the same. The grey line denotes “Input”, and the red line denotes “IP”. i Boxplot of log2fold change of peak score in PCIF1 knockdown and control mRNA. Enriched peaks are classified into three groups: m6Am peak (near TSS and without GGACH motif), m6A peak (not in TSS and with GGACH motif), and the rest (potentially m6Am + m6A). j A proposed model for mammalian mRNA m6Am modification mediated by PCIF1. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
To clearly identify the methyltransferase, we fractioned the cell lysates of HEK293 cells, which contain robust N6-methylation activity (Supplementary information, Fig.S1a). This activity was assayed by incubating the column fractions with a 25 nt, synthetic vaccinia virus RNA probe (Probe-1, see Supplementary information) that begins with m7GpppAm. We modified the purification route of cell lysates, based on the procedure originally reported3 (Supplementary information, Fig.S1b), and subjected the fractions of high N6-methylation activity to protein identification by sensitive mass spectrometry. We then searched for proteins with putative methyltransferase domain or sequence motif in the list of more than 100 proteins detected by MS, and found a protein named “phosphorylated CTD-interacting factor 1” (or PCIF1) (Fig. 1b; Supplementary information, Fig.S1c), which was bioinformatically proposed to be a DNA/RNA N6-adenosine methyltransferase.6 PCIF1 was originally identified and named due to its ability to directly bind to the phosphorylated C-terminal domain of RNA polymerase II via its WW domain7; hence it was speculated to play a role in mRNA biogenesis. However, no enzymatic activity has been reported for PCIF1.
To test whether PCIF1 possesses methyltransferase activity in vivo, we first knocked down PCIF1 in HEK293 cells by two independent siRNAs and confirmed the knockdown efficiency by qRT-PCR (Supplementary information, Fig.S2a). We then measured the level of m6Am in polyA + RNA fraction after decapping using LC-MS/MS. We were able to observe a reduction of m6Am level upon PCIF1 knockdown (Fig. 1c; Supplementary information, Fig.S2b); importantly, the level of the internal m6A modification remained unchanged (Fig. 1d), suggesting that PCIF1 is a specific methyltransferase for the terminal m6Am. Encouraged by the in vivo results, we then expressed and purified recombinant PCIF1 protein, and tested whether the single protein is capable of methylating RNA substrates under in vitro conditions (Supplementary information, Fig.S2c). The highest activity of PCIF1 was obtained with RNA Probe-1 beginning with a complete cap structure m7GpppAm; much lower activity was found with RNA beginning with GpppAm; and barely detectable activity was found with RNA beginning with pppAm or RNA Probe-2 with an internal Am (Fig. 1e). The above enzymatic preference was also supported by biochemical experiments using two different RNA probes (Probe-3 and Probe-4), which in addition showed that the ribose 2′-O-methylation is required for optimal methylation activity as well (Supplementary information, Fig.S2d). Moreover, we introduced point mutations in the highly conserved “NPPF” motif that is characteristic of adenosine methyltransferases, and found that the disruption of this motif reduced the methyltransferase activity of the mutant proteins (Fig. 1f; Supplementary information, Fig.S1d). Because PCIF1 is highly conserved among different species (Fig. 1b), we further tested whether the mouse PCIF1 protein is also functional. We knocked down mPcif1 by siRNA in mouse NIH-3T3 cells and also observed reduced m6Am level (Supplementary information, Fig.S3a, b). Additionally, mouse PCIF1 protein also exhibited a robust methylation activity in vitro (Supplementary information, Fig.S3c). Altogether, the evidence presented above demonstrated that PCIF1 is a novel mammalian m6Am writer, which is specific for the 5′-end capped RNA.
To identify the RNA targets of PCIF1, we performed m6A-seq experiments for PCIF1 knockdown and control cells using an anti-m6A antibody.8,9 Because the antibody recognizes m6Am and m6A, both types of modifications were enriched and hence detected simultaneously.10 m6A modifications are known to be enriched around 3′-UTR, with a small portion also present internally in the 5′-UTR; while m6Am modifications localized at the 5′-end of RNA. We envisioned that the cap-specific PCIF1 should selectively alter the m6Am modification at the 5′-terminal region of transcripts. Indeed, we observed a reduction of modification peaks at the 5′-end but not the 3′-UTR regions of mRNAs upon PCIF1 knockdown (Fig. 1g; Supplementary information, Fig.S3d). One example is the TBRG4 transcript, for which we found a 5′-end peak and a 3′-UTR peak by m6A-seq (Fig. 1h); only the former peak underwent a clear reduction while the latter remained the same. We then grouped the enriched peaks into three categories and again observed significantly decreased signals for the m6Am peaks after PCIF1 knockdown when comparing to the m6A and m6A + m6Am categories (Fig. 1i; Supplementary information, Table S1). We further adopted a different m6A-seq procedure that can preserve the 5′-end information of polyA+ RNA, and again found a decrease of m6Am peak intensity after PCIF1 knockdown (Supplementary information, Fig.S3e). In addition, a motif analysis revealed that m6Am modification occurs at the transcription start sites, in accordance with the known m6Am pattern (Supplementary information, Fig.S3f).4 Thus, results from our m6A-seq experiments revealed the direct mRNA targets of PCIF1 inside of human cells (Fig. 1j).
Taken together, in this study we revealed the exact identity of the m6Am writer protein, characterized its biochemical property and substrate preference, and profiled its cellular targets using an epitranscriptomic sequencing approach. PCIF1 recognizes the positively charged cap structure m7GpppAm for optimal activity and is a “stand-alone” RNA methyltransferase. In comparison, the internal m6A is installed by a methyltransferase complex, the core components of which are composed of METTL3, METTL14 and WTAP. The m6A methyltransferase complex also recognizes internal adenosines, with a preference for those located within a RRACH consensus motif. Hence, while m6Am and m6A share a common eraser protein FTO, the writer proteins for the two modifications are orthogonal. Manipulating the protein levels of the writers could potentially separate the differential roles of FTO in demethylating m6Am and m6A. The functional study of m6A is greatly facilitated by the discovery and characterization of its regulation system involving the writer, reader and eraser proteins; we envision that the identification of PCIF1 as the m6Am writer will pave the path toward functional and mechanistic dissection of this dynamic and reversible epitranscriptomic mark in the future.
Electronic supplementary material
Acknowledgements
The authors would like to thank Shengxian Gao and Shaokai Ning for helping with HEK293 suspension cell culture, Menghao Liu and Yuxiang Liu for technological advice of protein purification, Xushen Xiong for advice on bioinformatics analysis, Xiaoyu Li for discussions, Dong Liu for assistance with protein MS analysis, and the Core Facilities at School of Life Sciences, Peking university. Part of the analysis was performed on the High-Performance Computing Platform of the Center for Life Science. This work was supported by the National Natural Science Foundation of China (nos. 91740112 and 2182570 to C.Y.), the National Basic Research Foundation of China (no. 2016YFC0900301) and the Joint Laboratory of International Scientific and Technological Cooperation.
Author contributions
H.S. and M.Z. conceived all experiments under the guidance of C.Y., H.S. synthesized the RNA probes, purified the recombinant protein and performed in vitro methylation experiments. M.Z. developed the methylation assay, performed sequencing and cell biology experiments. K.L. performed the bioinformatics analysis with the help of C.Y., D.B. assisted in probe synthesis.
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0117-4.
References
- 1.Roundtree IA, Evans ME, Pan T, He C. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei C, Gershowitz A, Moss B. Nature. 1975;257:251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- 3.Keith JM, Ensinger MJ, Mose B. J. Biol. Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- 4.Mauer J, et al. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei J, et al. Mol. Cell. 2018;71:973–985. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Zhang D, Aravind L. Bioessays. 2016;38:27–40. doi: 10.1002/bies.201500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan H, et al. Biochem. Biophys. Res. Commun. 2003;301:378–385. doi: 10.1016/S0006-291X(02)03015-2. [DOI] [PubMed] [Google Scholar]
- 8.Dominissini D, et al. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y, et al. PLoS Biol. 2018;16:e2006092. doi: 10.1371/journal.pbio.2006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Xiong X, Yi C. Nat. Methods. 2016;14:23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.