Abstract
Background:
A number of case-control studies were conducted to investigate the association of angiotensin converting enzyme insertion/deletion (ACE I/D) polymorphism with breast cancer. But the results remain controversial. This meta-analysis aims to comprehensively evaluate the association of ACE I/D polymorphism with breast cancer.
Method:
A comprehensive literature search on PubMed, Google Scholar, SCOPUS and ISI Web of Knowledge databases for studies published up to June 01, 2018 was performed. Summary odds ratios (ORs) and 95% confidence intervals (CI) were estimated. Publication bias of literatures was evaluated using funnel plots and Egger’s test.
Results:
A total of 20 studies including 2846 breast cancer cases 9299 controls meeting the predefined criteria were involved in the meta-analysis. Overall, the ACE I/D polymorphisms was significantly associated with breast cancer under the allele model (I vs. D: OR= 0.803, 95% CI 0.647-0.996, p=0.046), the homozygote model (II vs. DD: OR= 0.662, 95% CI 0.462-0.947, p=0.024), the heterozygote model (ID vs. DD: OR= 0.707, 95% CI 0.528-0.946, p=0.020), the dominant model (II+ID vs. DD: OR= 0.691, 95% CI 0.507-0.941, p=0.019). In the subgroup analysis by ethnicity, a significant association was found among Asian and Caucasian populations, but not among mixed populations.
Conclusions:
This meta-analysis suggests that ACE I/D polymorphism may be associated with increased risk of breast cancer, especially among Asian and Caucasians. However, well-designed studies with larger sample size and more ethnic groups are needed to further validate the results.
Keywords: Breast cancer, ACE I/D, genetic polymorphism, cancer, Meta-analysis, susceptibility
Introduction
Breast cancer is the most frequently diagnosed cancer among women, which contributed to 25% of all cancer cases in women worldwide (Nedooshan et al., 2017; Yazdi et al., 2015). The etiology of breast cancer is still not fully understood. It is a heterogeneous disease in regard to its clinical, histological, and molecular profile (Kamali et al., 2017; Kamali et al., 2017). At present, genetic polymorphisms are increasingly recognized as contributors to breast cancer risk. Numerous breast cancer risk factors such as age, age at menarche, age at first childbirth, and number of first-degree female relatives with breast cancer have been identified (Shiryazdi et al., 2015; Shiryazdi et al., 2015). The heritable defects of BRCA1 and BRCA2 are identified in less than 50% of hereditary breast cancer cases (Forat-Yazdi et al., 2015; Forat-Yazdi et al.,2017; Neamatzadeh et al., 2015). Several epidemiological and molecular studies indicates ACE I/D gene polymorphism as a risk factor involved in the initiation and progression of different disease such as diabetic nephropathy, end-stage renal disease(ESRD) and cancer (Buraczynska et al., 2006; Singh et al., 2018).
The ACE, a zinc metalloenzyme, is a member of renin–angiotensin system (RAS) or the renin–angiotensin–aldosterone system (RAAS) involved in catalyzing the conversion of angiotensin I into a physiologically active peptide angiotensin II (Fagyas et al., 2014). The ACE gene (Gene ID: 1636; also known as: DCP; ACE1; DCP1; CD143) is localized in human chromosomes 17q23, comprises 26 exons, spans about 21 kb and more than 13 polymorphisms in this gene have been identified with susceptibility to different disease such as ACE I/D (rs 1799752), A240A>T (rs4291), 2350G>A (rs4344), and 17888C>T (rs4359) (Sayed-Tabatabaei et al., 2006). The ACE insertion/deletion (I/D) polymorphism is a nonsense and 287 bp Alu repeat sequence of DNA in the intron 16 of ACE gene, which represented by “Insertion” or “I”, and absence of the same denotes “Deletion” or “D” (Mahjoub et al., 2016).
To date, several studies have evaluated the association between ACE I/D polymorphism and breast cancer risk. However, the results are inconsistent and inconclusive, with some studies found significant association, while others were not. Although previous meta-analyses has tried to explore the whether the ACE I/D polymorphism is associated with increased risk of breast cancer, recently, new case-control studies in different ethnicities have been published. Therefore, we pooled all eligible studies in the present meta-analysis to evaluate the association of ACE I/D polymorphism with the risk of breast cancer.
Materials and Methods
Search Strategy
Electronic databases of PubMed (Medline), EMBASE and Google Scholar, ISI Web of Knowledge, Chinese National Knowledge Infrastructure database (CNKI), and Chinese Biomedical Literature Database (CBM) were used to identify all studies that evaluate the associations of ACE I/D polymorphism with breast cancer up to June 01, 2018. The key words used for search were: (breast, cancer, carcinoma, tumor, neoplasm, malignancy) and (angiotensin converting enzyme insertion/deletion, ACE I/D) and (polymorphism, mutation, variant, SNP). In addition, we have hand searched reference list of all eligible studies and related reviews for additional suitable studies. Studies in English, Persia, chinses and Portuguese languages were included to the meta-analysis. In addition, we have not set any restriction on time period, sample size, and ethnicity.
Data extraction
Two authors reviewed and extracted data independently in accordance with the inclusion criteria. The results were compared, and if any disagreement appeared, a third investigator was invited to evaluate such studies, and then the discrepancy was resolved by discussion. The following data were extracted: the name of first author, year of publication, country, ethnicity, sample size, allele numbers and genotype distributions in cases and controls, and the results of Hardy-Weinberg equilibrium (HWE) test.
Inclusion and exclusion criteria
The studies included in this meta-analysis had to meet the following criteria: (1) only case-control or cohort studies; (2) evaluation of the ACE I/D polymorphism and breast cancer risk; (3) provided sufficient genotyping data to calculate odds ratios (ORs) and their 95% confidence intervals (CIs). On the other side, the major reasons for study exclusion were: (1) studies were not relevant to ACE I/D polymorphism or breast cancer; (2) studies did not report genotype frequencies; (3) case only studies and control group including malignant tumor patients; (4) the design were based on sibling pairs or linkage studies; (5) reviews, case reports, abstracts, posters, letter and animal studies; and (6) overlapped or duplicated publications. When, more than one of the same populations was included in several studies, the most recently published study or the largest study with extractable data was selected. If original necessary data were unavailable in the selected studies, we have sent a request to the corresponding author for additional data.
Statistical analysis
Pooled ORs with corresponding 95% CIs were calculated to evaluate the association between ACE I/D polymorphism and breast cancer risk. We have used the Z-test to assess the significance of the pooled OR, in which P<0.05 was considered as statistically significant. Pooled ORs was calculated under the following five genetic models: the allele model (I vs. D), the homozygote model (II vs. DD), the heterozygote model (ID vs. DD), the dominant model (II+ID vs. DD), and the recessive model (II vs. ID+DD). The Q-test was used to evaluate between-study heterogeneity, was considered significant when p-value <0.05. Additionally, the I2 statistics was used to quantify or describe the percentage of between-study heterogeneity, which was considered as significant when I2>50% (I2 <25% indicating low heterogeneity; 25%-50% indicating moderate heterogeneity; and >50% indicating high heterogeneity) (Sobhan et al., 2018; Yazdi et al., 2017). Therefore, a fixed-effects model (the Mantel–Haenszel method) as used to calculate pooled OR in the absence of heterogeneity (Mantel and Haenszel, 1959). Otherwise, the random-effects model (the DerSimonian and Laird method) was used (DerSimonian and Laird 1986). The control genotypes were tested for Hardy- Weinberg equilibrium (HWE) using the Goodness of fit Chi-square test. The heterogeneity was adjusted by subgroup analysis and meta-regression. Sensitivity analyses were conducted to evaluate the effect of individual study on pooled ORs and assess the stability of results. The evidence of potential publication bias was evaluated by the funnel plot and Egger’s test, in which P<0.01 was considered as statistically significant. In addition, publication bias was assessed using visual inspection an asymmetric plot (Begg and Mazumdar, 1994; Egger et al., 1997). When publication bias observed, the Duval and Tweedie non-parametric ‘‘trim and fill’’ method was applied to adjust for it. The Comprehensive Meta-Analysis (CMA) Version 2.2 software program (Biostat, USA) was used to perform all statistical analysis, and all the P values were two sided.
Results
According to the present meta-analysis selection criteria, a total of 127 articles were retrieved by literatures search of online databases and hand searching. After review of titles and abstracts, 139 publications were excluded because were reviews, duplication and overlapping data, non-case-control studies, not relevant to ACE I/D polymorphism and breast cancer risk. Finally, a total of 20 case-control studies (Alves Corrêa et al., 2009; Felipe et al., 2011; Fishchuk and Gorovenko, 2013; Ghosh Roy et al., 2015; Gonzalez-Zuloeta Ladd et al., 2005; Haiman et al., 2003; van der Knaap et al., 2008; Koh et al., 2003; Kumar et al., 2016; Mendizábal-Ruiz et al., 2011; Namazi et al., 2010; El Sharkawy et al., 2014; Siddiqi et al., 2010; Singh et al., 2018; Xiaomei et al., 2014; Yaren et al., 2007, 2006) were included in our meta-analysis, consisting of 2,846 cases 9,299 controls. The detailed characteristics of the included studies are listed in Table 1. The original populations contain Singapore, Japan, Netherlands, Turkey, Brazil, Iran, India, Mexico, Colombia, Ukraine, china, and Egypt. There were 8 studies of subjects of Asians, 7 studies of subjects of Caucasian descent, 4 studies of subjects with mixed-ethnicity studies, and one study of subjects of African descent. All the genotype distributions of control subjects were in agreement with HWE for ACE I/D polymorphism except for 7 case-control studies (Table 1).
Table 1.
Characteristics of the Studies Included in the Meta-analysis
| First Author | Country | Ethnicity | Case | Control | Cases | Control | HWE | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Alleles | Genotypes | Alleles | ||||||||||||
| DD | ID | II | D | I | DD | ID | II | D | I | ||||||
| Koh 2003 | Singapore | Asian | 182 | 643 | 23 | 80 | 79 | 126 | 238 | 56 | 305 | 282 | 417 | 869 | 0.036 |
| Haiman 2003a | African Americans | 257 | 631 | 77 | 118 | 62 | 272 | 242 | 221 | 310 | 100 | 752 | 510 | 0.614 | |
| Haiman 2003b | Japan | Asian | 284 | 357 | 37 | 128 | 119 | 202 | 366 | 43 | 160 | 154 | 246 | 460 | 0.884 |
| Haiman 2003c | Mixed | 249 | 652 | 49 | 127 | 73 | 225 | 273 | 162 | 301 | 189 | 463 | 679 | 0.055 | |
| Haiman 2003d | Caucasians | 292 | 402 | 84 | 129 | 79 | 297 | 287 | 124 | 187 | 91 | 435 | 369 | 0.204 | |
| Ladd 2005 | Netherlands | Caucasians | 114 | 4203 | 37 | 55 | 22 | 129 | 99 | 1133 | 2192 | 878 | 4458 | 3948 | 0.002 |
| Yaren 2006 | Turkey | Caucasians | 44 | 46 | 25 | 17 | 2 | 67 | 21 | 28 | 12 | 6 | 68 | 24 | 0.028 |
| Yaren 2007 | Turkey | Caucasians | 57 | 52 | 31 | 24 | 2 | 85 | 28 | 33 | 12 | 7 | 78 | 26 | 0.005 |
| Van der 2008 | Netherlands | Caucasians | 153 | 655 | 54 | 67 | 32 | 175 | 131 | 185 | 329 | 141 | 699 | 470 | 0.815 |
| Alves Corrêa 2009 | Brazil | Mixed | 101 | 307 | 61 | 20 | 20 | 142 | 60 | 141 | 113 | 53 | 395 | 219 | ≤0.001 |
| Namazi 2010 | Iran | Asian | 70 | 70 | 20 | 42 | 8 | 82 | 58 | 29 | 34 | 7 | 92 | 48 | 0.514 |
| Siddiqi 2010 | India | Asian | 130 | 228 | 62 | 43 | 25 | 167 | 93 | 96 | 107 | 25 | 299 | 157 | 0.552 |
| Mendizábal-Ruiz 2011 | Mexico | Mixed | 63 | 288 | 53 | 6 | 4 | 112 | 14 | 63 | 151 | 74 | 277 | 299 | 0.394 |
| Felipe 2011 | Colombia | Mixed | 50 | 50 | 10 | 23 | 17 | 43 | 57 | 10 | 24 | 16 | 44 | 56 | 0.854 |
| Fishchuk 2013 | Ukraine | Caucasians | 131 | 102 | 41 | 53 | 37 | 135 | 127 | 21 | 50 | 31 | 92 | 112 | 0.918 |
| Xiaomei 2014 | China | Asian | 123 | 72 | 61 | 32 | 30 | 154 | 92 | 36 | 19 | 17 | 91 | 53 | ≤0.001 |
| El-Sharkawy 2014 | Egypt | African | 70 | 50 | 29 | 28 | 13 | 86 | 54 | 21 | 21 | 8 | 63 | 37 | 0.483 |
| Ghosh 2015 | India | Asian | 108 | 128 | 62 | 28 | 18 | 152 | 64 | 32 | 50 | 46 | 114 | 142 | 0.017 |
| Kumar 2016 | India | Asian | 213 | 213 | 35 | 86 | 92 | 156 | 270 | 24 | 77 | 112 | 125 | 301 | 0.061 |
| Singh 2018 | India | Asian | 155 | 150 | 86 | 59 | 10 | 231 | 79 | 29 | 74 | 47 | 132 | 168 | 0.989 |
HWE, Hardy Weinberg Equilibrium
Quantitative Data Synthesis
In the overall analysis, statistically significant association between ACE I/D polymorphism and breast cancer susceptibility was observed under the allele model (I vs. D: OR= 0.803, 95% CI 0.647-0.996, p=0.046, Figure 1A), the homozygote model (II vs. DD: OR= 0.662, 95% CI 0.462-0.947, p=0.024), the heterozygote model (ID vs. DD: OR= 0.707, 95% CI 0.528-0.946, p=0.020), the dominant model (II+ID vs. DD: OR= 0.691, 95% CI 0.507-0.941, p=0.019), but not under the recessive model (II vs. ID+DD: OR= 0.841, 95% CI 0.663-1.067, p=0.154, Figure 1B). When stratified, the association between ACE I/D polymorphism and breast cancer risk was observed among Asian populations under the heterozygote model (ID vs. DD: OR= 0.655, 95% CI 0.436-0.984, p=0.041) and Caucasian populations under the allele model (I vs. D: OR= 1.115, 95% CI 1.002-1.241, p=0.046), but not among mixed populations. In the subgroup analysis by studies quality (fulfilling HWE or not), a significant association between the ACE I/D polymorphism and breast cancer risk was observed under the heterozygote model (ID vs. DD: OR= 0.656, 95% CI 0.454-0.946, p=0.024).
Figure 1.
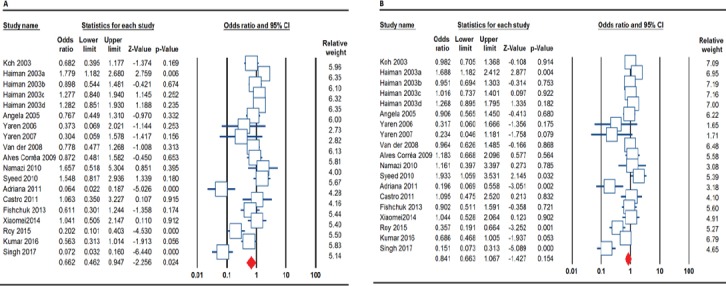
Forest Plots for Association between ACE I/D Polymorphism and Breast Cancer Risk. A, the allele model (I vs. D) and B, the recessive model (II vs. ID+DD).
Heterogeneity Test
We observed high to extreme heterogeneities under all genetic model including the allele model (I vs. D: I2= 88.84% and PH≤0.001), the homozygote model (II vs. DD: I2= 83.10% and PH≤0.001), the heterozygote model (ID vs. DD: I2= 81.79% and PH≤0.001), the dominant model (II+ID vs. DD: I2= 86.52% and PH≤0.001), and the recessive model (II vs. ID+DD: I2= 74.01% and PH≤0.001, Table 2). In the subgroup analysis by ethnicity, results were similar in the Asians and mixed population. While, in the Caucasians, between study heterogeneity was significantly reduced under the allele model (I vs. D: I2= 51.31% and PH=0.055), the heterozygote model (ID vs. DD: I2= 47.68% and PH=0.075), and the dominant model (II+ID vs. DD: I2= 46.81% and PH=0.080).
Table 2.
Summary Risk Estimates for Association between ACE I/D Polymorphism and Risk of Breast Cancer
| Subgroup | Genetic Model | Type of Model | Heterogeneity | Odds Ratio (OR) | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | ZOR | POR | PBeggs | PEggers | |||
| Overall | I vs. D | Random | 88.84 | ≤0.001 | 0.803 | 0.647-0.996 | -1.992 | 0.046 | 0.035 | 0.048 |
| II vs. DD | Random | 83.10 | ≤0.001 | 0.662 | 0.462-0.947 | -2.256 | 0.024 | 0.030 | 0.024 | |
| ID vs. DD | Random | 81.79 | ≤0.001 | 0.707 | 0.528-0.946 | -2.332 | 0.020 | 0.528 | 0.233 | |
| II+ID vs. DD | Random | 86.52 | ≤0.001 | 0.691 | 0.507-0.941 | -2.348 | 0.019 | 0.363 | 0.157 | |
| II vs. ID+DD | Random | 74.01 | ≤0.001 | 0.841 | 0.663-1.067 | -1.427 | 0.154 | 0.234 | 0.048 | |
| By Ethnicity Asian | I vs. D | Random | 90.25 | ≤0.001 | 0.735 | 0.512-1.055 | -1.668 | 0.095 | 0.710 | 0.606 |
| II vs. DD | Random | 86.73 | ≤0.001 | 0.578 | 0.303-1.103 | -1.663 | 0.096 | 0.710 | 0.657 | |
| ID vs. DD | Random | 74.70 | ≤0.001 | 0.655 | 0.436-0.984 | -2.040 | 0.041 | 0.265 | 0.494 | |
| II+ID vs. DD | Random | 84.71 | ≤0.001 | 0.628 | 0.388-1.015 | -1.899 | 0.058 | 0.536 | 0.705 | |
| II vs. ID+DD | Random | 82.10 | ≤0.001 | 0.739 | 0.483-1.131 | -1.394 | 0.163 | 0.265 | 0.513 | |
| Caucasian | I vs. D | Fixed | 51.31 | 0.055 | 1.115 | 1.002-1.241 | 1.992 | 0.046 | 0.133 | 0.232 |
| II vs. DD | Random | 62.26 | 0.014 | 0.913 | 0.620-1.343 | -0.462 | 0.644 | 0.367 | 0.064 | |
| ID vs. DD | Fixed | 47.68 | 0.075 | 0.925 | 0.778-1.099 | -0.885 | 0.376 | 1.000 | 0.587 | |
| II+ID vs. DD | Fixed | 46.81 | 0.080 | 0.964 | 0.821-1.132 | -0.443 | 0.658 | 0.548 | 0.800 | |
| II vs. ID+DD | Random | 54.24 | 0.041 | 1.044 | 0.772-1.414 | 0.282 | 0.778 | 0.035 | 0.018 | |
| Mixed | I vs. D | Random | 94.34 | ≤0.001 | 0.582 | 0.262-1.293 | -1.329 | 0.184 | 0.308 | 0.299 |
| II vs. DD | Random | 88.51 | ≤0.001 | 0.562 | 0.195-1.618 | -1.068 | 0.286 | 0.734 | 0.308 | |
| ID vs. DD | Random | 91.16 | ≤0.001 | 0.410 | 0.107-1.564 | -1.306 | 0.192 | 0.734 | 0.330 | |
| II+ID vs. DD | Random | 95.17 | ≤0.001 | 0.450 | 0.126-1.611 | -1.227 | 0.220 | 0.734 | 0.427 | |
| II vs. ID+DD | Random | 68.67 | 0.023 | 0.819 | 0.466-1.439 | -0.694 | 0.488 | 0.308 | 0.422 | |
Sensitivity Analysis
To evaluate the robustness of the association results, a meta-analysis was performed repeatedly with each study removed. The results indicated that the fixed-effect estimates before or after the deletion of any single study were generally similar, suggesting a high stability of the meta-analysis results (data not shown). After excluding studies which not fulfilling HWE the pooled ORs were materially altered under the allele model (I vs. D: OR= 0.814, 95% CI 0.608-1.090, p=0.167), the homozygote model (ID vs. DD: OR= 0.709, 95% CI 0.439-1.147, p=0.161), and the dominant model (II+ID vs. DD: OR= 0.672, 95% CI 00.438-1.029, p=0.064; data not shown). The sensitivity analyses substantially alter the pooled ORs except heterozygote model in overall populations, suggesting that the results of this meta-analysis were generally robust.
Publication bias
Publication bias were qualitatively assessed by Begg’s funnel plot and quantitatively assessed by Egger’s test. A significant publication bias was detected by Egger’s test under the allele model (PBeggs= 0.035, PEggers=0.048, Figure 2), the homozygote model (PBeggs= 0.030, PEggers=0.024) and the recessive model (PBeggs= 0.234, PEggers=0.048). In the stratified analysis by ethnicity, still obvious publication bias were found among Caucasians under the recessive model (PBeggs= 0.035, PEggers=0.018). Thus, we have repeated meta-analysis with the ‘‘trim and fill’’ method to adjust publication bias under the three genetic models. Adjusting these three genetic models by ‘‘trim and fill’’ method did not influence the conclusion, which indicated the robustness our results (data not shown).
Figure 2.
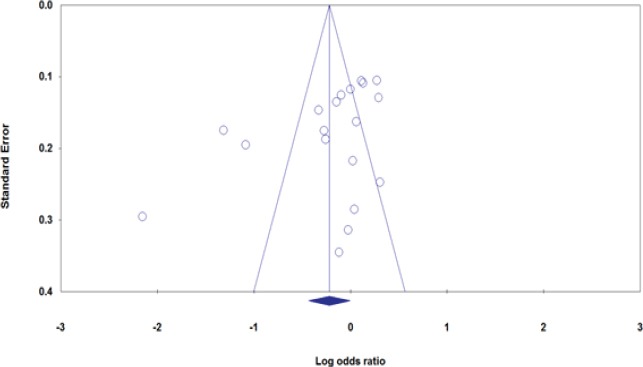
Begg’s Funnel Plots for the Association between ACE I/D Polymorphism and Breast Cancer under the Allele Model (I vs. D).
Discussion
ACE is well known to be a key part of the RAS that regulates blood pressure through conversion of the angiotensin I to the angiotensin II by removing the two C-terminal amino acids. The ACE I/D polymorphism, the most widely studied polymorphism within ACE gene, which individuals carrying the D allele have higher ACE activity. Rigat et al has initially detected ACE I/D polymorphism by restriction fragment length polymorphism (RFLP) analysis (Rigat et al., 1990). There have been a few meta-analyses focusing on association of ACE I/D polymorphism with breast cancer risk. However, most of these analyses wrongly not included all published studies and a new meta-analysis is needed to give a comprehensive conclusion due to the increasing data of case-control studies.
This present meta-analysis, including 2846 breast cancer cases and 9,299 controls from 20 case-control studies, explored the association between the ACE I/D polymorphism and breast cancer risk. This is the largest scale meta-analysis so far. Our results suggested that the ACE I/D polymorphism was significantly associated with increased risk of breast cancer. To further elucidate the association between the ACE I/D and the risk of breast cancer, we have performed subgroup analysis. In subgroup analysis by ethnicity, we found a marginal association of ACE I/D and breast cancer among Asian populations under the heterozygote model (ID vs. DD: OR= 0.655, 95% CI 0.436-0.984, p=0.041) and Caucasian populations under the allele model (I vs. D: OR= 1.115, 95% CI 1.002-1.241, p=0.046), but not among mixed populations. The inconsistency of genetic effects across the ethnicities was detected for breast cancer, and it showed that Asian populations had a greater genetic risk in developing depression on account of genetic variation in this locus than White populations. Results of this meta-analysis indicated that there was a significant association of ACE I/D polymorphism with increased risk of breast cancer. The present meta-analysis results are inconsistent with all previous the meta-analysis (Li et al., 2015; Pei and Li, 2012; Xi et al., 2011; Zhang et al., 2011). The inconsistent results between ours and previous meta-analyses might mainly be due to relevant smaller sample size from previous meta-analyses. Furthermore, most studies included in the present meta-analysis performed among Asians, while previous meta-analyses only had very limited Asian populations.
The great discrepancy among different studies indicated the existence of between-studies heterogeneity and it is generally introduced that meta-analyses should assess heterogeneity (Abedinzadeh et al., 2015; Nedooshan et al., 2017). There are several factors responsible for such heterogeneity such as the study designs, ethnicity of cases and controls, genotyping method, sample size, selection criteria and etc., (Mehdinejad et al., 2017; Sobhan et al., 2017). In the present meta-analysis, there was an extreme heterogeneity (I2 = 75% to 100%) under all five genetic models in overall estimations. Thus, we have performed stratified analysis by ethnicity to identify possible sources for the observed heterogeneity. However, after stratified analysis, there were still heterogeneities in Asian and mixed populations.
To our best knowledge, the present meta-analysis is the most comprehensive, latest one and has the largest sample size. Thus, the statistical power of our analyses was noticeably increased as a large sample size pooled from different studies, and has more statistical powerful than any single case-control study and previous meta-analyses. However, the conclusions of this meta-analysis should be interpreted cautiously due to some limitations. First, the number of published studies was not sufficiently large for stratified analyses by ethnicity. Because of limited available data for mixed population and Africans, our results should be interpreted with caution. Therefore, larger studies are required to explore the association ACE I/D polymorphism with susceptibility to the breast cancer in different ethnicities, especially among mixed population and Africans. Second, there was a significant between study heterogeneity under all five genetic models in overall estimations. One possible reason for such heterogeneity is a wide variation in various included ethnicities. Therefore, we have performed subgroup analysis by ethnicity to eliminate heterogeneity. However, the results showed that the heterogeneity did not reduced or disappeared significantly. Third, there was a significant publication bias under three genetic models which could influence the results of our meta-analysis. However, adjusting pooled ORs for these genetic models by ‘‘trim and fill’’ method did not influence the conclusion, which indicated the robustness our results. Forth, we included only published studies, so there was space for publication bias, which in fact was confirmed by formal statistical tests. Finally, lacking the original data for the included studies limited our further evaluation of potential interactions among gene–gene, gene–environment, or even different polymorphism loci of the ACE gene.
In summary, the present meta-analysis suggested that suggest that the ACE I/D polymorphism is significantly associated with risk of breast cancer. Considering the limited sample size among different ethnicities included in the present meta-analysis, further well-designed and unbiased studies with larger sample studies are needed to confirm our results.
References
- 1.Abedinzadeh M, Zare-Shehneh M, Neamatzadeh H, Abedinzadeh M, Karami H. Association between MTHFR C677T polymorphism and risk of prostate cancer:Evidence from 22 studies with 10,832 cases and 11,993 controls. Asian Pac J Cancer Prev. 2015;16:4525–30. doi: 10.7314/apjcp.2015.16.11.4525. [DOI] [PubMed] [Google Scholar]
- 2.Alves Corrêa SA, Ribeiro de Noronha SM, Nogueira-de-Souza NC, et al. Association between the angiotensin-converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms and breast cancer among Brazilian women. J Renin-Angiotensin-Aldosterone Syst. 2009;10:51–8. doi: 10.1177/1470320309102317. [DOI] [PubMed] [Google Scholar]
- 3.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 4.Buraczynska M, Ksiazek P, Drop A, et al. Genetic polymorphisms of the renin-angiotensin system in end-stage renal disease. Nephrol Dial Transplant. 1986;21:979–83. doi: 10.1093/ndt/gfk012. [DOI] [PubMed] [Google Scholar]
- 5.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 6.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagyas M, Úri K, Siket IM, et al. Karamyan V, editor. New perspectives in the Renin-Angiotensin-Aldosterone System (RAAS) I:Endogenous Angiotensin Converting Enzyme (ACE) Inhibition. PLoS One. 2014;9:e87843. doi: 10.1371/journal.pone.0087843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felipe A, Loango N, Ruíz B, Landázuri P. Asociación entre los polimorfismos de los genes de la enzima convertidora de angiotensina y los receptores AT1R y AT2R y El cáncer de mama. Estudio de Casos y Controles. 2011;62:37–44. [Google Scholar]
- 9.Fishchuk LE, Gorovenko NG. Genetic polymorphisms of the renin-angiotensin system in breast cancer patients. Exp Oncol. 2013;35:101–4. [PubMed] [Google Scholar]
- 10.Forat-Yazdi M, Neamatzadeh H, Sheikhha MH, Zare-Shehneh M, Fattahi M. BRCA1 and BRCA2 common mutations in Iranian breast cancer patients:A meta analysis. Asian Pac J Cancer Prev. 2015;16:1219–24. doi: 10.7314/apjcp.2015.16.3.1219. [DOI] [PubMed] [Google Scholar]
- 11.Forat-Yazdi M, Jafari M, Kargar S, et al. Association between SULT1A1 Arg213His (Rs9282861) polymorphism and risk of breast cancer:A systematic review and meta-analysis. J Res Health Sci. 2017;17:e00396. [PubMed] [Google Scholar]
- 12.Ghosh Roy A, Purkait P, Raha O, et al. Association between the polymorphism of the angiotensin - converting enzyme gene and breast cancer risk among the Bengalee caste hindu females of west Bengal, India. Int J Forensic Sci Pathol. 2015;3:79–88. [Google Scholar]
- 13.Gonzalez-Zuloeta Ladd AM, Arias Vásquez A, Sayed-Tabatabaei FA, et al. Angiotensin-converting enzyme gene insertion/deletion polymorphism and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2143–6. doi: 10.1158/1055-9965.EPI-05-0045. [DOI] [PubMed] [Google Scholar]
- 14.Haiman CA, Henderson SO, Bretsky P, Kolonel LN, Henderson BE. Genetic variation in angiotensin I-converting enzyme (ACE) and breast cancer risk:the multiethnic cohort. Cancer Res. 2003;63:6984–7. [PubMed] [Google Scholar]
- 15.Kamali M, Kargar S, Heiranizadeh N, et al. Lack of any association between the hoGG1 Ser326Cys polymorphism and breast cancer risk:A systematic review and meta-analysis of 18 studies. Asian Pacific J Cancer Prev. 2017;18:245–51. doi: 10.22034/APJCP.2017.18.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamali M, Hamadani S, Neamatzadeh H, et al. Association of XRCC2 rs321⅘polymorphism with susceptibility of breast and ovarian cancer:A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:1743–9. doi: 10.22034/APJCP.2017.18.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Knaap R, Siemes C, Coebergh J-WW, et al. Renin-angiotensin system inhibitors, angiotensin I-converting enzyme gene insertion/deletion polymorphism, and cancer. Cancer. 2008;112:748–57. doi: 10.1002/cncr.23215. [DOI] [PubMed] [Google Scholar]
- 18.Koh W-P, Yuan J-M, Sun C-L, et al. Angiotensin I-converting enzyme (ACE) gene polymorphism and breast cancer risk among Chinese women in Singapore. Cancer Res. 2003;63:573–8. [PubMed] [Google Scholar]
- 19.Kumar S, Rizwan Hussain S, Waseem M, et al. D allele frequency in insertion/deletion polymorphism of the angiotensin converting enzyme (ACE) gene is associated with development of breast cancer risk in Indian women. Curr Proteomics. 2016;13:297–304. [Google Scholar]
- 20.Li X-L, Zheng Z-J, Qu H-O. Lack of association of angiotensin-converting enzyme insertion/deletion polymorphism with breast cancer:An update meta-analysis based on 10405 subjects. J Renin-Angiotensin-Aldosterone Syst. 2015;16:1095–1100. doi: 10.1177/1470320314529357. [DOI] [PubMed] [Google Scholar]
- 21.Mahjoub SA, Abdelrhman E, El-Deen MEM, Mustafa MSE, Ali EW. Angiotensin-converting enzyme insertion/deletion polymorphism is not associated with vasoocclusive complications of sickle cell anemia. Int J Appl basic Med Res. 2016;6:267–70. doi: 10.4103/2229-516X.192594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 23.Mehdinejad M, Sobhan MR, Mazaheri M, et al. Genetic association between ERCC2, NBN, RAD51 gene variants and osteosarcoma risk:A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:1315–21. doi: 10.22034/APJCP.2017.18.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendizábal-Ruiz AP, Morales J, Castro Martinez X, et al. RAS polymorphisms in cancerous and benign breast tissue. J Renin-Angiotensin-Aldosterone Syst. 2011;12:85–92. doi: 10.1177/1470320310383735. [DOI] [PubMed] [Google Scholar]
- 25.Namazi S, Monabati A, Ardeshir-Rouhani-Fard S, Azarpira N. Association of angiotensin I converting enzyme (insertion/deletion) and angiotensin II type 1 receptor (A1166C) polymorphisms with breast cancer prognostic factors in iranian population. Mol Carcinog. 2010;49:1022–30. doi: 10.1002/mc.20685. [DOI] [PubMed] [Google Scholar]
- 26.Neamatzadeh H, Shiryazdi SM, Kalantar SM. BRCA1 and BRCA2 mutations in Iranian breast cancer patients:A systematic review. J Res Med Sci. 2015;20:284–93. [PMC free article] [PubMed] [Google Scholar]
- 27.Nedooshan JJ, Kargar S, Neamatzadeh H, et al. Lack of association of the fat mass and obesity associated (FTO) gene rs9939609 polymorphism with breast cancer risk:A systematic review and meta-analysis based on case - Control studies. Asian Pac J Cancer Prev. 2017;18:1031–7. doi: 10.22034/APJCP.2017.18.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nedooshan JJ, Yazdi MF, Neamatzadeh H, et al. Genetic association of XRCC1 gene rs1799782, rs25487 and rs25489 polymorphisms with risk of thyroid cancer:A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:263–70. doi: 10.22034/APJCP.2017.18.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei X-H, Li H-X. Insertion/deletion (I/D) in the angiotensin-converting enzyme gene and breast cancer risk:lack of association in a meta- analysis. Asian Pac J Cancer Prev. 2012;13:5633–6. doi: 10.7314/apjcp.2012.13.11.5633. [DOI] [PubMed] [Google Scholar]
- 30.Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayed-Tabatabaei FA, Oostra BA, Isaacs A, van Duijn CM, Witteman JCM. ACE polymorphisms. Circ Res. 2006;98:1123–33. doi: 10.1161/01.RES.0000223145.74217.e7. [DOI] [PubMed] [Google Scholar]
- 32.El Sharkawy RM, Zaki AM, El Fattah Kamel AA, Bedair RN, Ahmed AS. Association between the polymorphisms of angiotensin converting enzyme (Peptidyl-Dipeptidase A) INDEL mutation (I/D) and Angiotensin II type I receptor (A1166C) and breast cancer among post menopausal Egyptian females. Alexandria J Med. 2014;50:267–74. [Google Scholar]
- 33.Shiryazdi S, Kargar S, Nasaj H, Neamatzadeh H, Ghasemi N. The accuracy of Breastlight in detection of breast lesions. Indian J Cancer. 2015;52:513–6. doi: 10.4103/0019-509X.178389. [DOI] [PubMed] [Google Scholar]
- 34.Shiryazdi SM, Kargar S, Taheri-Nasaj H, Neamatzadeh H. BreastLight apparatus performance in detection of breast masses depends on mass size. Asian Pac J Cancer Prev. 2015;16:1181–4. doi: 10.7314/apjcp.2015.16.3.1181. [DOI] [PubMed] [Google Scholar]
- 35.Siddiqi M, Syeed N, Abdullah S, et al. ACE gene polymorphism in breast cancer patients of ethnic Kashmiri population. Chronicles Young Sci. 2010;1:40. doi: 10.4021/wjon261w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh A, Srivastava N, Amit S, et al. Association of AGTR1 (A1166C) and ACE (I/D) polymorphisms with breast cancer risk in North Indian population. Transl Oncol. 2018;11:233–42. doi: 10.1016/j.tranon.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sobhan MR, Mahdinezhad-Yazdi M, Jafari M, et al. Association of ESRαXbaI A and gt;G, PvuII T and gt;C and ESRβAlwNI T and gt;C polymorphisms with the risk of adolescent idiopathic scoliosis:a systematic review and genetic meta-analysis. Rev Bras Ortop (English Ed) 2018 doi: 10.1016/j.rboe.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobhan MR, Yazdi MF, Mazaheri M, et al. Association between the DNA repair gene XRCC3 rs↧39 polymorphism and risk of osteosarcoma:A systematic review and meta-analysis. Asian Pac J Cancer Prev. 2017;18:549–555. doi: 10.22034/APJCP.2017.18.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xi B, Zeng T, Liu L, et al. Association between polymorphisms of the renin–angiotensin system genes and breast cancer risk:a meta-analysis. Breast Cancer Res Treat. 2011;130:561–8. doi: 10.1007/s10549-011-1602-3. [DOI] [PubMed] [Google Scholar]
- 40.Xiaomei P, Halmurat U, Manshu S, Dilnur M, Dolikun M. Polymorphism and susceptibility of angiotensin converting enzyme (ACE) gene to breast cancer with abnormal hilit. Sci Technol Rev. 2014;32:65–8. [Google Scholar]
- 41.Yaren A, Turgut S, Kursunluoglu R, et al. Association between the polymorphism of the angiotensin-converting enzyme gene and tumor size of breast cancer in premenopausal patients. Tohoku J Exp Med. 2006;210:109–16. doi: 10.1620/tjem.210.109. [DOI] [PubMed] [Google Scholar]
- 42.Yaren A, Turgut S, Kursunluoglu R, et al. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene in patients with breast cancer and effects on prognostic factors. J Investig Med. 2007;55:255–61. doi: 10.2310/6650.2007.00006. [DOI] [PubMed] [Google Scholar]
- 43.Yazdi MF, Rafieian S, Gholi-Nataj M, et al. CYP2D6 genotype and risk of recurrence in tamoxifen treated breast cancer patients. Asian Pac J Cancer Prev. 2015;16:6783–7. doi: 10.7314/apjcp.2015.16.15.6783. [DOI] [PubMed] [Google Scholar]
- 44.Yazdi MM, Jamalaldini MH, Sobhan MR, et al. Association of ESRαgene Pvu II T>C, XbaI A>G and BtgI G>A polymorphisms with knee osteoarthritis susceptibility:A systematic review and meta-analysis based on 22 case-control studies. Arch Bone Jt Surg. 2017;5:351–62. [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, He J, Deng Y, et al. The insertion/deletion (I/D) polymorphism in the Angiotensin-converting enzyme gene and cancer risk:a meta-analysis. BMC Med Genet. 2011;12:159. doi: 10.1186/1471-2350-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]


