Abstract
NUTM1-rearranged tumors are defined by the presence of a gene fusion between NUTM1 and various gene partners and typically follow a clinically aggressive disease course with poor outcomes despite conventional multimodality therapy. NUTM1-rearranged tumors display histologic features of a poorly differentiated carcinoma with areas of focal squamous differentiation and typically express the BRD4–NUTM1 fusion gene defining a distinct clinicopathologic entity—NUT carcinoma (NC). NCs with mesenchymal differentiation have rarely been described in the literature. In this report, we describe the characterization of two cases of high-grade spindle cell sarcoma harboring a novel MGA–NUTM1 fusion. Whole-genome sequencing identified the presence of complex rearrangements resulting in a MGA–NUTM1 fusion gene in the absence of other significant somatic mutations. Genetic rearrangement was confirmed by fluorescence in situ hybridization, and expression of the fusion gene product was confirmed by transcriptomic analysis. The fusion protein was predicted to retain nearly the entire protein sequence of both MGA (exons 1–22) and NUTM1 (exons 3–8). Histopathologically, both cases were high-grade spindle cell sarcomas without specific differentiation markers. In contrast to typical cases of NC, these cases were successfully treated with aggressive local control measures (surgery and radiation) and both patients remain alive without disease. These cases describe a new subtype of NUTM1-rearranged tumors warranting expansion of diagnostic testing to evaluate for the presence of MGA–NUTM1 or alternative NUTM1 gene fusions in the diagnostic workup of high-grade spindle cell sarcomas or small round blue cell tumors of ambiguous lineage.
Keywords: MGA-NUT1, NUT carcinoma, NUTM1-rearranged tumor, spindle cell carcinoma, synovial sarcoma
INTRODUCTION
NUTM1-rearranged tumors represent a genetically defined group of clinically aggressive cancers characterized by recurrent chromosomal rearrangements involving the NUTM1 gene (French et al. 2004). Most NUTM1-rearranged tumors present as locally invasive and disseminated poorly differentiated carcinoma containing focal areas of squamous differentiation, usually occurring along the midline axis, and have been classified as NUT midline carcinomas (NCs) (French et al. 2004; Bauer et al. 2012; Chau et al. 2016). Given the rarity and overlapping histopathologic features with other disparate tumors, the incidence of NCs remains difficult to determine. Although NCs were originally thought to predominate in childhood, increasing reports document a wide distribution of ages through adulthood (Kees et al. 1991; Vargas et al. 2001; French et al. 2004; Bauer et al. 2012; French 2012). Perhaps the most clinically relevant feature of NC is its association with poor survival due to lack of durable responses despite conventional multimodality therapies (surgery, chemotherapy, and radiotherapy). Overall survival rates of 20%–30% underscore the need for accurate and timely diagnosis of NC and complete surgical resection of tumor, which has been associated with improved outcomes (French 2010; Bauer et al. 2012; Chau et al. 2016). Currently, use of adjuvant chemoradiotherapy has shown very limited benefit (Bauer et al. 2012; Chau et al. 2016).
The characteristic cytogenetic abnormality observed in NCs is the presence of a reciprocal chromosomal translocation involving the NUTM1 gene on Chromosome 15q14 with variant partner genes, most commonly the BET family gene BRD4 and less commonly BRD3, NSD3, ZNF532, MXD1, CIC, and BCORL1 (French et al. 2008, 2014; Alekseyenko et al. 2017; Dickson et al. 2018; Schaefer et al. 2018). Within the early descriptions of NC, cases included rarer variants that occurred outside of the anatomic midline, presenting instead as extremity tumors, resulting in initial alternative diagnoses ranging from Ewing sarcoma, pancreatoblastoma, neuroblastoma, to acute leukemia (Shehata et al. 2010; French 2012). Hence, the term NUT midline carcinoma was changed to NUT carcinoma by the World Health Organization classification of tumors (Thompson and Franchi 2018). In addition, rare cases of sarcomas demonstrating no epithelial differentiation have been reported (Dickson et al. 2018), suggesting that a subset of pure mesenchymal NUTM1-rearranged tumors are distinct from NC.
The advent of next-generation sequencing platforms that enable the identification of therapeutically relevant and targetable mutations and their incorporation into clinical diagnostics and treatment planning have offered renewed insight into the biology and potential curative approaches to rare and incurable cancers. In alignment with the concept of harnessing tumor genetic profiling to inform therapies, we developed a prospective program for screening patients with high-risk solid tumors with the goal of identifying drug-targetable mutations. As a result of this screening effort, we identified two cases with a primary diagnosis of high-grade spindle cell sarcoma harboring novel MGA–NUTM1 fusions. We describe the genetic and histologic characterization of this novel fusion and discuss the implications of this case on the pathologic classification of NUTM1-rearranged tumors.
CLINICAL PRESENTATIONS
Case 1
A previously healthy male presented at 10 yr of age with a history of leg pain and an enlarging thigh mass. The tumor was composed of a monomorphic spindle cell neoplasm arranged in vague sweeping fascicles. The tumor was associated with a peculiar dense hyalinized stroma, resembling amianthoid fibers. The tumor showed an increased mitotic activity of >10 MF/10 HFs, but necrosis was absent. The immunohistological profile was nonspecific, showing focal positivity for CD99, CD34, and BCL-2, rare desmin, and otherwise focal negativity for pan-cytokeratin, CK7, EMA, S-100, GFAP, and myogenin. Although the morphologic appearance suggested the possibility of a monophasic synovial sarcoma, FISH was negative for the SS18 gene rearrangement. A histological diagnosis of high-grade spindle cell sarcoma, not otherwise specified (NOS), was rendered. Radiographic workup did not demonstrate the presence of metastatic disease. The patient underwent surgical resection of the tumor but was found to have positive margins following pathologic review of resected tissue. The patient subsequently received adjuvant intensity-modulated radiation therapy (IMRT) (5580 cGy) but was found to have local recurrence proximal to the previous RT field ∼1 yr later. The recurrent tumor was biopsied showing histology similar to the initial tumor. The patient received neoadjuvant chemotherapy (ifosfamide and doxorubicin) followed by surgical resection. Consolidation with IMRT (5580 cGy) and adjuvant chemotherapy (ifosfamide/etoposide and additional cycles of ifosfamide/doxorubicin) followed. The patient was free of disease for >6½ yr when he presented with a second local recurrence. Radiographic imaging showed no evidence of metastatic disease, with tumor histopathologically reminiscent of the original tumor. The patient was enrolled on an NCI Phase I clinical trial utilizing autologous activated NK cells in combination with IL-15 and cyclophosphamide (NCT01875601) followed by radiation therapy. He had resection of residual tumor and has since remained disease-free for 11 yr following initial diagnosis.
Case 2
A previously healthy female presented at 10 yr of age with a 1-mo history of intermittent bilateral frontal headaches, followed by sudden onset vomiting and altered mental status. The patient was brought to a local emergency room for clinical workup, revealing a left-sided anterior intracranial mass and associated signs of increased intracranial pressure. She underwent an emergent left frontal craniotomy and was found to have a dural-based tumor that was completely resected. Subsequent diagnostic workup showed no evidence of metastatic disease. Pathology demonstrated a spindle cell sarcoma arranged in long intersecting fascicles. The tumor cells showed an isomorphic cytology, with variable mitotic activity and Ki-67 staining positive in only 10%–20% of cells. A variable collagenous stroma was present, ranging from scant to extensive. The immunohistologic profile was nonspecific for a line of differentiation, showing focal positivity for desmin and CD99, but negative for other markers including CD34, pan-cytokeratin, EMA, GFAP, STAT6, S-100, MUC4, BCOR, MYOD1, and SATB2. FISH testing was negative for rearrangements of EWSR1, FUS, and SS18. Given these results, a diagnosis of high-grade spindle cell sarcoma, NOS was made. The patient underwent focal RT (5580 cGy) to the resection cavity and has since been alive and well, with no evidence of disease, 15 mo following her initial diagnosis.
RESULTS
Genomic Analysis
Case 1
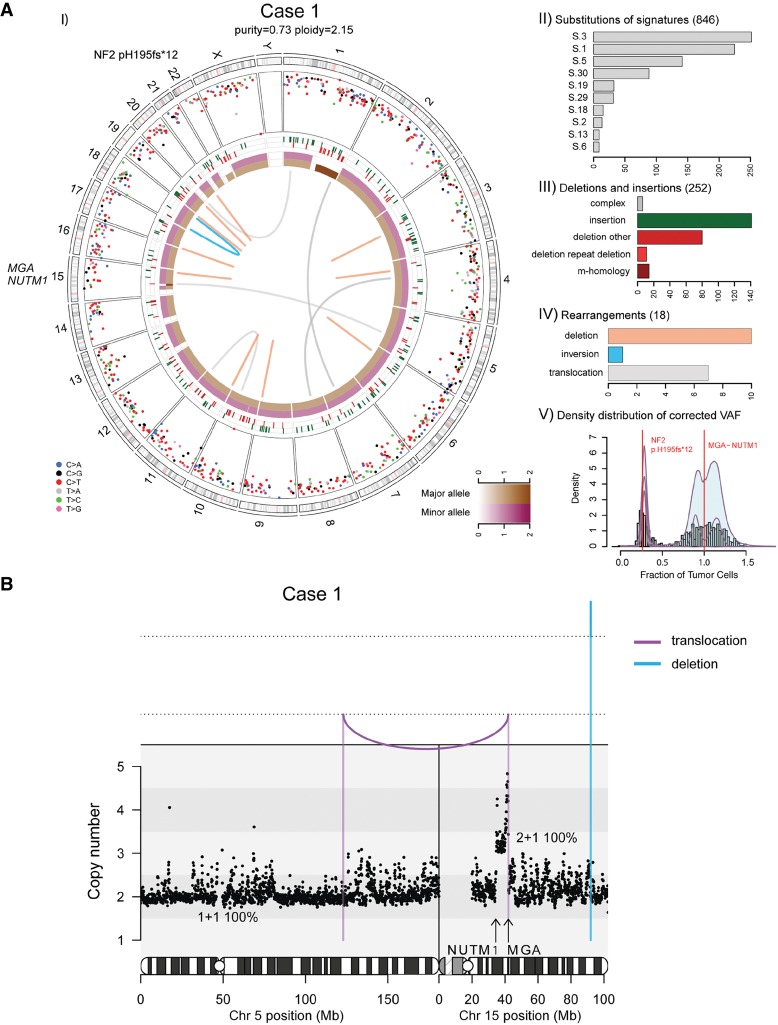
We performed whole-genome sequencing (WGS) of the patient's primary tumor and matched normal tissue to a mean haploid coverage of 92× and 49×, respectively (Fig. 1A; Supplemental Table 3). The tumor sample was diploid and had high tumor content (73%). Analysis of the WGS data identified a total of 1116 somatic alterations (846 single-nucleotide substitutions, 252 indels, and 18 rearrangements) indicating a relatively low mutation burden (2.9 × 10−4 subs/Mb). Assessment of the contribution of known mutation signatures from COSMIC (http://grch37-cancer.sanger.ac.uk/cosmic/signatures) in the mutation spectrum of the substitution data showed that a majority of mutations were assigned to S-1 (aging-related, clock-like), S-3 (associated with biallelic inactivation of BRCA1 and BRCA2), and S-8 (unknown etiology) (Alexandrov et al. 2013; Forbes et al. 2017). However, the tumor did not exhibit other genomic features indicative of BRCA1/BRCA2 deficiency—namely, an excess of microhomology-mediated deletions, short <10-kilobases (kb), or long >100-kb tandem duplications—precluding the possibility of a “BRCAness” phenotype (Davies et al. 2017).
Figure 1.
(A) Whole-genome analysis identifies a novel MGA–NUTM1 fusion in Case 1. (I) Circos plot summarizing the whole-genome sequencing (WGS) data. The two innermost tracks depict the integer copy-number changes for the major (brown) and minor (dark pink) allele. The outermost track shows the intermutation distance for substitutions each plotted according to the type of nucleotide change. The middle track shows the genomic positions of the small insertions (green) and deletions (red) along the genome. Rearrangements are plotted as arcs inside the Circos plot. Genes affected by potential oncogenic changes are annotated. (II) Mutation signature analysis of the substitutions using Mutational Patterns. Only the signatures with the 10 highest exposures are shown. (III) Summary of the indel data also showing the contribution of repeat- or microhomology-mediated mechanisms among deletions. (IV) Summary of rearrangement data. (V) Statistical analysis of the corrected variant allele frequency of substitution by Bayesian Dirichlet process–based clustering. Empiric histogram of substitutions is shown in gray together with the density from clustering in pale green and the fitted distribution in dark pink. Potential oncogenic alterations are annotated. (B) Integrated copy-number/rearrangement plots showing the WGS-based absolute copy number (y-axis) across the indicated genomic region (x-axis). Rearrangements are depicted as lines perpendicular to the x-axis.
Case 1 revealed only two somatic changes of potential functional impact: a clonal copy-number gain on Chromosome 15 from intron 2/7 of NUTM1 (Chr 15:34,639,000) to intron 22/24 of MGA (Chr 15:42,057,000), and a subclonal frameshift deletion in NF2 (p.H195fs*12) (Table 1). Rearrangement analysis of WGS data identified an unbalanced translocation within a few bases of the breakpoint in intron-22 of the MGA locus (Chr 5:122919228:+ → Chr 15:420571 93:−) and supported by 14 reads, but failed to identify any aberrant read groups in intron 2 of NUTM1 (Table 1; Fig. 1B).
Table 1.
Molecular characteristics of the two patients with MGA–NUTM1 fusion
| Tumor Type | Patient ID | Chr | Location | Wild type | Mutant | Allele frequency in tumor | Coverage in tumor | Allele frequency in normal | Coverage in normal | Variant Type | Genes affected | HGVS DNA change | HGVS protein change | Predicted effect | ClinVar ID | IHC | Allele origin | Clinical significance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HGSCS | Case 1 | 22 | 30051645 | AGAG CACC |
A | 0.13 | 67 | 0 | 48 | Deletion | NF2 | c.580_586del GAGCACC |
p.H195fs*12 | Frameshift | SCV000804554 | Unknown | Somatic | Pathogenic |
| HGSCS | Case 1 | 5 | 122919229 | T | T]15:42057194] | 0.09 | 152 | 0 | 66 | Translocation |
MGA, NUTM1 |
t(5;15)(q23.2;q15.1) (g.71436_104584) |
MGA/NUTM1 | Fusion | SCV000804552 | Retained | Somatic | Pathogenic |
| HGSCS | Case 2 | 15 | 34638819 | A | [15:42055110[A | 0.13 | 123 | 0 | 70 | Translocation |
MGA, NUTM1 |
t(15;15)(q14;15q.1)(g.753_102500) | MGA/NUTM1 | Fusion | SCV000804554 | Retained | Somatic | Pathogenic |
| HGSCS | Case 2 | 15 | 42054977 | T | [15:34638819[T | 0.17 | 114 | 0 | 71 | Translocation |
MGA, NUTM1 |
t(15;15)(q14;15q.1)(g.1016_102367) | MGA/NUTM1 | Fusion | SCV000804553 | Retained | Somatic | Pathogenic |
Genomic coordinates, wild type, and mutant alleles are reported in GRCh37 and according to VCF specifications.
HGSCS, high-grade spindle cell sarcoma.
Case 2
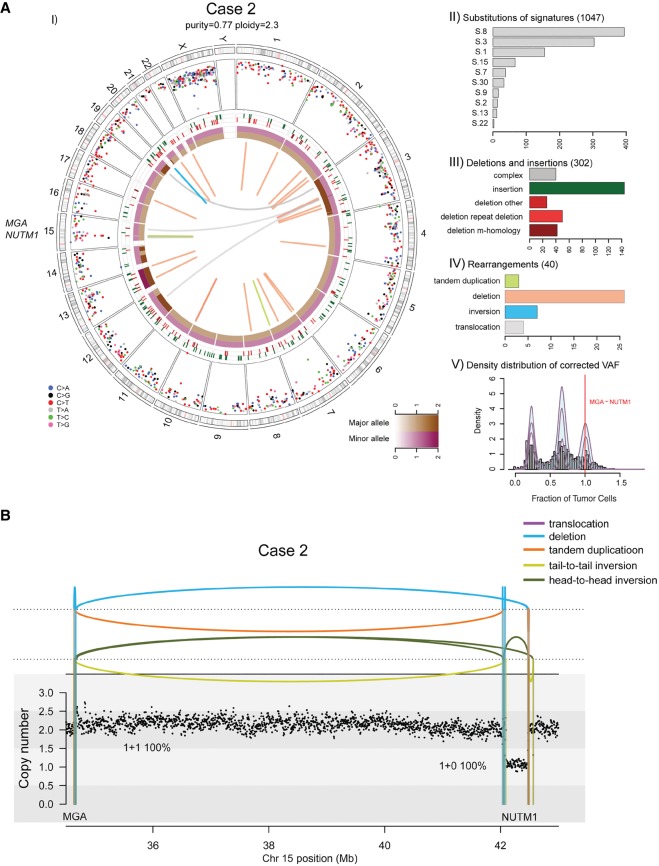
The tumor sample had high tumor content (77%), and WGS of the patient's primary tumor and matched normal tissue to a mean haploid coverage of 91× and 49× was performed (Supplemental Table 3). Analysis of the WGS data identified a total of 1389 somatic alterations (1047 single-nucleotide substitutions, 302 indels, and 40 rearrangements) confirming a relatively low mutation burden (3.6 × 10−4 subs/Mb) in this tumor (Fig. 2A). Assessment of the contribution of known mutation signatures from COSMIC shows a similar signature profile distribution to Case 1 (Fig. 2A).
Figure 2.
(A) Whole-genome analysis identifies a novel MGA–NUTM1 fusion in Case 2. (I) Circos plot summarizing the whole-genome sequencing (WGS) data. (II) Mutation signature analysis of the substitutions using Mutational Patterns. (III) Summary of the indel data also showing the contribution of repeat- or microhomology-mediated mechanisms among deletions. (IV) Summary of rearrangement data. (V) Statistical analysis of the corrected variant allele frequency of substitution by Bayesian Dirichlet process–based clustering. For details, see legend to Figure 1. (B) Integrated copy-number/rearrangement plots showing the WGS-based absolute copy number (y-axis) across the indicated genomic region (x-axis). Rearrangements are depicted as lines perpendicular to the x-axis.
After reviewing all genomic alterations of Case 2, the only somatic alteration of potential functional impact was the MGA–NUTM1 fusion created by a complex structural variant including multiple rearrangement breakpoints between intron 2/7 of NUTM1 and intron 22/24 of MGA (Table 1; Fig. 2B). The same rearrangements were associated with a clonal deletion indicating that the fusion gene is found in 100% of the tumor cells.
Fluorescence In Situ Hybridization (FISH)
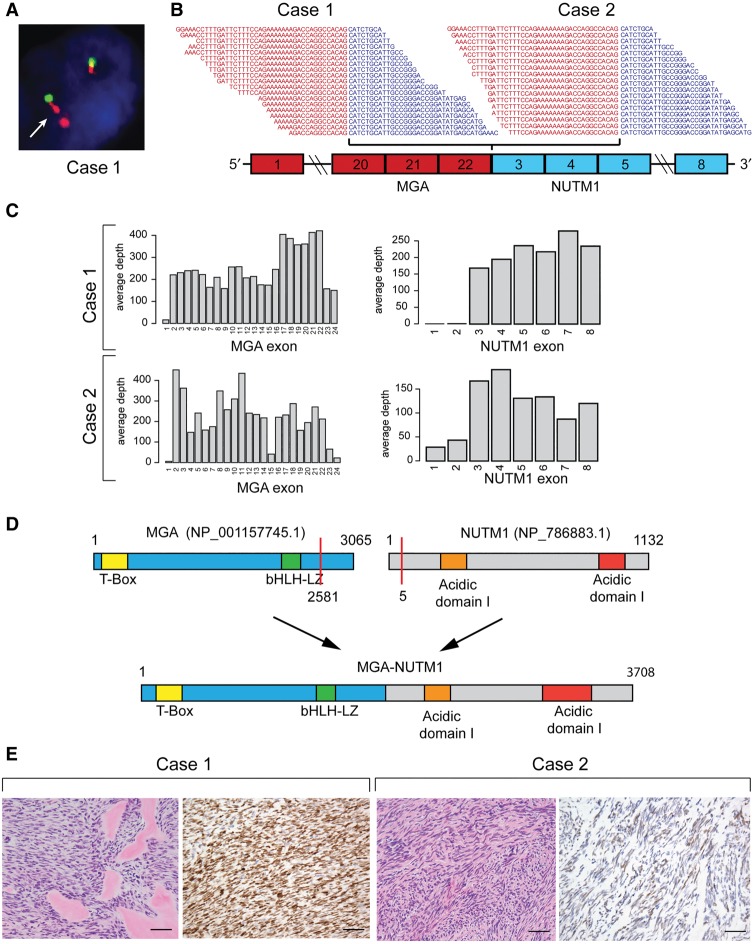
Dual-color break-apart probes for the NUTM1 gene were applied to tumor tissue samples from Case 1 (Fig. 3A). No tissue from Case 2 was available for FISH testing. Tissue from two different resections of Case 1 showed satisfactory FISH signals with minimal background. Tissue from initial diagnosis exhibited a complex break-apart signal pattern consistent with a rearrangement at the NUTM1 locus. Interestingly 60% of tumor cells showed a doublet of the NUTM1 3′ probe (orange signal). The same abnormal signal pattern was observed in tissue obtained from the patient's second local recurrence, ranging from 60% to almost 100% in various tumor areas (data not shown). In both samples, gain of extra signals were observed in a small percentage of cells. The signal patterns observed in these two samples were indicative of a complex rearrangement involving the 3′ NUTM1 locus and consistent with the WGS findings.
Figure 3.
(A) Case 1: Fluorescence in situ hybridization assay showing evidence of complex NUTM1 rearrangements. The arrow indicates examples of abnormal doublet signals of the 3′ NUTM1 locus (orange). (B) RNA-sequencing reads spanning the junction between MGA exon 22 and NUTM1 exon 3. (C) RNA-seq coverage of MGA and NUTM1 exons. (D) Schematic representation of MGA and NUTM1 protein domains and resulting MGA–NUTM1 fusion protein structure. (E) H&E staining of both cases showed a monomorphic spindle cell morphology arranged in fascicles with an associated collagenous stroma. Case 1 showed dense hyalinized material, the so-called “amianthoid fibers,” resembling osteoid matrix deposition. Immunohistochemically Case 1 showed strong, diffuse nuclear reactivity for NUTM1, whereas Case 2 showed a multifocal weak staining pattern. Scale bars, 500 µM.
RNA-Sequencing Analysis
Analysis of RNA-seq data for fusion genes unequivocally identified an in-frame chimeric transcript containing exons 1–22 of MGA (NM_001164273.1) and exons 3–8 of NUTM1 (NM_175741.2) in both cases (Fig. 3B). The fusion was detected independently by three different fusion caller algorithms with high read support in both cases. RNA-seq coverage of all MGA exons and all but the first two exons of NUTM1 confirm that expression of the fusion is driven by the MGA promoter (Fig. 3C). The predicted MGA–NUTM1 fusion protein (3708 amino acids) is composed of almost the entire MGA protein and maintains both DNA binding domains of MGA (the Max interacting bHLH leucine zipper domain [bHLHLZ] and the T-box domain), as well as NUTM1 domains normally included in other NUT fusion proteins (Fig. 3D). Analysis of the protein coding sequence identified two nuclear localization signals (NLSs) suggesting the fusion protein should localize to the nucleus.
Immunohistochemical Analysis
To confirm and localize the expression of MGA–NUTM1, we performed immunohistochemical analysis for NUTM1. As shown in Figure 3D, the Case 1 primary tumor showed a diffuse (100% positive nuclei) intense nuclear staining for NUTM1, confirming the expression of the fusion at the protein level. Positive staining for NUTM1 was also confirmed in Case 1 recurrence and metastatic samples (Supplemental Fig. 1). The Case 2 primary tumor had diffuse (60% positive nuclei) but weaker staining. In the former sample the staining was less intense and diffusely distributed than typical NUT carcinoma cases. These results may be due to the sensitivity of the IHC assay, which was optimized to detect levels typical in the testes and NUT carcinoma samples (Haack et al. 2009).
Supplemental Table 2 summarizes the results of all assays performed on the various surgical specimens. Through multiple orthogonal assays, we confirmed the presence and expression of a novel MGA–NUTM1 fusion gene in two cases of pediatric high-grade sarcoma.
DISCUSSION
We describe the genetic and histologic characterization of two cases of high-grade spindle cell sarcoma harboring a novel MGA–NUTM1 fusion. Most prior descriptions of NUTM1-rearranged tumors represent poorly differentiated carcinomas with variable degrees of squamous differentiation (French et al. 2004). Rare cases of NC with mesenchymal differentiation have been reported, with one case having the appearance of a poorly differentiated carcinoma with squamoid differentiation along with a malignant mesenchymal (chondroid) component positive for the BRD4–NUTM1 fusion (den Bakker et al. 2009). There appear to be a group of spindle cell sarcomas harboring NUTM1 rearrangement that are completely distinct from NCs. These tumors harbored BCORL1–NUTM1 and CIC–NUTM1 rearrangements (Dickson et al. 2018; Mangray et al. 2018; Watson et al. 2018). Likewise, the two cases presented here lack any evidence of epithelial differentiation and show a high-grade sarcoma phenotype with a monomorphic/primitive cytomorphology. Notably, the expression of MGA–NUTM1 fusion is driven by the MGA gene promoter. During mouse embryonic development, MGA expression profiles overlap with those of several T-box transcription factors, and although it is widely expressed throughout the mouse embryo, it reaches its highest levels of expression in the limbs, branchial arches, and tail, suggesting its involvement in the induction of mesoderm (Hurlin et al. 1999; Burn et al. 2018). Therefore, it is plausible that expression of NUTM1-rearranged fusions, in particular MGA–NUTM1, within a progenitor cell of either neural crest or mesenchymal origin can drive the malignant transformation into a high-grade sarcoma.
The overall survival outcomes for NC patients have historically been poor with only a small percentage of survivors despite aggressive multimodality treatment (French 2010; Bauer et al. 2012; Chau et al. 2016). However, our two described pediatric cases of MGA–NUTM1 sarcomas are both alive without evidence of disease. Surgical resection and radiotherapy were utilized in three cases (two cases described in this report and one case described by den Bakker et al. [2009]), and systemic cytotoxic chemotherapy, utilizing a regimen used in the treatment of soft tissue and bone sarcomas, was used in two cases. Case 2 did not incorporate systemic therapy highlighting the importance of local control, via complete surgical resection and radiotherapy, in the management of this subset of NUTM1-rearranged tumors. Although the sum cases of NUTM1-rearranged sarcomas remain small, the curative outcomes associated with these tumors following multimodality treatment bear a striking and clinically notable contrast to patients with more classical features of NCs. Therefore, it will be imperative to fully characterize tumors of ambiguous lineage for the presence of NUTM1 gene rearrangements and tissue lineage markers in order to identify this unique subset of NUTM1-rearranged tumors with associated mesenchymal markers and institute prompt multimodality therapies to maximize the chances of long-term cure.
In summary, we describe two cases of NUTM1-rearranged tumors with a novel MGA–NUTM1 fusion gene occurring within the context of a histologically defined high-grade spindle cell sarcoma and in the absence of epithelial differentiation. We propose that the limited series of patients described herein and reported represent a new and distinct subset of NUTM1 fusion-positive sarcomas that may be associated with a favorable prognosis presuming prompt institution of multimodality therapy. Given the unclear prevalence of NUTM1-rearranged tumors, screening for the presence of MGA–NUTM1 or alternative NUTM1 gene fusions should be considered in the routine diagnostic evaluation of high-grade spindle cell tumors and tumors of small round blue cell histology.
METHODS
Nucleic Acid Extraction
DNA Extraction
DNA extraction was performed following the manufacturer's protocol using DNeasy Blood & Tissue kit (QIAGEN).
RNA Extraction
Tissues were homogenized in 1 mL TRIzol (Invitrogen) using TissueLyzer (QIAGEN) and RNA extraction was performed following the manufacturer's protocol.
DNA- and RNA-Sequencing Preparation
DNA-sequencing libraries were prepared using the KAPA Library preparation kit (Kapa Biosystems). This process includes shearing the DNA, repairing the ends of the fragments, adding an A-base to the 3′ ends, ligating Illumina adapters, and amplifying the DNA to prepare samples for sequencing. The DNA was sheared to an average base pair size of 450 bp using the Covaris LE220 instrument (Covaris) under default settings. Following shearing, the DNA end repair step uses T4 DNA polymerase and Klenow DNA polymerase to remove 3′ overhangs and fill in the 5′ overhangs. As the adapter ligation requires the presence of a 3′ A-base on the double-stranded DNA fragments, the adenylation step uses dATPs and Exo(−) Klenow to adenylate the DNA fragments. Illumina NEXTflex-96 DNA adapters (Bioo Scientific) are attached to the 3′ ends using DNA ligase followed by PCR enrichment. The final libraries are then assayed for quality using the Agilent 2100 Bioanalyzer using the DNA 1000 chip (Agilent Technologies). Final libraries that passed QC have a concentration of >2 ng/µl and a library size of >200 bp with average peak height of 400 bp or greater.
For RNA preparation, samples used Kapa's Stranded RNA-Seq Kit with RiboErase (HMR) combined with Agilent's SureSelectXT Target Enrichment Kit for Illumina Multiplex Sequencing. RNA samples are fragmented using heat and magnesium and converted into cDNA. A-tailing is performed to add dAMP to the 3′ ends of the dscDNA library fragments. dsDNA adapters with 3′-dTMP overhangs are ligated to the A-tailed library insert fragments. Library fragments carrying appropriate adapter sequences at both ends were amplified using high-fidelity, low-bias PCR. Samples were then hybridized with biotinylated RNA library baits and the targeted regions are selected using magnetic streptavidin beads before amplification. The library quality was confirmed on an Agilent 2100 Bioanalyzer using the Agilent High Sensitivity chip and the quantity was confirmed using Thermo Fisher's Qubit 4 Fluorometer with the dsDNA BR Assay kit (Thermo Fisher Scientific). Final libraries passed QC with a concentration of >2 ng/µl and an average peak height of >300 bp.
DNA and RNA Sequencing
DNA sequencing was performed on the Illumina HiSeq X 2 × 150 bp run (Illumina). The final DNA library was diluted, denatured, and introduced into the lanes of the flow cell using the Illumina cBot 2 system according to the manufacturer's protocol. The libraries were loaded at a 2:1 tumor:normal ratio to reach coverage (average read depth) of 80× for the tumor sample and 40× for the normal sample. RNA libraries were sequenced on the Illumina HiSeq 2500 2 × 50 bp Rapid Run platform multiplexing a total of seven samples onto one flow cell giving a minimum of 40,000 reads per sample.
Whole-Genome Sequencing and RNA Sequencing
Alignment
Short-insert paired-end reads were aligned to the GRCh37 reference human genome with 1000 Genomes decoy contigs using BWA-MEM (Li and Durbin 2010).
Substitution
Single base substitutions were called using CaVEMan (Cancer Variants through Expectation Maximisation) (http://cancerit.github.io/CaVEMan/). As described previously (Nik-Zainal et al. 2012), the algorithm compares sequence data from each tumor sample to its own matched noncancerous sample and calculates a mutation probability at each genomic locus. Copy-number and cellularity information for CaVEMan were predicted with the Battenberg algorithm (Nik-Zainal et al. 2012) using 1000 Genomes (Genomes Project Consortium et al. 2012) loci within the WGS data. Additional filtering criteria are described in Supplemental Materials and Methods.
Small Insertions and Deletions
Small somatic insertions and deletions (indels) were identified using a modified version of Pindel (https://github.com/cancerit/cgpPindel) (Raine et al. 2015). Variant annotation was done in Ensembl v74 using VAGrENT (Menzies et al. 2015). Additional filtering criteria are described in Supplemental Materials and Methods.
Structural Rearrangements
Structural rearrangements were detected by an in-house algorithm, BRASS (Breakpoints via assembly; https://github.com/cancerit/BRASS), which first groups discordant read pairs that span the same breakpoint and then uses Velvet de novo assembler performs local assembly within the vicinity to reconstruct and determine the exact position of the breakpoint to nucleotide precision (Zerbino and Birney 2008).
Copy-Number Changes
Segmental copy-number information was derived for each sample using the Battenberg algorithm as previously described (Nik-Zainal et al. 2012). Additional filtering criteria are described in Supplemental Materials and Methods.
Mutational Signature Analysis
Mutational signature analysis of the substitutions were performed using the R package mutational-signatures (Gehring et al. 2015). Small insertion/deletions were interrogated for the presence of either short tandem repeat or microhomology at the breakpoints as described previously (Nik-Zainal et al. 2016). Complex indels were excluded from this analysis.
Clonality Analysis
For each mutation we calculated corrected VAF (aka cancer cell fraction) as previously described (Bolli et al. 2014), using the mutant allele burden, tumor purity, and locus-specific copy number in the tumor and matched normal. Subclones were identified by clustering the cancer cell fraction with Dirichlet process–based clustering as described previously (Gundem et al. 2015).
Fusion Detection
RNA-seq data was interrogated for potential fusion genes using three different fusion callers: FusionCatcher (Nicorici et al. 2014), SOAPFuse (Jia et al. 2013), and STAR-Fusion (Haas et al. 2017).
Immunohistochemistry
Immunohistochemical analysis was performed as previously described (Haack et al. 2009) using 4-µm formalin-fixed paraffin-embedded (FFPE) tissue sections using antibody to NUT (Cell Signaling; clone C52B1; 1:200).
FISH Analysis
FFPE tissue sections of 4 µm thickness with tumor areas marked were used for FISH following standard protocols. Briefly, the tissue sections were de-paraffinized in CitriSolv solution (Fisher Scientific), followed by dehydration in 100% ethanol. After heating in 20% citrate buffer, the tissue sections were treated with pepsin (0.5 mg/ml in 0.1 N HCl) for 10–25 min at 40°C, followed by fixation in 10% formalin, and dehydration in a series of 70%, 85%, and 100% ethanol. The NUTM1 dual color break-apart probes (ZytoVision) contain a 5′ probe of 590 kb, labeled in green, and the 3′ probe of 455 kb, labeled in orange. After applying the FISH probes to the tissue areas, both tissue and probes were co-denatured for 7 min at 94°C, and then incubated overnight at 37°C, followed by posthybridization washing in 2× SSC/0.3% NP-40 for 2 min at 77°C. Tissue sections were counterstained with DAPI. Signal analysis was performed in combination with morphology correlation, and at least 100 interphase cells within the marked tumor area were evaluated. A positive result is determined if >10% of cells show a split or other abnormal signal pattern.
ADDITIONAL INFORMATION
Data Deposition and Access
The results of whole-genome sequencing have been deposited into EGA (https://www.ebi.ack.uk/ega/home) under accession number EGAS00001003341, and RNA-seq data has been made available in the Gene Expression Omnibus (GEO) database. The genomic variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000804554, SCV000804552, and SCV000804553.
Ethics Statement
Informed and signed consent was obtained and archived for the research performed and publication of the results. The patients were enrolled onto the Memorial Sloan Kettering Cancer Center (MSKCC) targeted gene sequencing research study (Genomic profiling in cancer patients; NCT01775072) with approval from the MSKCC Institutional Review Board under protocol IRB# 12-245.
Acknowledgments
We would like to thank Dr. Vaidehi Jobanputra, New York Genome Center (NYGC) Clinical Laboratory Director, who leads the NYGC clinical laboratory. The patients presented in this manuscript were sequenced as participants in the Cancer Alliance Study at the NYGC.
Author Contributions
D.D., F.D.C., G.G., N.B., M.B., Y.Z., R.D., C.A,. C.F., E.P., A.K., and N.S. designed the study and analyzed the data. F.D.C., N.B., M.B., A.C., I.D., and N.S. collected clinical data. Y.Z., C.A., and C.F. reviewed pathology slides and collected pathology data. G.G., R.D., and E.P. performed the genomic analyses. K.W., V.F., R.S., M.S., H.G., and S.R. analyzed WGS and RNA data. D.D., F.D.C., G.G., Z.Y., I.D., C.A., C.F., E.P., A.K., and N.S. wrote and/or edited the manuscript. All authors reviewed the manuscript.
Funding
The genomic studies described were supported by the Memorial Sloan Kettering (MSK) Cancer Center Support Grant (P30 CA008748). F.D.C. would like to thank and acknowledge research support by the Willens Family Fund, the Paulie Strong Foundation, and the Alan B. Slifka Foundation. F.D.C. and A.K. would like to acknowledge the generous support of this research by the Sohn Conference Foundation. This study was supported in part by a grant from IBM.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Alekseyenko AA, Walsh EM, Zee BM, Pakozdi T, Hsi P, Lemieux ME, Dal Cin P, Ince TA, Kharchenko PV, Kuroda MI, et al. 2017. Ectopic protein interactions within BRD4-chromatin complexes drive oncogenic megadomain formation in NUT midline carcinoma. Proc Natl Acad Sci 114: E4184–E4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Mitchell CM, Strait KM, Lathan CS, Stelow EB, Lüer SC, Muhammed S, Evans AG, Sholl LM, Rosai J, et al. 2012. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res 18: 5773–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, et al. 2014. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5: 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn SF, Washkowitz AJ, Gavrilov S, Papaioannou VE. 2018. Postimplantation Mga expression and embryonic lethality of two gene-trap alleles. Gene Expr Patterns 27: 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau NG, Hurwitz S, Mitchell CM, Aserlind A, Grunfeld N, Kaplan L, Hsi P, Bauer DE, Lathan CS, Rodriguez-Galindo C, et al. 2016. Intensive treatment and survival outcomes in NUT midline carcinoma of the head and neck. Cancer 122: 3632–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, Ramakrishna M, Martin S, Boyault S, Sieuwerts AM, et al. 2017. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 23: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker MA, Beverloo BH, van den Heuvel-Eibrink MM, Meeuwis CA, Tan LM, Johnson LA, French CA, van Leenders GJ. 2009. NUT midline carcinoma of the parotid gland with mesenchymal differentiation. Am J Surg Pathol 33: 1253–1258. [DOI] [PubMed] [Google Scholar]
- Dickson BC, Sung YS, Rosenblum MK, Reuter VE, Harb M, Wunder JS, Swanson D, Antonescu CR. 2018. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol 42: 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA. 2010. Demystified molecular pathology of NUT midline carcinomas. J Clin Pathol 63: 492–496. [DOI] [PubMed] [Google Scholar]
- French CA. 2012. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol 7: 247–265. [DOI] [PubMed] [Google Scholar]
- French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, Nose V, Vargas SO, Moschovi M, Tzortzatou-Stathopoulou F, et al. 2004. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol 22: 4135–4139. [DOI] [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. 2008. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27: 2237–2242. [DOI] [PubMed] [Google Scholar]
- French CA, Rahman S, Walsh EM, Kühnle S, Grayson AR, Lemieux ME, Grunfeld N, Rubin BP, Antonescu CR, Zhang S, et al. 2014. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov 4: 928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring JS, Fischer B, Lawrence M, Huber W. 2015. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics 31: 3673–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Högnäs G, Annala M, et al. 2015. The evolutionary history of lethal metastatic prostate cancer. Nature 520: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack H, Johnson LA, Fry CJ, Crosby K, Polakiewicz RD, Stelow EB, Hong SM, Schwartz BE, Cameron MJ, Rubin MA, et al. 2009. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol 33: 984–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, Bankapur A, Ganote C, Doak T, Pochet N, et al. 2017. STAR-Fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv 10.1101/120295. [DOI] [Google Scholar]
- Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN. 1999. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J 18: 7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, Yu Y, Zhu D, Nickerson ML, Wan S, et al. 2013. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol 14: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kees UR, Mulcahy MT, Willoughby ML. 1991. Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19). Am J Pediatr Hematol Oncol 13: 459–464. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangray SK, Guellec SL, Fridman E, Shago M, Matoso A, Li R, Lombardo K, Coindre J, Ali S, Somer G, et al. 2018. USCAP 2018 abstracts: genitourinary pathology (894–1126). Mod Pathol 31: 323.29551799 [Google Scholar]
- Menzies A, Teague JW, Butler AP, Davies H, Tarpey P, Nik-Zainal S, Campbell PJ. 2015. VAGrENT: variation annotation generator. Curr Protoc Bioinformatics 52: 15 18 11–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kilkku O. 2014. FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing date. bioRxiv 10.1101/011650. [DOI] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. 2012. The life history of 21 breast cancers. Cell 149: 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. 2016. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine KM, Hinton J, Butler AP, Teague JW, Davies H, Tarpey P, Nik-Zainal S, Campbell PJ. 2015. cgpPindel: identifying somatically acquired insertion and deletion events from paired end sequencing. Curr Protoc Bioinformatics 52: 15 17 11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer IM, Dal Cin P, Fletcher CDM, Hanna GJ, French CA. 2018. CIC-NUTM1 fusion: a case which expands the spectrum of NUT-rearranged epithelioid malignancies. Genes Chromosomes Cancers 10.1002/gcc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata BM, Steelman CK, Abramowsky CR, Olson TA, French CA, Saxe DF, Ricketts RR, Katzenstein HM. 2010. NUT midline carcinoma in a newborn with multiorgan disseminated tumor and a 2-year-old with a pancreatic/hepatic primary. Pediatr Dev Pathol 13: 481–485. [DOI] [PubMed] [Google Scholar]
- Thompson LDR, Franchi A. 2018. New tumor entities in the 4th edition of the World Health Organization classification of head and neck tumors: nasal cavity, paranasal sinuses and skull base. Virchows Arch 472: 315–330. [DOI] [PubMed] [Google Scholar]
- Vargas SO, French CA, Faul PN, Fletcher JA, Davis IJ, Dal Cin P, Perez-Atayde AR. 2001. Upper respiratory tract carcinoma with chromosomal translocation 15;19: evidence for a distinct disease entity of young patients with a rapidly fatal course. Cancer 92: 1195–1203. [DOI] [PubMed] [Google Scholar]
- Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, Guinebretière JM, Freneaux P, Le Loarer F, Bouvet M, et al. 2018. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol 245: 29–40. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.