Abstract
Endometrial cancer is the most common gynecologic malignancy in industrialized countries, and both its incidence and its associated mortality are increasing. The “liquid biopsy” is becoming an important transformative precision oncology tool, but barriers intrinsic to blood sampling have limited its use in early cancer detection. We hypothesized that using a more targeted sample for analysis—namely, a uterine lavage—should provide a more sensitive and specific diagnostic test for endometrial cancer. Using a custom 12-gene endometrial cancer panel, molecular analysis of uterine lavage fluid from an asymptomatic 67-yr-old female without histopathologic evidence of premalignant lesions or cancer in her uterine tissue revealed two oncogenic PTEN mutations. Ten months later, the patient returned with postmenopausal bleeding and a single microscopic focus of endometrial cancer. DNA isolated and sequenced from laser-capture microdissected tumor tissue revealed the same two PTEN mutations. These mutations were unlikely to occur by chance alone (P < 3 × 10−7). This illustrative case provides the first demonstration that future, tumor-specific mutations can be identified in an asymptomatic individual without clinical or pathologic evidence of cancer by using already established sequencing technologies but targeted sampling methods. This finding provides the basis for new opportunities in early cancer screening, detection, and prevention.
Keywords: endometrial carcinoma
INTRODUCTION
The “liquid biopsy” is becoming an increasingly important tool in precision oncology's goal of transforming cancer care, quickly moving from the research setting into the clinic (Wan et al. 2017). Overwhelmingly, however, the power of this technology has been focused on issues related to advanced disease in already diagnosed patients under active treatment. For example, and following initial treatment, liquid biopsies are used to detect the presence of minimal residual disease, predict disease recurrence, assess the emergence of drug resistance, and provide guidance for tailored salvage treatment (Garcia-Murillas et al. 2015; Pereira et al. 2015; Tie et al. 2016; Mok et al. 2017). By marked contrast, advances in early cancer detection remain both limited and limited in degrees of success (Phallen et al. 2017). One of the key constraints cited is the implicit rarity of finding circulating tumor DNA (ctDNA) molecules and circulating tumor cells (CTCs) when a tumor is small and still in an early stage (Thierry et al. 2010; Bettegowda et al. 2014). Beyond this, identifying ctDNA or CTCs by liquid biopsy provides no definitive information on tumor type or origin.
Although analytic techniques for early detection will require improvements for overcoming these intrinsic barriers to blood sampling, a parallel possibility exists for developing alternative sampling strategies (Wan et al. 2017). We and others have recently explored the use of a uterine lavage to detect endometrial and ovarian cancers (Maritschnegg et al. 2015; Nair et al. 2016). Indeed, sequencing cellular and cell-free DNA (cfDNA) isolated from uterine lavage fluid in a prospective study allowed detection of 100% of all endometrial cancers, including even microscopic stage IA cancers (Nair et al. 2016). The ability to detect microscopic-sized cancers by this targeted sampling approach immediately suggested the potential for early cancer detection and a broader-based approach toward endometrial cancer screening and prevention.
Endometrial cancer is the most common gynecologic malignancy in industrialized countries, and both its incidence, most likely linked to the rise in obesity, and associated mortality are increasing (Morice et al. 2016). The genomic landscape of primary endometrial cancers and their metastases have been detailed (The Cancer Genome Atlas Research Network et al. 2013; Gibson et al. 2016) and morphologically defined premalignant tissue lesions, each with different attributable risks of progression to carcinoma, have been defined (Bandyopadhyay and Ali-Fehmi 2017). Despite these well-recognized genetic and pathological associations, there are no screening tests.
Here, we present proof of principle for early endometrial cancer detection using a targeted liquid biopsy technique. This illustrative case provides the first demonstration that future, tumor-specific mutations can be identified in an asymptomatic individual without clinical or pathologic evidence of cancer. In this individual, mutations were identified nearly 1 yr before symptoms developed, and a single, microscopic focus of cancer was eventually diagnosed.
RESULTS
We report the case of an asymptomatic 67-yr-old female with a BMI of >34 who had initially been referred to gynecologic oncology for follow-up evaluation of an abnormal endometrial stripe detected as an incidental finding during a pelvic sonography for kidney stone evaluation. Family history was negative for any evidence of breast, pancreatic, ovarian, or endometrial cancers. On examination, the patient was found to have a stenotic cervix that prevented an in-office endometrial biopsy. She therefore underwent hysteroscopy and dilation and curettage in the operating room under anesthesia. The patient was enrolled in our study to sequence cellular DNA and cfDNA isolated from uterine lavage fluid collected during hysteroscopy as a potential cancer diagnostic (Nair et al. 2016). Ten milliliters of saline fluid hysteroscopy lavage was collected for molecular analysis and DNA was sequenced at ultra-high coverage (>7000×) using a 12-gene, custom endometrial cancer typing panel, as previously described (Nair et al. 2016).
Intraoperative findings revealed no gross evidence of malignancy in the uterine cavity that was scraped aggressively. Histopathologic analysis revealed benign endometrium with no evidence of hyperplasia or cancer. In contrast, molecular analysis of the lavage cell pellet identified a total of five driver mutations in four genes. The gene mutations and their allele frequencies (AFs) and sequencing coverage were PTEN R130G (1.3%/107 reads) and S338fs (1.1%/349 reads); PIK3CA N345I (4.9%/379 reads); FBXW7 R505C (5.0%/467 reads); and FGFR2 S252W (5.1%/355 reads).
Comparison of these mutations against The Cancer Genome Atlas (TCGA) endometrial cancer data set, using the cBioPortal tool (Gao et al. 2013), revealed each mutation to be previously associated with this cancer. Strikingly, the PTEN R130 site is mutated in nearly 48% of all tumors. In order, the comparative frequencies of the other uterine lavage-detected mutations in the TCGA data set were FGFR2 S252W, 7.4%; FBXW7 R505C, 5.0%; PIK3CA-N345I, 4.1%, and PTEN S338fs, 0.8%.
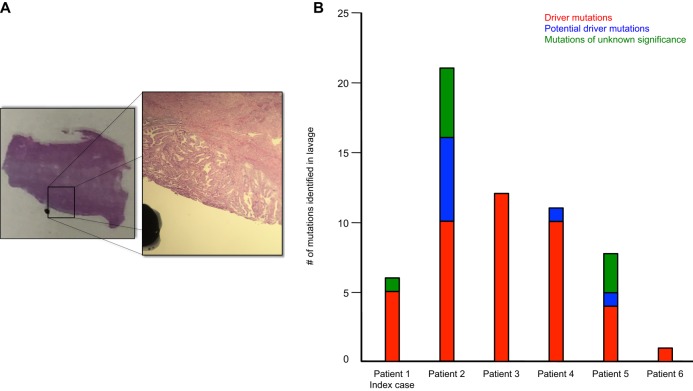
Ten months later, the patient returned with a chief complaint of postmenopausal bleeding. In-office biopsy revealed complex hyperplasia with atypia, a known preneoplastic tissue lesion, suspicious for low-grade endometrioid adenocarcinoma. The patient was brought to the OR for total laparoscopic hysterectomy, bilateral salpingo-oophorectomy, bilateral pelvic sentinel lymph node mapping, and resection. Pathology revealed a single microscopic focus of endometrioid endometrial adenocarcinoma without any evidence of myometrial invasion (Fig. 1A).
Figure 1.
(A) A section of the H&E-stained biopsy containing normal tissue and tumor is shown with a superimposed and magnified image highlighting the tumor from which laser-capture microdissection (LCM) was performed. Diagnosis revealed the single focal grade 1 endometrioid endometrial adenocarcinoma, stage IA, without evidence of myometrial invasion. (B) An overview of the mutational profile of the six patients, including the index case, possessing PTEN 130 mutations. The sum of all mutations—driver, potential driver, and unknown significance—is shown for each patient.
Laser-capture microdissection (LCM) was used to isolate tumor tissue for molecular analysis. DNA was extracted and sequenced. The two oncogenic somatic PTEN mutations, R130G and Ser338fs, first identified 10 mo earlier in the uterine lavage cell pellet, were present in the tumor at AFs now >10%. We then calculated the probability that the shared PTEN mutations could occur by chance alone. To estimate this likelihood, we made the assumption that all positions interrogated by our 12-gene sequencing panel could be mutated with equal probability. Under this assumption, it is highly unlikely that the two identical PTEN mutations identified in two samples—preclinical lavage and LCM—would occur by chance alone (P < 3 × 10−7).
DISCUSSION
Using an integrated liquid biopsy–based and cancer-targeted sampling approach, we have established a benchmark for earliest endometrial cancer detection. Previous studies by others had demonstrated that occult cancers can be detected through blood-based sampling. These studies included incidental detection of various cancers in women following unexplained chromosomal aneuploidies identified during noninvasive prenatal testing (Amant et al. 2015; Bianchi et al. 2015; Dharajiya et al. 2018) and nasopharyngeal cancer through the targeted tracking of Epstein–Barr virus (EBV) DNA in an endemic but asymptomatic population (Chan et al. 2017). Meanwhile, nearly 25 years ago, a retrospective study of cytology samples from lung cancer patients revealed that cancer cells can be present in the sputum prior to detection by chest X-ray diagnosis (Mao et al. 1994). Taken together, we believe all these results establish both proof of principle for cancer screening by liquid biopsy and the paradigm for collection and analysis of relevantly targeted samples beyond blood.
For more than 70 years, it has been appreciated that endometrial and ovarian cancers and their precursors exfoliate cells into the uterine cavity (Traut and Papanicolaou 1943), and for almost as long, attempts have focused on obtaining these cancerous/precancerous cells for diagnostic purposes (McGowan et al. 1974). Nonetheless, screening for these cancers is an unmet clinical need. In our studies, we coupled the advances in next-generation sequencing (NGS) technologies and bioinformatics to an almost abandoned technique first introduced 60 years ago, concurrent with the Papanicolaou (Pap) test for detecting cervical cancer, the uterine lavage (Morton et al. 1957). In the future, the uterine lavage and molecular analysis could be performed in-office. It would avoid the time and costs associated with uterine sonography and the costs, pain/discomfort, risks, time, and need for anesthesia associated with an in–operating room hysteroscopy. Beyond this, a simpler, more sensitive, and cost-effective in office procedure for screening for these cancers in all women including those at increased risk because of age, BMI, or inherited predisposition could have major effects on mortality and morbidity and could also remove barriers that have kept minorities and women of lower socioeconomic status from receiving equal care and the opportunity for improved outcomes (Collins et al. 2014).
Although the potential paradigm shift for early cancer detection is obvious, the clinical dilemma introduced on molecular diagnosis of a precancer is also illustrated by this case. The driver mutations identified in the patient's uterine lavage were known to be bona fide signatures of endometrial cancer as revealed by comparison with TCGA data set. The presence of these mutations in cells isolated from the lavage would be expected to impart not only a possible proliferative advantage but also some degree of cancer risk notwithstanding the fact that the patient was asymptomatic and without evidence of premalignant changes on biopsy. However, no estimate of the likelihood or timing of future cancer onset could be made. Specifically, two independent studies have now demonstrated a high prevalence of cancer driver mutations in endometrial cells from women in the absence of endometrial cancer or morphologically recognized premalignant lesions (Nair et al. 2016; Anglesio et al. 2017). PTEN mutations have long been recognized as being relatively common findings in histologically normal premenopausal endometrium isolated during the proliferative phase of the menstrual cycle (e.g., see Mutter et al. 2001, 2014). In addition, an ever-increasing list of studies have demonstrated an accumulation of age-related, genetic driver mutations in blood and tissues in individuals with benign conditions and no evidence of cancer (Kato et al. 2016; Wan et al. 2017). Taken together, these findings suggest that what we have defined as cancer “driver mutations” may in actuality be necessary but not sufficient for cancer development (Brock and Huang 2017).
Given that the two PTEN mutations identified in the patient's lavage were tumor-specific (i.e., present in the original tumor), we therefore interrogated our original study of 102 women to identify other women who also possessed these specific PTEN mutations in their uterine lavage (Nair et al. 2016). A total of five other cases with PTEN mutations at the R130 site were identified (patients 2–6; see Tables 1 and 2). These five patients, in order, were previously noted as PT468, PT395, PT476, PT459, and PT513). A histogram of the number and classifications of all mutations, including the PTEN R130 mutation, identified in the lavages from all the patients is shown in Figure 1B.
Table 1.
Patients detected with mutations at the PTEN R130 site
| Patient | Age | BMI | Chief complaint at presentation | Final pathology | Gene—PTEN | PTEN detection status in lavage | Follow-up period | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome: position | CDS change | AA change | dbSNP | Cell pellet (AF) | cfDNA (AF) | ||||||
| 1 | 67 | 34 | Pelvic pain; abnormal uterine sonogram detected as an incidental finding during workup for renal stone | Initial presentation: benign endometrium with no evidence of hyperplasia or cancer | 10:87933147-87933147 | c.388C>G | R130G | 121909224 | +(1.3%) | - | 14 mo |
| On return: single microscopic focus of FIGO grade 1, stage IA endometrioid endometrial adenocarcinoma | |||||||||||
| 2 | 67 | 39 | Postmenopausal bleeding; thickened endometrium detected on sonogram | FIGO grade 3, stage IA endometrioid endometrial adenocarcinoma; mixed type histology | 10:87933148-87933148 | c.389G>A | R130Q | 121909229 | +(21.9%) | +(38.8%) | NA |
| 3 | 51 | 22 | Postmenopausal bleeding | Proliferative endometrium with polypoid fragments and glandular/stromal breakdown | 10:87933147-87933147 | c.388C>G | R130G | 121909224 | +(2.4%) | - | 30 mo |
| 4 | 37 | 34 | Dysmenorrhea; endometriosis | Proliferative endometrium; bilateral benign endometriotic cysts; pelvic peritoneum and uterosacral ligament biopsies showing endometriosis | 10:87933148-87933148 | c.389G>A | R130Q | 121909229 | +(1.3%) | - | 26 mo |
| 5 | 47 | 23 | Postmenopausal bleeding; endometriosis | Benign endometrium, superficial adenomyosis, early secretory endometrium | 10:87933147-87933147 | c.388C>G | R130G | 121909224 | +(1.6%) | - | 26 mo |
| 6 | 41 | 19 | Abnormal uterine bleeding; polyp on sonogram | Polyp with proliferative glands and focal simple hyperplasia without atypia, proliferative endometrium | 10:87933147-87933147 | c.388C>T | R130* | 121909224 | - | +(1.7%) | 26 mo |
BMI, body mass index; dbSNP, Single Nucleotide Polymorphism database; AF, allele frequency; NA, not available; FIGO, International Federation of Gynecology and Obstetrics.
Table 2.
PTEN R130 site variant information
| Gene | Chr:pos | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP | Genotype | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| PTEN | 10:87933147-87933147 | c.388C>G | R130G | Missense | Pathogenic | 121909224 | Heterozygous | 375958 |
| 10:87933148-87933148 | c.389G>A | R130Q | Missense | Pathogenic | 121909229 | Heterozygous | 7829 | |
| 10:87933147-87933147 | c.388C>T | R130* | Nonsense | Pathogenic | 121909224 | Heterozygous | 7819 |
HGVS, Human Genome Variation Society; dbSNP, Single Nucleotide Polymorphism database.
The eldest of the cases (patient 2) was a 67-yr-old female with a BMI > 38 who was originally evaluated for postmenopausal bleeding and thickened endometrium with endometrial nodules detected on sonography. A stage IA mixed histology cancer, one component being high-grade serous adenocarcinoma and the other being grade 3 endometrioid adenocarcinoma, was identified at the time of hysteroscopy and surgery. Ten driver gene mutations in five genes (ARID1A, RB1, PIK3R1, PIK3CA, and PTEN) were detected in her lavage fluid, including a PTEN R130Q mutation in both the cell pellet (AF 22%) and cfDNA (AF 39%) (Nair et al. 2016). Tumor DNA was isolated, and the PTEN R130Q mutation was orthogonally confirmed by Sanger sequencing (AF 32%).
The four other cases (patients 3–6) were all younger than the two with endometrial cancer by >20 yr (age range: 37–51) and as a group had a lower average BMI (average: 25; range: 19–34). Three of the women were originally evaluated for abnormal uterine bleeding and the fourth for dysmenorrhea. Two of the women also had endometriosis. PTEN uterine lavage mutation status is shown in Tables 1 and 2. At the time of their surgical exploration, none of these women had morphologic evidence of precancer lesions or cancer. Unlike patients 1 and 2 who developed cancer, none of these four women have demonstrated evidence of cancer symptoms during the >2 yr follow-up since their original hysteroscopy procedure and lavage.
At the very outset of the patient care spectrum, precision oncology offers the holy grail—early cancer detection with the possibility of prevention. Our case report highlights the fact that although the opportunity exists, there remains an unexpected but surmountable bottleneck in immediately translating these findings to patients. As recently highlighted by a blue ribbon panel convened to guide the NCI's future U.S. cancer research priorities, “[t]he rate-limiting step in developing and implementing precision-based prevention approaches has been our limited understanding of precancer biology, which stands in contrast with the extensive knowledge of advanced disease” (Jaffee et al. 2017). We have no guidance in understanding the order in which and at what point, if ever, somatic mutations identified prior to the development of morphologically defined preneoplastic lesions result in cancer.
This knowledge gap has engendered a call for a concerted effort to create a Pre-Cancer Genome Atlas (PCGA) (Jaffee et al. 2017; Spira et al. 2017). Moving beyond the previously successful goal of TCGA in cataloging the genomic landscape of advanced disease, the future goal would be to integrate the multiomics landscape and order the sequence of events and immunologic features that result in cancer. By way of a marathon analogy, interviewing only the elite runners as they cross the finish line (TCGA) reveals nothing about the larger total field of runners, those who dropped out and why, and whether we can reasonably predict if the remaining runners will complete or abandon the course (PCGA).
In conclusion, the sequencing technology and targeted sample collection methods are now available to facilitate detection of microscopic-sized tumors and even cells containing tumor-specific mutations before morphologically recognized tissue changes are diagnosed. In general, early detection would have marked benefits on patient survival, quality of life, and health-care costs. We now need to establish the road map detailing the steps leading from precancer to malignancy, and the converse, preventing precancer from developing into malignancy, so that we can make the giant leap from laboratory to clinically relevant prevention and detection of earliest cancers.
METHODS
Patient Enrollment
All samples used in this study were collected in accordance with the Institutional Review Board of the Icahn School of Medicine at Mount Sinai (GCO# 10–1166) at the time of the original diagnostic procedure as we have previously described, and all patients provided written consent (Nair et al. 2016). All experimental protocols were approved by the Icahn School of Medicine at Mount Sinai in accordance with our research protocol (GCO# 10–1166). All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from each patient.
Uterine Lavage and DNA Isolation
Uterine lavage specimens were collected in the operating room at the time of a patient's hysteroscopy, as we have previously described (Nair et al. 2016). Immediately upon entering the uterine cavity with the hysteroscope, the initial 20–30 ml of saline fluid, used to dilate the uterus, was then collected using a 40-ml specimen trap device (Medline Mucus Specimen Trap 40cc, No. DYND44140, Venture Respiratory Inc) attached to suction. Following the lavage collection, the patient underwent the remainder of their procedure as per their surgeon's discretion while the lavage samples in the specimen trap device were placed on ice and taken to the laboratory and processed within 1 h of collection.
In the laboratory, the lavage specimens were transferred to 50-ml centrifuge tubes and centrifuged. The acellular supernatant was separated from the cell pellet using a pipette and recentrifuged to remove any remaining cellular material and debris. This fraction was then collected and stored at −80°C until final DNA extraction. The cell pellet was washed with red blood cell lysis solution (5 Prime, No. 2301310), resuspending by gentle pipetting, incubating at room temperature, and then centrifuging in a tabletop centrifuge. The RBC lysis supernatant was then discarded, leaving behind the cell pellet. This was repeated until the cell pellet was cleared of visible red cell contamination. The cell pellet was stored at −80°C until DNA isolation was performed.
cfDNA was first concentrated from the acellular portion using a centrifugal filter (Amicon Ultra-15 30 kiloDalton Filter Units, EMD Millipore, No. UFC903096) into smaller volumes. The concentrated cfDNA was then extracted (Circulating Nucleic Acid Kit, QIAGEN). Cellular DNA was extracted from the cell pellets (ArchivePure DNA Kit, 5 Prime) using a modified protocol to account for low cell density. Germline DNA was isolated from peripheral blood samples collected from each patient at the time of their hysteroscopy (ArchivePure DNA Kit, 5 Prime). DNA concentrations of all fractions were determined by QuBit fluorometry (Thermo-Fischer Scientific).
For LCM, paraffin-embedded tumor samples were cut into two 10 µm thickness sections and subjected to microdissection (PixCell IIe Laser Capture Microdissection System microscope, Arcturus). Genomic DNA was extracted from the isolated caps by following the protocol in the Applied Biosystems PicoPure DNA Extraction Kit.
Library Preparation and Next-Generation Sequencing
As we have previously described these methods in great detail (Nair et al. 2016), we describe them briefly herein. For all the patients described in the study, a set of sample trios from germline PBMC DNA and lavage cellular and acellular were sequenced to an average of 5000× coverage using a targeted amplicon panel (Swift Biosciences). Samples were either sequenced using the Accel-Amplicon 56G Oncology Panel (Swift Biosciences) or a specifically designed smaller custom endometrial tumor amplicon panel. The 12 genes in this smaller panel were PTEN, PIK3CA, TP53, CTNNB1, KRAS, FGFR2, FBXW7, RB1, ATM, APC, ARID1A, and PIK3R1. The genomic target regions were designed to cover both hotspot loci and contiguous full-coding exons, including the full exonic coverage of TP53 (see Supplemental Table 1 in Nair et al. 2016 for a complete listing of all genomic loci covered).
Resulting targeted NGS libraries were sequenced on an Illumina MiSeq using v2 chemistry. Somatic variant calling was performed using MuTect, VarScan, and LoFreq after following GATK Best Practices. A target of 5000× coverage and 10-ng inputs enabled the lower limit of detection to be set to the 1% fraction.
ADDITIONAL INFORMATION
Data Deposition and Access
The variants described in this paper have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000803739 and SCV000808062.
Ethics Statement
This study was approved by the Icahn School of Medicine at Mount Sinai's Program for the Protection of Human Subjects and for WCHN through the BRANY Institutional Review Board Services. Written informed consent was obtained from all patients prior to sample collection.
Acknowledgments
The authors thank the patients for their participation in the study and members of the Department of Obstetrics, Gynecology, and Reproductive Sciences, Icahn School of Medicine at Mount Sinai, New York, for their assistance in sample collection and annotation.
Funding
This study was supported in part by funding from the Gordon family, the Ruttenberg family, the Goldstone family, and the Varadi Ovarian Initiative in Cancer Education (VOICE). The funders had no role in study design, data collection, or its analysis.
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Amant F, Verheecke M, Wlodarska I, Dehaspe L, Brady P, Brison N, Van Den Bogaert K, Dierickx D, Vandecaveye V, Tousseyn T, et al. 2015. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol 1: 814–819. [DOI] [PubMed] [Google Scholar]
- Anglesio MS, Papadopoulos N, Ayhan A, Nazeran TM, Noë M, Horlings HM, Lum A, Jones S, Senz J, Seckin T, et al. 2017. Cancer-associated mutations in endometriosis without cancer. N Engl J Med 376: 1835–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Ali-Fehmi R. 2017. Endometrial carcinoma: precursor lesions and molecular profiles. In Precision molecular pathology of uterine cancer (ed. Deavers MT, Coffey DM), pp. 3–18. Springer International Publishing, Cham. [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, Garber JE, Wilkins-Haug L, Vora NL, Warsof S, et al. 2015. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 314: 162–169. [DOI] [PubMed] [Google Scholar]
- Brock A, Huang S. 2017. Precision oncology: between vaguely right and precisely wrong. Cancer Res 77: 6473–6479. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network; Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. 2013. Integrated genomic characterization of endometrial carcinoma. Nature 497: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, Chu SWI, Mak C, Tse IOL, Leung SYM, et al. 2017. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 377: 513–522. [DOI] [PubMed] [Google Scholar]
- Collins Y, Holcomb K, Chapman-Davis E, Khabele D, Farley JH. 2014. Gynecologic cancer disparities: a report from the Health Disparities Taskforce of the Society of Gynecologic Oncology. Gynecol Oncol 133: 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharajiya NG, Grosu DS, Farkas DH, McCullough RM, Almasri E, Sun Y, Kim SK, Jensen TJ, Saldivar JS, Topol EJ, et al. 2018. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem 64: 329–335. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa I, et al. 2015. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 7: 302ra133. [DOI] [PubMed] [Google Scholar]
- Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, Holst F, Zack TI, Werner HM, Staby KM, et al. 2016. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet 48: 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee EM, Dang CV, Agus DB, Alexander BM, Anderson KC, Ashworth A, Barker AD, Bastani R, Bhatia S, Bluestone JA, et al. 2017. Future cancer research priorities in the USA: a Lancet Oncology Commission. Lancet Oncol 18: e653–e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Lippman SM, Flaherty KT, Kurzrock R. 2016. The conundrum of genetic “drivers” in benign conditions. J Natl Cancer Inst 108: djw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D. 1994. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res 54: 1634–1637. [PubMed] [Google Scholar]
- Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, Vergote I, Heitz F, Sehouli J, Kinde I, Diaz LA Jr, et al. 2015. Lavage of the uterine cavity for molecular detection of Müllerian duct carcinomas: a proof-of-concept study. J Clin Oncol 33: 4293–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan L. 1974. Cytologic methods for the detection of endometrial cancer. Gynecol Oncol 2: 272–278. [DOI] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, Shepherd FA, He Y, Akamatsu H, Theelen WS, et al. 2017. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 376: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. 2016. Endometrial cancer. Lancet 387: 1094–1108. [DOI] [PubMed] [Google Scholar]
- Morton DG, Moore JG, Chang N. 1957. Endometrial lavage as an aid in the diagnosis of carcinoma of the endometrium. West J Surg Obstet Gynecol 65: 113–125. [PubMed] [Google Scholar]
- Mutter GL, Ince TA, Baak JP, Kust GA, Zhou XP, Eng C. 2001. Molecular identification of latent precancers in histologically normal endometrium. Cancer Res 61: 4311–4314. [PubMed] [Google Scholar]
- Mutter GL, Monte NM, Neuberg D, Ferenczy A, Eng C. 2014. Emergence, involution, and progression to carcinoma of mutant clones in normal endometrial tissues. Cancer Res 74: 2796–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, Schumacher CA, Irish JC, Harkins TT, Freeman E, Garcia I, et al. 2016. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med 13: e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, Nair N, Moshier E, Wooten M, Uzilov A, et al. 2015. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One 10: e0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, et al. 2017. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 9: eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira A, Yurgelun MB, Alexandrov L, Rao A, Bejar R, Polyak K, Giannakis M, Shilatifard A, Finn OJ, Dhodapkar M, et al. 2017. Precancer atlas to drive precision prevention trials. Cancer Res 77: 1510–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F. 2010. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 38: 6159–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al. 2016. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8: 346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut HF, Papanicolaou GN. 1943. Cancer of the uterus: the vaginal smear in its diagnosis. Cal West Med 59: 121–122. [PMC free article] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17: 223–238. [DOI] [PubMed] [Google Scholar]