Abstract
Investigators have utilized the CRISPR/Cas9 gene-editing system to specifically target well-conserved regions of HIV, leading to decreased infectivity and pathogenesis in vitro and ex vivo. We utilized a specialized extracellular vesicle termed a “gesicle” to efficiently, yet transiently, deliver Cas9 in a ribonucleoprotein form targeting the HIV long terminal repeat (LTR). Gesicles are produced through expression of vesicular stomatitis virus glycoprotein and package protein as their cargo, thus bypassing the need for transgene delivery, and allowing finer control of Cas9 expression. Using both NanoSight particle and western blot analysis, we verified production of Cas9-containing gesicles by HEK293FT cells. Application of gesicles to CHME-5 microglia resulted in rapid but transient transfer of Cas9 by western blot, which is present at 1 hr, but is undetectable by 24 hr post-treatment. Gesicle delivery of Cas9 protein preloaded with guide RNA targeting the HIV LTR to HIV-NanoLuc CHME-5 cells generated mutations within the LTR region and copy number loss. Finally, we demonstrated that this treatment resulted in reduced proviral activity under basal conditions and after stimulation with pro-inflammatory factors lipopolysaccharide (LPS) or tumor necrosis factor alpha (TNF-α). These data suggest that gesicles are a viable alternative approach to deliver CRISPR/Cas9 technology.
Keywords: gesicle, Cas9 ribonucleoprotein, HIV
In this manuscript, Dr. Campbell and colleagues utilized small, “virus-like” particles called gesicles to deliver Cas9 ribonucleoprotein and target integrated HIV genomes. This pharmacological approach to transiently provide CRISPR-based genome editing to cells resulted in mutation and excision of an HIV provirus in a microglial cell line.
Introduction
The CRISPR/Cas9 gene-editing system has become a versatile and efficient technique to manipulate genomic DNA. This method has been utilized in basic science studies to correct the pathogenic phenotype in a number of disease models stemming from genetic deviations, including muscular dystrophy,1, 2, 3 hemophilia,4, 5 autism,6, 7 cancers,8 and others. Furthermore, the CRISPR/Cas9 system has been shown to have efficacy in infectious diseases, including infection by HIV.9, 10 Investigators have successfully targeted well-conserved regions of the integrated HIV provirus (e.g., long terminal repeat [LTR] regions, gag-pol, and env regions) to cause mutations, leading to a decrease in proviral activity and mature virus production, along with an “immunization” of cells expressing the CRISPR/Cas9 components in vitro. Nevertheless, evidence of guide RNA (gRNA)-specific, CRISPR/Cas9-resistant HIV has been reported, which may occur from constitutive expression of CRISPR/Cas9 components in conjunction with active viral production.11, 12 These data, in addition to numerous studies showing that continuous expression of the CRISPR/Cas9 components can increase the occurrence of off-target mutation events,13, 14, 15 signify the need for alternative methods of delivery that would enable transient, limited CRISPR/Cas9 activity.
A well-established strategy for transient CRISPR/Cas9 activity is the delivery of a Cas9 ribonucleoprotein (RNP) complex (i.e., Cas9 already coupled to its guide RNA).16, 17 The most common methods to accomplish this involves electroporation18 or encapsulating the complex with cationic lipids.19 An alternative approach is to use extracellular vesicles (EVs) containing the Cas9 RNP complex for direct delivery to the recipient cell. EVs have become a topic of focus because of their implications as disease biomarkers in cancer and immunological disorders,20, 21 functions in normal physiology such as cell-to-cell communication,22, 23 and bioengineering-restorative capabilities.24, 25, 26, 27 EVs are endogenously produced membranous vessels that are heterogeneous in size, shape, and cargo. In general, EVs can be characterized into three subpopulations based on size and process of formation: microvesicles, exosomes, and apoptotic bodies (reviewed in Kalra et al.,28 Momen-Heravi et al.,29 and Akyurekli et al.30). Microvesicles are formed from budding of the outer membrane of the cell, and typically range from 50 to 1,000 nm in diameter.31 Exosomes are smaller and arise from the packaging of inner cellular membrane bodies, resulting in a multi-vesicular structure. The inner individual vesicles of the multi-vesicular structure range from 40 to 100 nm.32, 33 Finally, apoptotic bodies are formed when programmed cell death occurs, and contain degrading organelles and cellular debris. These are the largest of the EVs, with sizes ranging from 50 to 5,000 nm.34, 35 Cells can be engineered to increase their production of EVs. A recent study described a microvesicle that is produced through overexpression of vesicular stomatitis virus G glycoprotein termed a “gesicle.”36 The investigators showed that gesicles can be utilized for direct transfer of membrane, cytoplasmic, and nuclear proteins. Due to the relatively high and ubiquitous expression of the cellular receptor for vesicular stomatitis virus G protein (VSV-G), the low-density lipoprotein (LDL) receptor,37, 38 gesicles have a broad tropism. Recently, gesicles have been engineered to contain Cas9 as an RNP complex,39 creating the potential for gesicle-mediated delivery of CRISPR to target cells.
In this study, we explored the efficacy of gesicle-mediated delivery of Cas9 RNPs for inactivating HIV proviral activity in a previously described microglial cell line.40 The features of this characterized cell line allowed us to examine proviral mutation, copy number loss, and functional activity in a stable, reproducible setting. We first showed that gesicles can package Cas9 RNP complexes and confer acute, transient delivery of this functional RNP complex to CHME-5 cells. Using a Cas9 RNP containing a gRNA to the HIV LTR, we observed both mutation and copy number loss of the HIV provirus, along with decreased activity as measured by NanoLuc luminescence and the protein analysis of viral protein Nef. Therefore, the use of gesicles may be an alternative strategy to deliver CRISPR/Cas9 in protein form. The transient nature of protein delivery is aimed to reduce the potential deleterious effects of constitutive Cas9 activity that may arise from virus-mediated delivery methods.
Results
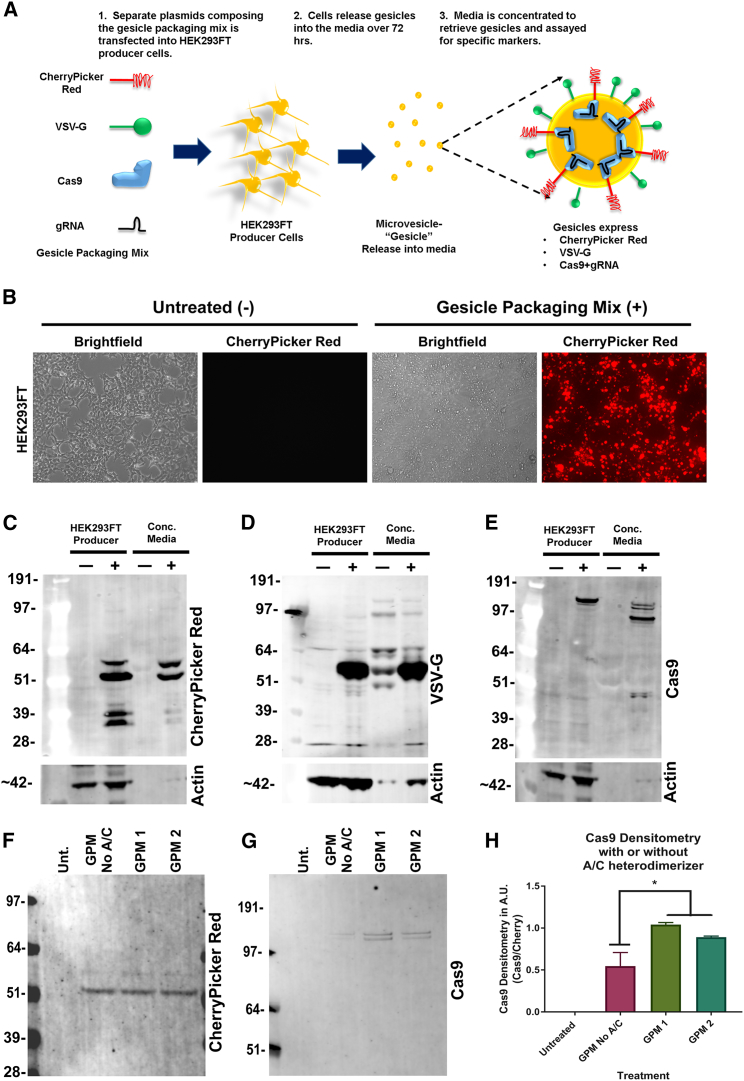
Confirmation of CherryPicker Red, VSV-G, and Cas9 Expression by Gesicles
Gesicles are produced using a packaging system comprised of four components: CherryPicker Red, VSV-G, Cas9, and the chosen gRNA (Figure 1A). After transfection into HEK293FT producer cells, VSV-G promotes gesicle formation through membrane fusion and eventual budding from the cellular membrane. CherryPicker Red is a membrane-associated protein that contains a “DmrA” domain that physically associates with the DmrC domain present on the Cas9 RNP in an inducible manner.41, 42 This occurs by the addition of the A/C heterodimerizer molecule, facilitating Cas9 RNP packaging into the gesicle. Additionally, Cas9 dissociation occurs by dilution of the A/C heterodimerizer once the gesicle fuses with the membrane of the recipient cell. We first confirmed the presence of CherryPicker Red, VSV-G, and Cas9 in both the HEK293FT producer cells and the gesicles released into the media after transfection with the gesicle-forming components. Untransfected producer cells and media were used as controls. Epifluorescent imaging of live HEK293FT cells transfected to produce gesicles showed robust expression of CherryPicker Red (Figure 1B). Cell lysates from HEK293FT producer cells were then prepared, and western blot analysis confirmed expression of CherryPicker Red, VSV-G, and Cas9 (Figures 1C–1E, HEK293FT Producer Cell columns). Gesicles isolated from the producer cells by filtration and ultracentrifugation were probed for the same markers. Western blot analysis confirmed the presence of CherryPicker Red, VSV-G, and Cas9 within the sedimented material (Figures 1C–1E, Conc. Media columns). An additional gesicle preparation was made in the absence of A/C heterodimerizer. Western blot analysis confirmed the presence of CherryPicker Red in all groups prepared with the gesicle packaging mix (Figure 1F). As expected, the presence of A/C heterodimerizer increased the levels of Cas9 associated with the isolated gesicle material (Figures 1G and 1H).
Figure 1.
Production and Characterization of Cas9 Gesicles
(A) HEK293FT producer cells were transfected with the gesicle packaging mix containing CherryPicker Red, VSV-G, Cas9, and a chosen gRNA. Gesicles were released over the course of 48 hr and were collected by media filtration and ultracentrifugation. (B) Live-cell images of HEK293FT cells either untreated (−) or transfected with the gesicle packaging mix (+). The gesicle packaging mix condition showed strong CherryPicker Red fluorescence in comparison with the untreated condition. Cell lysates were prepared from HEK293FT producer cells from untreated (−) and gesicle packaging mix (+) conditions, and a western blot was run to identify protein expression of CherryPicker Red (C), VSV-G (D), and Cas9 (E). Additionally, media were taken and concentrated to obtain gesicles from untransfected (−) or gesicle packaging mix (+) cells (Conc. Media column). We observed an increased expression of CherryPicker Red (B), VSV-G (C), and Cas9 (D) in the preparation from cells transfected with the gesicle packaging mix. A parallel preparation of gesicles was prepared using the gesicle packaging mix without the presence of A/C heterodimerizer. We observed CherryPicker Red protein expression in all preparation conditions using the gesicle packaging mix (F). The absence of A/C heterodimerizer significantly decreases the expression of Cas9 as observed by western blot (G) and densitometry (H). Data are the mean + SEM of three experiments, analyzed using a one-way ANOVA with post hoc test; *p < 0.05 versus no A/C treatment group.
Characterizing Size and Concentration of Gesicles
Gesicles were originally characterized as membranous, vesicle-like structures that measured 100 nm in diameter on average.36 We measured the size and concentration of particles using NanoSight technology43, 44 for both the media from untransfected cells (media control) and cells transfected to produce gesicles. Preparations were conducted using EV-depleted fetal bovine serum (FBS) and normal FBS. For transfected cells, we compared Gesicle Packaging Mix with Xfect or Lipofectamine to compare the impact of the transfection reagent. The CherryPicker Red fluorescent particles ranged from 50 to 700 nm (Figure 2A, bottom panels). The most numerous populations of CherryPicker Red fluorescent particles (peaks ranging from ∼50 to 100 nm) are consistent with the published size of a gesicle.36 The larger fluorescent particles (400–700 nm) may include a mixed population of EVs (e.g., microvesicles and apoptotic bodies).45 Media from untransfected cells did not have detectable fluorescent microparticles (Figure 2A, bottom left). Light scatter detected EVs in all samples regardless of conditions (Figure 2, top panels). The dominant peaks for fluorescent and light scatter readings did not coincide and may be the result of aggregates or objects derived from the FBS or EVs produced from HEK293FT cells. Sample videos captured during the NanoSight analysis are available (Video S1). We observed that the transfection reagents produced comparable particles as measured by fluorescence and light scatter, and both reagents increased total particles by a 3- to 6-fold increase over untreated cells. Preparations that used EV-depleted media contained almost twice as many fluorescent particles in the total EV population compared with normal serum containing media (0.38% versus 1%; Figure 2B). These data support the use of EV-depleted media in the production of gesicles.
Figure 2.
Assaying Gesicle Size and Concentration
(A and B) Media from untreated HEK293FT producer cells or cells transfected with either the gesicle packaging mix or Lipofectamine were prepared in EV-depleted FBS or normal FBS and analyzed to determine size distribution and concentration of particles after ultracentrifugation and resuspension. (A) Top panels: in cells grown using EV-depleted FBS, light scatter analysis of media control from HEK293FT producer cells and producer cells transfected with the gesicle packaging mix or Lipofectamine + plasmids confirm that all conditions produce extracellular vesicles. Bottom panels: only the gesicle packaging mix and Lipofectamine conditions contain fluorescent particles. The fluorescent particles ranged between 50 and 700 nm in size. (B) Preparations of media control, gesicle packaging mix, and Lipofectamine conditions were prepared and analyzed for particle number. Transfection with the gesicle packaging mix or Lipofectamine increases the presence of extracellular vesicles in the media by 3- to 6-fold over untreated cells. Within the total population of extracellular vesicles, ∼1% are CherryPicker Red+ under EV-depleted conditions while normal FBS conditions exhibit only 0.38% of particles.
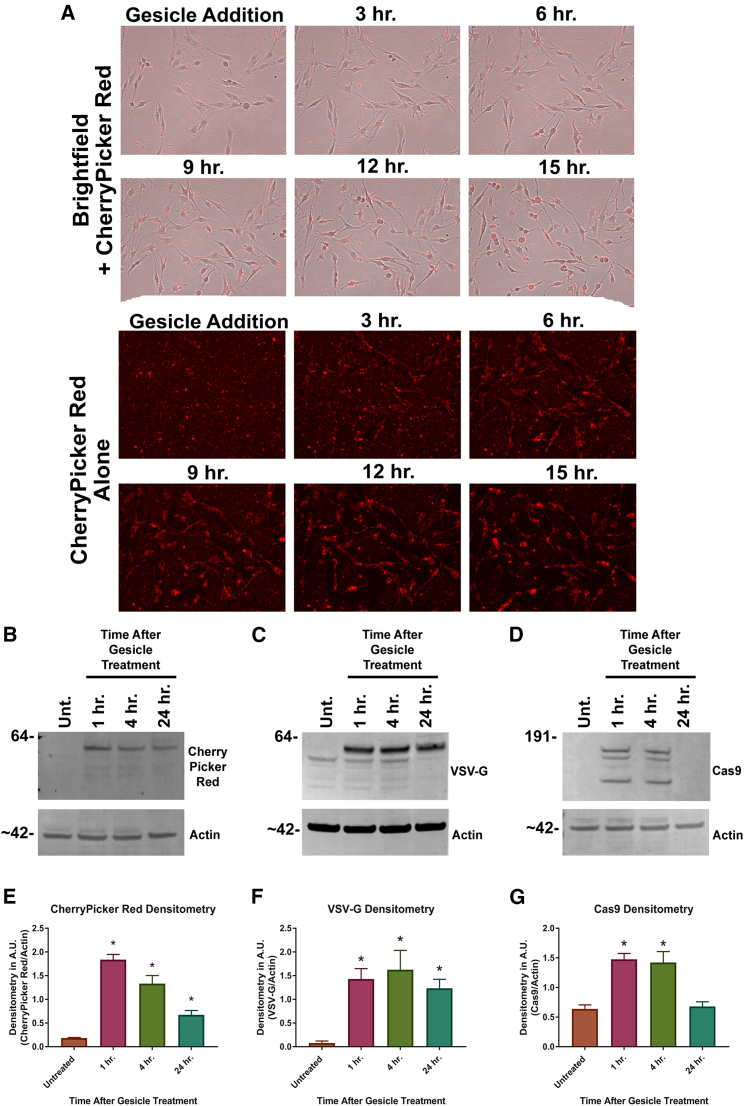
Determining Transfer of Gesicle Proteins to CHME-5 Cells
Gesicles provide rapid and direct transfer of cytoplasmic, membrane-associated, and nuclear proteins to recipient cells.36 After gesicle treatment, we observed CherryPicker Red fluorescence associated with CHME-5 cells. The fluorescence remained associated with the cellular morphology during cellular migration and division (Figure 3A). A full time-course video is shown in Video S2. Because gesicle-mediated transfer of CherryPicker Red and Cas9 protein has not been characterized, we performed western blot analysis on lysates prepared from CHME-5 cells after gesicle treatment. Gesicles were applied to CHME-5 microglia cells by centrifugation, and cells were harvested at 1, 4, and 24 hr post-centrifugation. Western blot analysis for CherryPicker Red, VSV-G, and Cas9 confirm the direct and rapid transfer of these proteins to their recipient cells (Figures 3B–3D). CherryPicker Red, VSV-G, and Cas9 proteins associate with CHME-5 cells as early as 1 hr after centrifugation. CherryPicker Red and VSV-G protein were detectable for at least 24 hr (Figures 3E and 3F). However, there is no detection of Cas9 at 24 hr when compared with untreated control (Figure 3G), suggesting its half-life is shorter than the other proteins delivered by gesicles.
Figure 3.
Gesicle Treatment Rapidly Delivers Protein to CHME-5 Microglia
(A–G) CHME-5 microglia were treated with gesicles by centrifugation and placed on an EVOS microscope system of longitudinal imaging of live cells and CherryPicker Red fluorescence. (A) A combination of bright-field and CherryPicker Red fluorescence showed co-localization over time. Cell lysates were prepared at 1, 4, and 24 hr post-centrifugation. Cell lysates were run on a western blot and probed for CherryPicker Red (B), VSV-G (C), and Cas9 (D) in comparison with untreated cells. Densitometry was then performed for CherryPicker Red (E), VSV-G (F), and Cas9 (G). We observed a rapid yet transient delivery of Cas9 by gesicles. Data are the mean ± SEM of three experiments, analyzed using a one-way ANOVA with post hoc test; *p < 0.05 versus untreated cells.
Reducing HIV Proviral Activity Using Gesicle-Mediated Delivery of Cas9
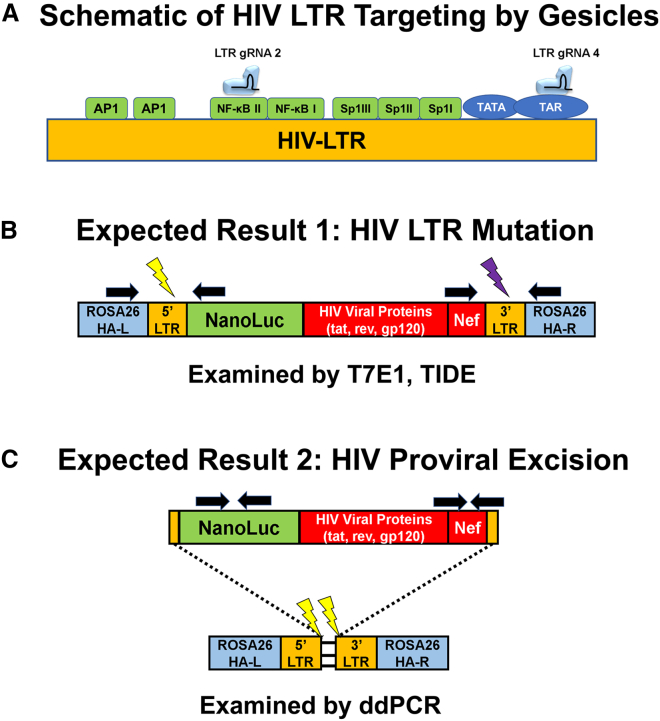
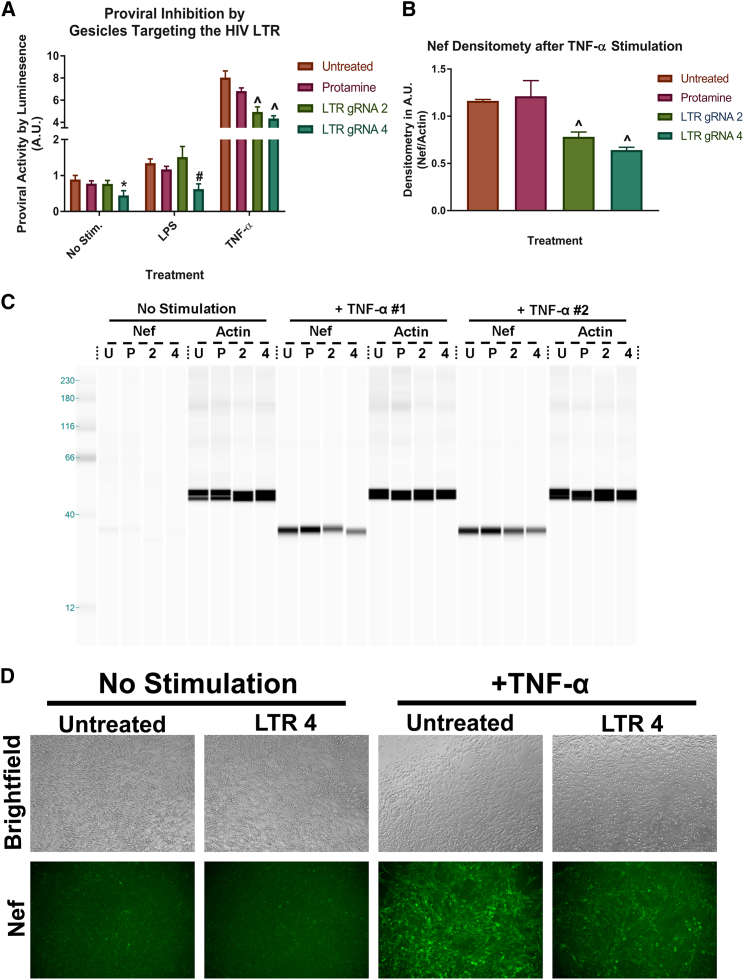
To examine the efficacy of gesicle-mediated delivery of Cas9 RNPs, we targeted the LTR of HIV in a microglial cell line HIV-NanoLuc CHME-5, which contains a modified HIV provirus with a NanoLuciferase reporter construct under control of the HIV LTR.40 LTR-mediated expression can be enhanced by nuclear factor κB (NF-κB) activating agents including lipopolysaccharide (LPS) and tumor necrosis factor alpha (TNF-α). The cell line also produces the HIV viral protein Nef and contains a stable copy number of integrated proviruses. Two gRNAs designated LTR gRNA 2 and 4 were used for gesicle production. LTR gRNA 2 targets the NF-κB II site of the HIV LTR, and LTR gRNA 4 targets the 3′ end of the LTR near the TAR region (Figure 4A). After gesicle delivery, the expected results include mutation of the HIV proviral LTR regions (Figure 4B) and/or proviral excision, resulting in copy number loss (Figure 4C).
Figure 4.
Schematic of the HIV Long Terminal Repeat and Expected Products after CRISPR/Cas9 Targeting by Gesicles
(A) Two gRNAs were developed to target the HIV LTR. LTR gRNA 2 targeted the NF-κB II transcription site, whereas LTR gRNA 4 targeted the TAR region. (B) The HIV provirus utilizes two LTR regions at the 5′ and 3′ ends. If CRISPR/Cas9 occurs at separate instances (denoted by the light and dark lightning bolts), mutations in these regions can occur. Black arrows denote the primer binding sites to assay mutations within the LTR regions. (C) Additionally, if a double-stranded break occurs at the same instance, there is a possibility of whole provirus excision resulting in a loss of HIV proviral copies. Black arrows denote primers used to assay the loss of integrated proviruses by ddPCR.
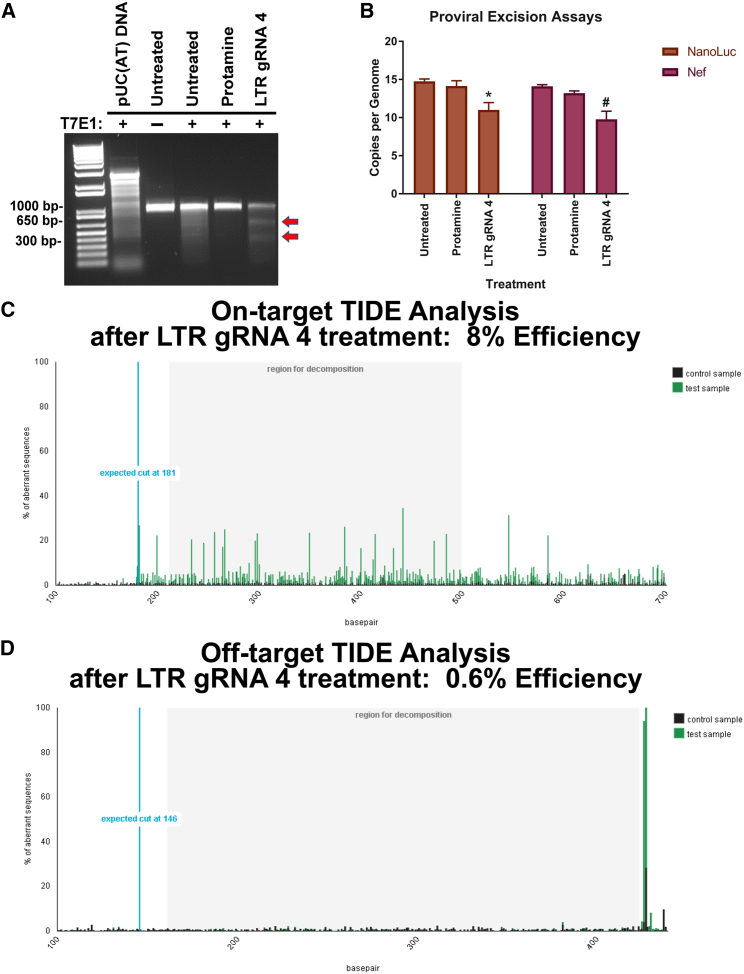
We first verified mutation within the LTR region using T7E1 assay, which produced the expected cleavage products (Figure 5A; Figure S1B). We used protamine sulfate to reduce non-specific binding of gesicles and therefore used protamine sulfate treatment alone as “vehicle control. We next examined genomic DNA for evidence of gesicle-mediated proviral copy number loss using droplet digital PCR (ddPCR) analysis of both NanoLuc and Nef (Figure 4C). The cells treated with gesicles showed a decrease in the copy number of HIV proviruses (Figure 5B; Figure S1C). Direct sequencing of the PCR products, followed by tracking of indels by decomposition (TIDE) analysis,46 further confirmed mutational events at the HIV LTR using gRNA 4, with approximately 8% of amplified products harboring mutations (Figure 5C). A primary concern when using CRISPR/Cas9 technology is off-target mutational events. We found that no sites were completely homologous to our chosen gRNA sequences, and putative off-target sites were not located in coding regions of genes. Two off-targets for LTR gRNAs 2 and 4 were examined by TIDE; however, no mutational events were observed above 2% background46 (Figure 5D; Figure S1D).
Figure 5.
Gesicle Treatment Causes Specific Mutation Events at the HIV LTR
HIV-NanoLuc CHME-5 microglia were assayed to determine mutation and a loss of proviral copy number. (A) T7E1 of the 5′ LTR amplified region showed positive products for mutation by LTR gRNA 4. (B) Droplet digital PCR was performed using a probe for the NanoLuc and the Nef regions of the provirus relative to a GGT1, a single copy gene. We observed a decrease in proviral copies per genome by LTR gRNA 4. (C) TIDE analysis was performed for LTR gRNA 4, confirming mutational events near the designated gRNA target site. The average efficiency of mutation events was 8%. (D) Two of the top off-target sites for LTR gRNA 4 were amplified and assayed for mutational events by TIDE analysis. Off-target 1 showed little to no variation from the control sample with an efficiency of 0.6%. Data are the mean ± SEM of three experiments, analyzed using a two-way ANOVA with post hoc test; *p < 0.05 versus untreated cells for NanoLuc ddPCR; #p < 0.05 versus untreated cells for Nef.
We next determined whether HIV proviral activity and cell phenotype were altered by gesicle treatment. NanoLuciferase activity, both basal and stimulated, was decreased following gesicle treatment (Figure 6A). Additionally, a decrease in the viral protein Nef was observed by WES analysis (Figures 6B and 6C) and immunofluorescence (Figure 6D).
Figure 6.
Gesicle Treatment Reduces HIV Proviral Activity
(A) Cell populations treated with LTR gRNA 2 and LTR gRNA 4 were stimulated with pro-inflammatory factors, and cell lysates were taken for the luciferase assay. LTR gRNA 2 and 4 significantly decreased proviral activity after stimulation with TNF-α (50 ng/mL). Additionally, LTR gRNA 4 significantly decreased proviral activity under both basal conditions and LPS (100 ng/mL) stimulation. (C) Lysates were also run on a WES and probed for the HIV protein Nef. (B) Densitometry analysis confirms that HIV viral protein Nef is decreased using LTR gRNAs 2 and 4 after TNF-α treatment. (D) Immunohistochemistry also showed decreased immunoreactivity of Nef in cells treated with LTR gRNA 4. Data are the mean ± SEM of two stable cultures, with three experiments each, analyzed using a two-way ANOVA with post hoc test for each stimulation group (A) or one-way ANOVA with post hoc test (B). *p < 0.05 versus untreated cells: unstimulated; #p < 0.05 versus untreated cells: LPS stimulated; ˆp < 0.05 versus untreated cells: TNF-α stimulated. 2, LTR gRNA 2; 4, LTR gRNA 4; P, protamine; U, untreated.
Finally, we compared gesicle delivery with Lipofectamine-based transfection of plasmids expressing Cas9 and gRNAs. We observed that a single transfection was not sufficient to result in decreased proviral activity in the same time duration as a gesicle treatment. HIV-NanoLuc CHME-5 cells required two rounds of transfection to decrease proviral activity (Figure S2A). Incidentally, we observed significant proviral copy number loss for LTR gRNA 2 after two rounds of transfection as well (Figure S2B). When off-target analysis was performed by TIDE, we continued to observe low off-target effects (Figure S2C), although LTR 4 was higher by plasmid transfection (2%) compared with gesicles (0.6%).
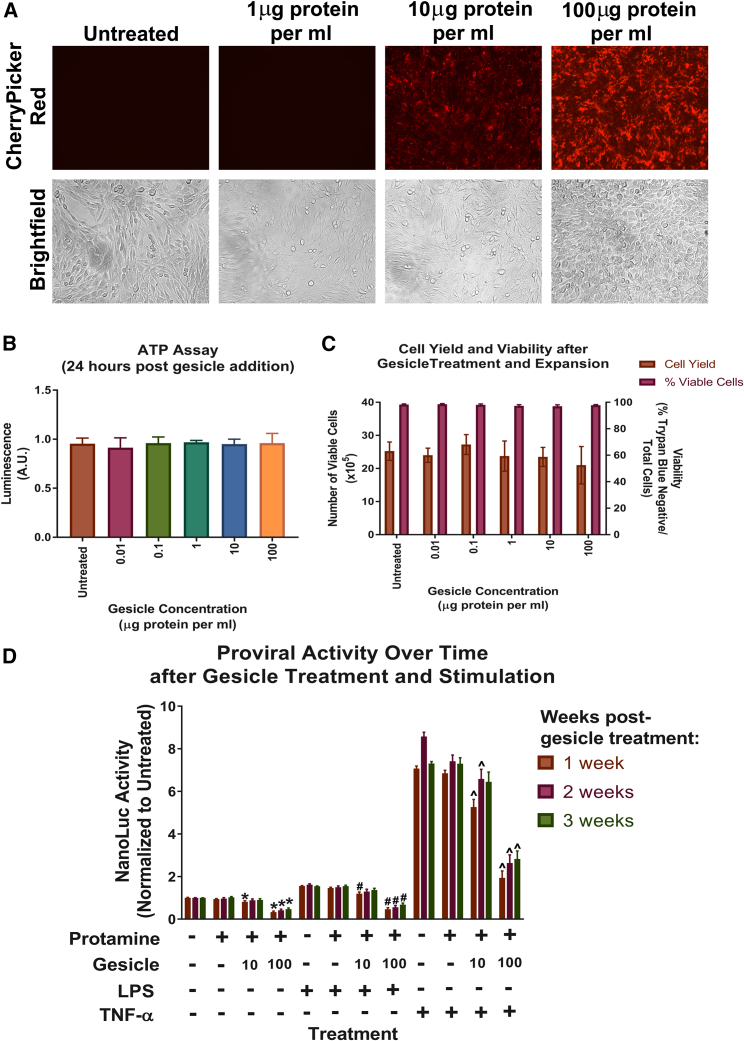
Gesicle Dosage and Viability
We tested various concentrations of gesicles ranging from 0.01 to 100 μg protein/mL for effects on viability. Gesicles were added by centrifugation, and differences in CherryPicker Red fluorescence were observed 24 hr after addition (Figure 7A). We measured viability both acutely and after cellular expansion and passaging. No loss of viability as measured by ATP assay was detectable acutely at 24 hr for all doses of gesicles (Figure 7B). For longer experiments in which cells were grown to confluency and passaged for 3 weeks, both cell yield and viability remained unchanged (Figure 7C). LTR gRNA 4 produced significant proviral inhibition at both 10 and 100 μg protein/mL concentrations, with 100 μg protein/mL maintaining significant proviral inhibition for up to 3 weeks (Figure 7D).
Figure 7.
Gesicle Dosage and Viability
(A–D) HIV-NanoLuc CHME-5 microglia were treated with gesicles containing LTR gRNA 4 at varying concentrations (0.01–100 μg protein/mL). (A) Live-cell images 24 hr post-centrifugation indicated CherryPicker Red fluorescence increases as gesicle dose increases. (B) Viability 24 hr post-gesicle treatment was assayed using the ATP viability assay. No significant differences were detected between the different gesicle concentrations. (C) Long-term viability after cellular expansions and culture was measured by cell yield and % viability using trypan blue. No significant differences were observed between treatment conditions. (D) Cells treated with 10 and 100 μg protein/mL concentrations of LTR gRNA 4 or protamine alone. After expansion, cells were plated and stimulated with pro-inflammatory factors LPS (100 ng/mL) and TNF-α (50 ng/mL), which was repeated for 2 additional weeks. A significant decrease in proviral activity was observed using 100 μg/mL at all conditions and time points versus control samples. Cells treated with 10 μg/mL showed significant proviral inhibition after LPS and TNF-α stimulation at weeks 1 and 2, but no significant differences were observed at week 3 versus control samples. Data are the mean + SEM of three experiments (B), or three stable cultures with three experiments each (C and D), analyzed using a one-way ANOVA with post hoc test (B) or analyzed using a two-way ANOVA with post hoc test for each stimulation group (C and D); *p < 0.05 versus untreated cells: unstimulated; #p < 0.05 versus untreated cells: LPS stimulated; ˆp < 0.05 versus untreated cells: TNF-α stimulated.
Discussion
The versatility and selectivity that comes with CRISPR/Cas9 gene editing has opened the possibility for translation into human therapies. However, many issues related to safety, efficacy, and specificity remain before such therapies are feasible. Incremental advances to address these issues will facilitate the translation of CRISPR/Cas9 into a viable approach for the treatment of disease. Here, we demonstrate that gesicles can be used to rapidly and transiently deliver Cas9 RNP to a microglial cell line. Furthermore, we showed that gesicle-mediated delivery of the Cas9 RNP complex containing gRNAs targeting the HIV LTR can produce site-specific mutations, proviral copy number loss, and decreased HIV proviral activity, consistent with other findings.9, 10 To our knowledge, this is the first instance where a gesicle and Cas9 RNP complex were used in conjunction to inactivate HIV. Additionally, the use of gesicles may serve as an alternative strategy to deliver functional Cas9 in comparison with gene delivery vehicles such as adeno-associated viruses and lentiviruses.
Gesicles as Delivery Vehicles of Protein
In the manuscript first describing gesicles by Mangeot, Lotteau, and colleagues,36 the authors showed that several types of proteins including cytosolic, membrane bound, and nuclear localized can be loaded and subsequently transferred to recipient cells by gesicles. Here we confirmed that membrane-bound CherryPicker Red and nuclear Cas9 can be packaged into VSV-G-produced gesicles (Figure 1). Interestingly, the proteins packaged into gesicles may be structurally different than in the producer cell line. For example, the cherry fluorescent protein is expressed as an ∼30-kDa monomer,47 and addition of the DmrA domain to form CherryPicker Red is ∼60 kDa. We observed four protein bands ranging from ∼30 to 60 kDa in the HEK293FT producer cell lysates. However, only the 50- to 60-kDa molecular weight bands were present in isolated gesicles (Figure 1C). The data suggest that the mature CherryPicker Red protein is present at the membrane, but the cell-associated protein is incompletely processed or cleaved. For example, a significant portion of the overexpressed CherryPicker Red may be misfolded and targeted for degradation before ever reaching the membrane. Additionally, a portion of the membrane containing CherryPicker Red may be recycled and degraded instead of becoming part of a gesicle. We also observed a difference in apparent size of Cas9 between producer cells (∼180 kDa) and gesicles (three proteins at 180 kDa and lower). Based on expected sizes of the modified Cas9, it appears that it may be cleaved once associated with gesicles, or the other forms are a result of degradation. Further studies are needed to understand the nature of this process and whether it affects the specific activity of a Cas9-containing gesicle.
Mangeot et al.36 measured gesicle size using gold labeling and found the size of the gesicle to be roughly 100 nm. Hung and Leonard48 similarly performed a size characterization by both electron microscopy and NanoSight analysis, revealing gesicles to be 50–200 nm in diameter. Our analysis of cherry particles by NanoSight confirms the presence of 50- to 200-nm particles (Figure 2A). In addition, we observed fluorescent particles with diameters greater than 200 nm. Currently, we are unable to determine whether these are separate and distinct gesicles or a possible clustering and aggregation of membrane-fused gesicles. Using the NanoSight analysis, we were able to examine two previously uncharacterized facets of gesicle biology: the extent to which the transfection of gesicle components facilitates vesicle formation and the percentage of particles that are CherryPicker Red+ (and potentially Cas9+) after purification. We found that transfection increases the presence of microparticles in the media by 91%–140% over the untransfected control. Although we observed an increase in the presence of microparticles, only 1% of these particles were CherryPicker Red+ (Figure 2B). The relatively low efficiency of CherryPicker Red incorporation into microparticles may be improved by altering the time course of expression between VSV-G and CherryPicker Red. Because VSV-G is overexpressed to produce gesicles, the formation of gesicles may occur before CherryPicker Red has fully trafficked to the membrane. Such an approach might also improve the packaging of Cas9 by allowing it to associate with the membrane before being incorporated into gesicles. Our study provides a basis for further optimization of the production of Cas9-containing gesicles.
Transient Delivery of Cas9
Studies targeting HIV provirus using CRISPR/Cas9 have shown that gene editing may result in the development of CRISPR/Cas9-resistant strains of HIV.11, 12 Furthermore, the potential of off-target mutations is widely discussed regarding CRISPR/Cas9 gene editing. One strategy to reduce off-target mutational effects is to limit the duration of Cas9 activity, which can be achieved by delivering Cas9 in a RNP form. Work by Kim et al.17 demonstrated that Cas9 RNP complexes have a more rapid time course of mutational events when compared with CRISPR/Cas9 delivered by DNA, with detectable mutations starting at 3 versus 12 hr, respectively. Similar to our findings (Figure 3), Kim et al.17 also found that the presence of Cas9 RNP in recipient cells after delivery is transient, with protein detection of Cas9 nearly undetectable at 24 hr. Furthermore, Cas9 RNP delivery resulted in decreased off-target mutations and cellular toxicity when compared with plasmid delivery of CRISPR/Cas9. This is consistent with studies reporting that Cas9 RNP delivery results in high-fidelity mutational events with minimal cell mortality.49, 50 In the current study, we found no evidence of off-target effects, but we did observe mutations within the LTR region, as well as a loss of copy number likely resulting from the presence of two target sites (one in each LTR). The relatively close proximity of the provirus’s remaining LTR may also serve as a donor template for the repair of the first LTR that obtains a Cas9-mediated double-strand break. This type of repair would also result in a deletion of the proviral genome, leaving a single LTR (which can be acted upon further) in its place. Importantly, we also found no significant changes in viability as a consequence of gesicle treatments, both acutely and through long-term cell passaging (Figure 7). These results suggest that gesicle-mediated Cas9 RNP delivery maintains the characteristics of high mutation efficiency, low toxicity, and low off-target effects. Gesicles may potentially be used as an alternative CRISPR/Cas9 delivery system to target integrated HIV proviruses.
Enhancing Gesicle Production and Future Directions
We observed that a relatively low percentage of isolated microparticles were CherryPicker Red+ (1%; Figure 2B), but our gesicle preparation produced phenotype-altering mutations in the target cells. Given that we could induce functional mutations with our gesicle preparations, it is feasible to employ them for in vivo applications. However, the delivery of the extraneous materials for in vivo systems warrants concern for potential adverse effects including immunogenic and cytotoxicity. Improving the homogeneity of the resulting gesicle suspension would be beneficial to decrease these unwanted effects, while increasing efficacy. Mangeot et al.36 showed that gesicle preparations can be separated using an iodixanol gradient. Combining these gesicles with capture antibodies to VSV-G or CherryPicker Red may result in a highly purified gesicle product. Further optimization of production methods may improve gesicle yield and reduce the relative portion of non-Cas9-carrying gesicles. Overall, new efforts should be explored to enhance gesicle production and purification to aid in the feasibility of translation into a possible human therapeutic method.
In these experiments, we evaluated gesicles as a delivery vehicle for CRISPR/Cas9 to target and inactivate HIV proviral DNA. We used a previously characterized microglial cell line that contains a modified HIV provirus, HIV-NanoLuc-CHME-5.40 We showed that gesicle-mediated delivery of Cas9 RNP with gRNA targeted to LTR could reduce copy number of proviral genomes and reduce viral protein expression. These studies establish the gesicle as a capable delivery vehicle of CRISPR/Cas9 for HIV targeting. Future studies using additional cell models of HIV infection are needed to establish efficacy and off-target effects. This would include lymphocyte cell lines such as the Jurkat51, 52 and SupT153, 54 cell lines, or peripheral blood mononuclear cells55 isolated from patient samples that are transduced with replication-competent HIV. While there are data showing that T cell populations lack the VSV-G receptor,56 there may be other membrane modifications that can be made to facilitate gesicle-mediated transduction of this cell type including co-pseudotyping with other virus glycoproteins.57, 58 These data would add to the feasibility of gesicle-mediated Cas9 RNP delivery in T cell populations that are commonly infected with the virus. Lastly, in vivo delivery of Cas9 RNPs using gesicles needs to be tested. Previous work showed that Cas9 RNP complexes fused with Simian vacuolating virus 40 nuclear localization sequences can be delivered through intracranial injection.59 Overall, gesicle-mediated Cas9 RNP delivery in vitro inhibited proviral activity in the HIV-NanoLuc CHME-5 cell line. Future studies of these unique EVs may lead to alternative therapeutic approaches for controlled CRISPR/Cas9 activity.
Materials and Methods
Gesicle Production
Gesicles were prepared two ways. Unless otherwise noted, gesicles were prepared using the Guide-it CRISPR/Cas9 Gesicle Production System (catalog [Cat.] no. 631613; Clontech/Takara, Mountain View, CA, USA) according to the manufacturer’s instructions. In brief, two guide RNA sequences (LTR gRNA 2: 5′-TTCTACAAGGGACTTTCCGC-3′ and LTR gRNA 4: 5′-GCCCGTCTGTTGTGTGACTC-3′) were developed to target the HIV-1 (pYU2 GenBank: M93258.1) LTR region using http://crispr.mit.edu. Oligonucleotide duplexes encoding the gRNA sequences were ligated into the pGuide-it-sgRNA vector and confirmed by sequencing. A single LTR gRNA sequence was used in one gesicle preparation. The chosen LTR gRNA plasmid (10 μg) was diluted in nuclease-free water to a volume of 600 μL and added to the gesicle packaging mix. The entire mix was added dropwise onto HEK293FT producer cells, which were plated at 4.5 × 106 cells in a 10-cm dish. Forty-eight hours post-transfection, the media were collected, filtered through a 0.45-μm syringe, and spun at 8,000 relative centrifugal force (RCF), 4°C for 16 hr in an ultracentrifuge (Beckman-Coulter, Brea, CA, USA). The pellet was resuspended in PBS and aliquoted for use. Regarding Figure 2, we utilized Lipofectamine 2000 (Thermo Fisher, Waltham, MA, USA) as the transfection reagent to compare the particle size and concentration of gesicles prepared using this method with those produced by the Gesicle Production System (Clontech/Takara). The transfection mix was prepared following the manufacturer’s instructions for a 10-cm dish, using the following plasmid ratios: CherryPicker Red, 3 μg; VSV-G, 12 μg; Cas9, 3 μg; and chosen LTR gRNA plasmid, 3 μg. After transfection, the media were prepared as previously described. Using the current isolation methods, all gesicle preparations contain a heterogenous mix of exosomes, microvesicles, and apoptotic bodies.
Western Blot
Cell lysates for western blot analysis were produced using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris HCl [pH 7.5], 0.25% sodium deoxycholate, 150 mM NaCl, and 1 mM EDTA) with 1% NP-40 detergent (Thermo Fisher) and protease inhibitor (Sigma-Aldrich, Allentown, PA, USA). Cells were lysed for 20 min on a rotating plate at 4°C, after which lysates were collected and spun down for 10 min at 10,000 revolutions/min (RPM; 9,400 RCF) in a microcentrifuge. The supernatant was extracted, and protein levels were read using the detergent compatible (DC) assay (Thermo-Fisher). Protein was normalized to 20 μg per treatment and separated on a NuPage 4%–12% Bis-Tris gel (Thermo-Fisher). Protein was transferred to a Novex 0.45-μm polyvinylidene fluoride (PVDF) membrane, washed with PBS, and blocked with Rockland Blocking Buffer for Fluorescent Western (Rockland Immunochemicals, Limerick, PA, USA) for 1 hr at 24°C. Primary antibodies were diluted in Rockland Blocking Buffer at a 1:1,000 dilution and incubated overnight at 4°C on a rotating shaker. Primary antibodies used were as follows: Living Colors mCherry (Cat. no. 632543; Clontech/Takara), VSV-G (Cat. no. v4888; Sigma-Aldrich), Cas9 (Cat. no. MAC133; Millipore, Burlington, MA, USA), Nef (the following reagent was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: Nef Antibody Cat. no. 3689 from Dr. James Hoxie), β-actin-mouse (Cat. no. ab3280; Abcam, Cambridge, UK), and β-actin-rabbit (Cat. no. A5060; Sigma-Aldrich). After primary antibody addition, membranes were washed with PBS three times, and secondary antibody was applied at a 1:10,000 dilution in Rockland Blocking Buffer for 1 hr. Secondary antibodies used were Rockland Dyelight anti-mouse IR680 (Cat. no. 610-144-002; Rockland) and Rockland Dyelight anti-rabbit IR800 (Cat. no. 661-145-002; Rockland). After secondary addition, membranes were washed three times with PBS and imaged using a LI-COR Odyssey Scanner (LI-COR Biosciences, Lincoln, NE, USA). Additionally, samples were prepared for examination by Wes (ProteinSimple, San Jose, CA, USA). Samples were prepared as previously described (RIPA buffer) and used at a 400 μg/mL concentration in conjunction with buffers and antibodies provided by the manufacturer (Cat. no. PS-MK15; Master Kit with Split Buffer) to detect Nef protein expression.
Immunocytochemistry
HIV-NanoLuc CHME-5 cells were washed with 1× PBS and fixed with 4% paraformaldehyde solution for 30 min. Cells were then permeabilized using blocking buffer (2.5% bovine serum albumin with 0.01% Triton X-100 for 30 min), after which the primary antibody (Nef, 1:500 dilution) was added and incubated overnight at 4°C in on a shaker. The following reagent was obtained through the AIDS Reagent Program, Division of AIDS, NIAID, NIH: Nef Antibody Cat. no. 3689 from Dr. James Hoxie. After which, cells were washed three times with 1× PBS and a 1:1,000 dilution of secondary antibody Alexa Fluor 488 (Cat. no. Z25002; Thermo Fisher) was added for 1 hr to determine fluorescence.
NanoSight Analysis
Analysis of size and concentration was performed on different gesicle preparations using the NanoSight NS500 (NanoSight/Malvern, Salisbury, UK). Light scatter mode was used to detect all particles in the solution. Gesicle preparations were diluted at a 200 μg/mL concentration and infused into the instrument. Six screen captures of 30 s each were used for particle analysis. Fluorescent mode using a 565-nm laser was utilized to detect CherryPicker Red+ particles only. Camera level was used at 16 (nanoparticle tracking analysis [NTA] 3.0 levels), and detection threshold was set at 3.
Cell Culture and Treatments
CHME-5 microglia or HIV-NanoLuc CHME-5 microglia were grown in microglia growth media: high-glucose DMEM (GIBCO/Thermo-Fisher) supplemented with 5% FBS (Hyclone/GE, Logan, UT, USA) and 1% penicillin and streptomycin (GIBCO). Cells were passaged every 4 days using 0.05% trypsin (GIBCO). HIV-NanoLuc CHME-5 were stimulated with LPS 100 ng/mL (Cat. no. L3012; Sigma-Aldrich) or TNF-α 50 ng/mL (Cat. no. T5944; Sigma-Aldrich) diluted in microglia growth media.
Gesicle Application
For time-dependent protein expression experiments (Figure 3), CHME-5 microglia were plated on a 24-well plate at 0.5 × 105 cells/well in 1 mL of media the day before experimentation. On the day of gesicle application, media were changed to 600 μL of growth media supplemented with protamine sulfate (8 μg/mL). Gesicles were added (50 μg protein/mL) and centrifuged at 1,150 RCF for 30 min as per the gesicle kit instructions. Cells were lysed, and protein processed at 1, 4, and 24 hr post-centrifugation. For HIV-LTR targeting experiments (Figures 4, 5, and 6), HIV-NanoLuc CHME-5 cells were plated and treated as previously described (with protamine supplement and centrifugation). After gesicle addition, cells were allowed to expand for 3 days. The entire cell population was then trypsinized and transferred to a 12-well plate for 48 hr. After this time the entire population was trypsinized again and transferred to a 10-cm culture dish as a stable cell line stock. For dosage and viability experiments (Figure 7), HIV-NanoLuc CHME-5 cells were plated in a 96-well plate at 5.6 × 104 cells/well in 200 μL of media, and gesicles were applied at varying concentrations (0.01–100 μg protein/mL). Two days after gesicle addition, cells were trypsinized and transferred to a 12-well plate for a stable cell line stock. This stock of cells was maintained for 3 weeks for repeated experimentation. Gesicles prepared using normal FBS were applied in Figures 3, 4, 5, and 6, and gesicles prepared in EV-depleted FBS were applied in Figures 2 and 7.
Transient Transfection of CRISPR/Cas9
HIV-NanoLuc CHME-5 cells were plated on a 24-well plate at 0.5 × 105 cells/well in 1 mL of media. The plasmids utilized were FLAG-tagged NLS-Cas9-T2A-PuroR (48139; Addgene), and the LTR gRNAs were ligated into the pGuide-it-sgRNA vector (previously described). Lipofectamine 2000 (Thermo Fisher) was used as the transfection reagent, and the concentration for each treatment are as follows: Cas9, 1.6 μg/mL + pBluescript II SK1.6 (212205; Addgene); Cas9, 1.6 μg/mL + LTR; gRNA 2, 1.6 μg/mL; Cas9, 1.6 μg/mL + LTR; gRNA 4, 1.6 μg/mL. Cells were treated with either one round of transfection or two rounds of transfection after the first cellular expansion and plating procedure.
EVOS Live-Cell Imaging
For live-cell time-course imaging of gesicle application, HIV-NanoLuc CHME-5 microglia were plated on a 24-well plate with 0.5 × 105 cells/well in 1 mL of media the day before experimentation. The day of experimentation, media were changed to 600 μL of FluorBrite Imaging Media (GIBCO) supplemented with 5% FBS (Hyclone), 1% penicillin and streptomycin (GIBCO), and 8 μg/mL protamine sulfate. Gesicles were applied by centrifugation as described, and the plate was placed in an EVOS FL Auto2 incubator-microscope (EVOS/Thermo Fisher). Bright-field and red fluorescent images were taken every 15 min for 16 hr.
Determining Off-Target Mutation by CRISPR/Cas9
Potential off-target regions utilizing HIV LTR gRNAs were determined using http://crispor.tefor.net. The HIV LTR gRNA sequences were scanned against the rat genome due to CHME-5 cells originating from the rat.60 The top two predicted off-target sites for LTR gRNAs 2 and 4 were chosen, and primers were developed to amplify this region to be used for the resolvase assay. LTR gRNA 2 off-target 1 is located on chromosome 1, and off-target 2 is located on chromosome 17. LTR gRNA 4 off-target 1 is located on chromosome 14, and off-target 2 is located on chromosome 17. After comparing the off-target sites with the gRNA sequence, we confirmed that no gRNA sequence had 100% homology to the paired off-target. Furthermore, no off-target areas are located within exon regions. Primers used for analysis are as follows: LTR gRNA 4 off-target 1, Fwd: 5′-GCGTGAGGGCTTTGTAGAGCTG-3′, Rev: 5′-GCTAGCAAACATCACCACAG-3′, TIDE primer: 5′-GACTTTCATCAGCCAGGGCAC-3′; LTR gRNA 4 off-target 2, Fwd: 5′-GACCAAGCCATCTTCTGACAC-3′, Rev: 5′-GTTTGGGTTGCAGCCTTTCTCC-3′, TIDE primer 5′-GGGTTGGTGTGGTTGGTAGAG-3′; LTR gRNA 2 off-target 1, Fwd: 5′-GGAGCAACTGGTGTGATTCTG-3′, Rev: 5′- GGTTCCATTCTGCGAAGATGAG-3′, TIDE primer 5′-CATAGGGACAGGCATTATGG-3′; LTR gRNA 2 off-target 2: Fwd: 5′-GGCTTCAGAAGCCTCAGAATG-3′, Rev: 5′- GCTCTCCTTCCTGCCAGTGTAG-3′, TIDE primer 5′-GCTTTCACTTTCCCAGTGCC-3′.
PCR
The genomic DNA were isolated using the NucleoSpin Tissue Column (Macherey-Nagel, Bethlehem, PA, USA) according to the manufacturer’s instructions. DNA concentrations were approximated by A260 reading, using a NanoDrop 2000 (Thermo Fisher). PCRs were prepared using Phusion High Fidelity GC Buffer (New England Biolabs, Ipswich, MA, USA) with 5 μM forward and reverse primers and 1 μL of DNA sample. PCR was carried out in a Bio-Rad C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the following cycle. For amplification of HIV-LTR: 98°C for 30 s, 40× cycle of 98°C → 72°C, 72°C 5 min, 12°C hold. PCR products were run on a 1% agarose gel made with 1× TAE (VWR, Radnor, PA, USA) with SYBR Safe DNA gel stain (Thermo Fisher) for 45 min. LTR amplification primers used were Fwd: 5′-CTCTGCTGCCTCCTGTCTTCTG-3′, Rev: 5′-GTTTCAGAATCTCGGGGTGTCCG-3′, and TIDE primer: 5′-CCTTCTAGCCTCCGCTAGTC-3′.
T7 Endonuclease 1
The T7 Endonuclease 1 enzyme “T7E1” (New England Biolabs) was used to detect mutations in the HIV-LTR PCR-amplified region.40, 61 Amplified products were denatured and slowly annealed in NEBuffer 2 (New England Biolabs) using the following cycle: 95°C for 5 min, 85°C for 30 s → ramp 2°C per 5 s, 25°C 30 s → ramp 0.1°C per second, 25°C hold. T7E1 was then added to the reannealed products and incubated at 37°C for 1 hr. DNA were run on a 1% agarose gel, and presence of T7E1 digestion was used to verify CRISPR/Cas9-mediated mutation.
ddPCR
The relative copy number per genome of the modified HIV provirus was determined using ddPCR (QX200; Bio-Rad) to quantify the number of provirus templates per microliter for the NanoLuc transgene relative to the autosomal reference gene GGT1. Primers and probes used are as follows: NanoLuc ddPCR, Fwd: 5′-ATTGTCCTGAGCGGTGAAA-3′, Rev: 5′-CACAGGGTACACCACCTTAAA-3′, Probe: FAM-5′-TGGGCTGAAGATCGACATCCATGT-3′-Iowa Black; GGT1 ddPCR, Fwd: 5′-CCACCCCTTCCCTACTCCTAC-3′, Rev: 5′-GGCCACAGAGCTGGTTGTC-3′, Probe: HEX-5′-CCGAGAAGCAGCCACAGCCATACCT-3′-Iowa Black; Nef EVAGreen assay, Nef Fwd: 5′-GGCTGGATGGCCTACTGTAAGG-3′, Nef Rev 5′-GTCTTTCCAGGTCTCGAGATACTGC-3′. Reaction conditions consisted of a master mix containing 1× ddPCR Supermix for Probes (no dUTP) (Bio-Rad), 450 nM forward and reverse primers, 50 nM probe, 0.1 U MseI restriction enzyme with 50 ng of genomic DNA. Reactions were run analyzing NanoLuc and GGT1 simultaneously as a duplex reaction. Reactions for Nef were performed using EVAGreen Dye (Bio-Rad). Equation for calculating copy number is ([copies/μL NanoLuc]/[copies/μL GGT1])*(2) = copy number provirus per genome.
Luciferase Assay
NanoLuciferase was assayed as previously described.40 Cells that were prepared for luciferase assay were lysed directly in an opaque 96-well plateusing RIPA lysis buffer/1% NP-40 with protease inhibitor. Luminescence was measured in the opaque plate using the substrate coelenterazine (Regis Technologies Morton Grove, IL, USA) in a Bio-Tek Synergy 2 plate reader (Winooski, VT, USA).
Cell Viability
Cell viability was assessed by two methods. For acute viability, gesicles were applied to HIV-NanoLuc CHME-5 cells supplemented with protamine by centrifugation, and viability was assayed 24 hr post-treatment by using the CellTiter-Glo ATP assay (Promega Fitchburg, WI, USA) according to the manufacturer’s instructions. For long-term viability and cell yield, gesicle were applied to cells and expanded for 1 week. HIV-NanoLuc CHME-5 cells were split, with cell yield and viability assayed using trypan blue in the Countess automated cell counter (Invitrogen, Waltham, MA, USA).
Statistical Analysis
All analyses were evaluated by Prism GraphPad (GraphPad Software, La Jolla, CA, USA). Data were analyzed with one-way ANOVA with Bonferroni’s multiple comparison post-test, or with two-way ANOVA with Tukey’s multiple comparison post-test. Data are expressed as the mean ± SEM. Statistically significant differences were considered as p < 0.05.
Author Contributions
L.A.C. designed and performed the majority of experiments, and wrote the manuscript. L.M.C. performed DDCPR experiments and revised the manuscript. C.T.R. developed LTR amplification primers and revised the manuscript. L.V.F. worked to develop parameters for EVOS live-cell imaging and revised the manuscript. A.Y.P. performed experiments for LTR gRNAs 1 and 2 and revised the manuscript. B.K.H. is the corresponding author, designed experiments, and did a detailed revision of the manuscript.
Acknowledgments
This work was supported by the Intramural Research Program at the National Institute on Drug Abuse. The authors would like to thank the AIDS Reagent Program, Division of AIDS, NIAID, NIH for the use of antibodies. The authors would like to thank Dr. Maja Mustapic for assistance with the NanoSight.
Footnotes
Supplemental Information includes two figures, Supplemental Materials and Methods and two videos and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.10.002.
Contributor Information
Lee A. Campbell, Email: lee.campbell@nih.gov.
Brandon K. Harvey, Email: bharvey@mail.nih.gov.
Supplemental Information
References
- 1.Long C., Li H., Tiburcy M., Rodriguez-Caycedo C., Kyrychenko V., Zhou H., Zhang Y., Min Y.L., Shelton J.M., Mammen P.P.A. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap9004. eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salmaninejad A., Valilou S.F., Bayat H., Ebadi N., Daraei A., Yousefi M., Nesaei A., Mojarrad M. Duchenne muscular dystrophy: an updated review of common available therapies. Int. J. Neurosci. 2018;128:854–864. doi: 10.1080/00207454.2018.1430694. [DOI] [PubMed] [Google Scholar]
- 3.Amoasii L., Long C., Li H., Mireault A.A., Shelton J.M., Sanchez-Ortiz E., McAnally J.R., Bhattacharyya S., Schmidt F., Grimm D. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan8081. eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohmori T., Nagao Y., Mizukami H., Sakata A., Muramatsu S.I., Ozawa K., Tominaga S.I., Hanazono Y., Nishimura S., Nureki O., Sakata Y. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 2017;7:4159. doi: 10.1038/s41598-017-04625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang P., Mokhtari R., Pedrosa E., Kirschenbaum M., Bayrak C., Zheng D., Lachman H.M. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie N., Gong H., Suhl J.A., Chopra P., Wang T., Warren S.T. Reactivation of FMR1 by CRISPR/Cas9-mediated deletion of the expanded CGG-repeat of the fragile X chromosome. PLoS ONE. 2016;11:e0165499. doi: 10.1371/journal.pone.0165499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S.M., Yang Y., Oh S.J., Hong Y., Seo M., Jang M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. USA. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Pan Q., Gendron P., Zhu W., Guo F., Cen S., Wainberg M.A., Liang C. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 12.Yoder K.E., Bundschuh R. Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci. Rep. 2016;6:29530. doi: 10.1038/srep29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosicki M., Rajan S.S., Lorenzetti F.C., Wandall H.H., Narimatsu Y., Metzakopian E., Bennett E.P. Dynamics of indel profiles induced by various CRISPR/Cas9 delivery methods. Prog. Mol. Biol. Transl. Sci. 2017;152:49–67. doi: 10.1016/bs.pmbts.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uemura T., Mori T., Kurihara T., Kawase S., Koike R., Satoga M., Cao X., Li X., Yanagawa T., Sakurai T. Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in utero electroporation techniques. Sci. Rep. 2016;6:35861. doi: 10.1038/srep35861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack M., Kleinschmidt A., Brühl H., Klier C., Nelson P.J., Cihak J., Plachý J., Stangassinger M., Erfle V., Schlöndorff D. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat. Med. 2000;6:769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- 21.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins P.D., Dorronsoro A., Booker C.N. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J. Clin. Invest. 2016;126:1173–1180. doi: 10.1172/JCI81131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins P.D., Morelli A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno S., Grange C., Collino F., Deregibus M.C., Cantaluppi V., Biancone L., Tetta C., Camussi G. Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS ONE. 2012;7:e33115. doi: 10.1371/journal.pone.0033115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 26.EL Andaloussi S., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 27.Lee J., Lee H., Goh U., Kim J., Jeong M., Lee J., Park J.H. Cellular engineering with membrane fusogenic liposomes to produce functionalized extracellular vesicles. ACS Appl. Mater. Interfaces. 2016;8:6790–6795. doi: 10.1021/acsami.6b01315. [DOI] [PubMed] [Google Scholar]
- 28.Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borràs F.E., Breakefield X., Budnik V. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momen-Heravi F., Balaj L., Alian S., Mantel P.Y., Halleck A.E., Trachtenberg A.J., Soria C.E., Oquin S., Bonebreak C.M., Saracoglu E. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013;394:1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akyurekli C., Le Y., Richardson R.B., Fergusson D., Tay J., Allan D.S. A systematic review of preclinical studies on the therapeutic potential of mesenchymal stromal cell-derived microvesicles. Stem Cell Rev. 2015;11:150–160. doi: 10.1007/s12015-014-9545-9. [DOI] [PubMed] [Google Scholar]
- 31.McVey M.J., Spring C.M., Semple J.W., Maishan M., Kuebler W.M. Microparticles as biomarkers of lung disease: enumeration in biological fluids using lipid bilayer microspheres. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016;310:L802–L814. doi: 10.1152/ajplung.00369.2015. [DOI] [PubMed] [Google Scholar]
- 32.Jung M.K., Mun J.Y. Sample preparation and imaging of exosomes by transmission electron microscopy. J. Vis. Exp. 2018;(131):e56482. doi: 10.3791/56482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lankford K.L., Arroyo E.J., Nazimek K., Bryniarski K., Askenase P.W., Kocsis J.D. Intravenously delivered mesenchymal stem cell-derived exosomes target M2-type macrophages in the injured spinal cord. PLoS ONE. 2018;13:e0190358. doi: 10.1371/journal.pone.0190358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 35.Trotta T., Panaro M.A., Cianciulli A., Mori G., Di Benedetto A., Porro C. Microglia-derived extracellular vesicles in Alzheimer’s disease: a double-edged sword. Biochem. Pharmacol. 2018;148:184–192. doi: 10.1016/j.bcp.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 36.Mangeot P.E., Dollet S., Girard M., Ciancia C., Joly S., Peschanski M., Lotteau V. Protein transfer into human cells by VSV-G-induced nanovesicles. Mol. Ther. 2011;19:1656–1666. doi: 10.1038/mt.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2013;110:7306–7311. doi: 10.1073/pnas.1214441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolic J., Belot L., Raux H., Legrand P., Gaudin Y., A Albertini A. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat. Commun. 2018;9:1029. doi: 10.1038/s41467-018-03432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takara. Gesicles enable CRISPR/Cas9-mediated gene editing with high efficiency and no additional footprint. https://www.takarabio.com/learning-centers/gene-function/gene-editing/gene-editing-technology-overviews/crispr/cas9-gesicles-overview.
- 40.Campbell L.A., Richie C.T., Zhang Y., Heathward E.J., Coke L.M., Park E.Y., Harvey B.K. In vitro modeling of HIV proviral activity in microglia. FEBS J. 2017;284:4096–4114. doi: 10.1111/febs.14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellano F., Montcourrier P., Guillemot J.C., Gouin E., Machesky L., Cossart P., Chavrier P. Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr. Biol. 1999;9:351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- 42.Graef I.A., Holsinger L.J., Diver S., Schreiber S.L., Crabtree G.R. Proximity and orientation underlie signaling by the non-receptor tyrosine kinase ZAP70. EMBO J. 1997;16:5618–5628. doi: 10.1093/emboj/16.18.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNicholas K., Michael M.Z. Immuno-characterization of exosomes using nanoparticle tracking analysis. Methods Mol. Biol. 2017;1545:35–42. doi: 10.1007/978-1-4939-6728-5_3. [DOI] [PubMed] [Google Scholar]
- 44.Tian X., Nejadnik M.R., Baunsgaard D., Henriksen A., Rischel C., Jiskoot W. A comprehensive evaluation of nanoparticle tracking analysis (NanoSight) for characterization of proteinaceous submicron particles. J. Pharm. Sci. 2016;105:3366–3375. doi: 10.1016/j.xphs.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 45.Willms E., Johansson H.J., Mäger I., Lee Y., Blomberg K.E., Sadik M., Alaarg A., Smith C.I., Lehtiö J., El Andaloussi S. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 48.Hung M.E., Leonard J.N. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles. 2016;5:31027. doi: 10.3402/jev.v5.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meccariello A., Monti S.M., Romanelli A., Colonna R., Primo P., Inghilterra M.G., Del Corsano G., Ramaglia A., Iazzetti G., Chiarore A. Highly efficient DNA-free gene disruption in the agricultural pest Ceratitis capitata by CRISPR-Cas9 ribonucleoprotein complexes. Sci. Rep. 2017;7:10061. doi: 10.1038/s41598-017-10347-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hain A., Krämer M., Linka R.M., Nakhaei-Rad S., Ahmadian M.R., Häussinger D., Borkhardt A., Münk C. IL-2 inducible kinase ITK is critical for HIV-1 infection of Jurkat T-cells. Sci. Rep. 2018;8:3217. doi: 10.1038/s41598-018-21344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abner E., Stoszko M., Zeng L., Chen H.C., Izquierdo-Bouldstridge A., Konuma T., Zorita E., Fanunza E., Zhang Q., Mahmoudi T. A new quinoline BRD4 inhibitor targets a distinct latent HIV-1 reservoir for re-activation from other ‘shock’ drugs. J. Virol. 2018;92 doi: 10.1128/JVI.02056-17. e02056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santos S., Obukhov Y., Nekhai S., Bukrinsky M., Iordanskiy S. Virus-producing cells determine the host protein profiles of HIV-1 virion cores. Retrovirology. 2012;9:65. doi: 10.1186/1742-4690-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuhl B.D., Sloan R.D., Donahue D.A., Bar-Magen T., Liang C., Wainberg M.A. Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology. 2010;7:115. doi: 10.1186/1742-4690-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rozera G., Fabbri G., Lorenzini P., Mastrorosa I., Timelli L., Zaccarelli M., Amendola A., Vergori A., Plazzi M.M., Cicalini S. Peripheral blood HIV-1 DNA dynamics in antiretroviral-treated HIV/HCV co-infected patients receiving directly-acting antivirals. PLoS ONE. 2017;12:e0187095. doi: 10.1371/journal.pone.0187095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amirache F., Lévy C., Costa C., Mangeot P.E., Torbett B.E., Wang C.X., Nègre D., Cosset F.L., Verhoeyen E. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 57.Carpentier D.C., Vevis K., Trabalza A., Georgiadis C., Ellison S.M., Asfahani R.I., Mazarakis N.D. Enhanced pseudotyping efficiency of HIV-1 lentiviral vectors by a rabies/vesicular stomatitis virus chimeric envelope glycoprotein. Gene Ther. 2012;19:761–774. doi: 10.1038/gt.2011.124. [DOI] [PubMed] [Google Scholar]
- 58.Geng X., Doitsh G., Yang Z., Galloway N.L., Greene W.C. Efficient delivery of lentiviral vectors into resting human CD4 T cells. Gene Ther. 2014;21:444–449. doi: 10.1038/gt.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staahl B.T., Benekareddy M., Coulon-Bainier C., Banfal A.A., Floor S.N., Sabo J.K., Urnes C., Munares G.A., Ghosh A., Doudna J.A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia-Mesa Y., Jay T.R., Checkley M.A., Luttge B., Dobrowolski C., Valadkhan S., Landreth G.E., Karn J., Alvarez-Carbonell D. Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J. Neurovirol. 2017;23:47–66. doi: 10.1007/s13365-016-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.