Abstract
Diamond–Blackfan Anemia (DBA) is a rare polygenic disorder defined by congenital hypoplastic anemia with marked decrease or absence of bone marrow erythroid precursors. Identifying the specific genetic etiology is important for counseling and clinical management. A 6-yr-old boy with a clinical diagnosis of DBA has been followed by our pediatric hematology team since birth. His clinical course includes transfusion-dependent hypoplastic anemia and progressive autoimmune cytopenias. Genetic testing failed to identify a causative mutation in any of the classical DBA-associated genes. He and his parents underwent trio whole-exome sequencing (WES) with no genetic etiology identified initially. Clinical persistence and suspicion led to testing for adenosine deaminase 2 (ADA2) activity and whole-genome sequencing (WGS) that identified compound heterozygous pathogenic mutations in the ADA2-encoding CECR1 gene, a recently appreciated etiology for congenital hypoplastic anemia. This case illustrates current challenges in genetic testing and how they can be overcome by multidisciplinary expertise in clinical medicine and genomics.
Keywords: congenital hypoplastic anemia, congenital neutropenia
CASE PRESENTATION
A former full-term, 2-mo-old Caucasian male presented with tachypnea, pallor, an otherwise normal physical exam, and severe anemia (hemoglobin [Hb] 1.5 g/dl, mean corpuscular volume [MCV] 91.8 fl, and reticulocyte level 0.5%). White blood cell and platelet counts were normal. Direct and indirect Coombs tests were negative. Renal function was normal. Anemia persisted after several red blood cell transfusions. Bone marrow biopsy at age 6 mo showed 80%–90% cellularity with marked erythroid hypoplasia, consistent with congenital pure red cell aplasia (Diamond–Blackfan anemia [DBA]), which is most commonly caused by autosomal dominant mutations in one of 17 ribosomal protein (RP) genes (Mirabello et al. 2017). Commercial genetic testing using Sanger sequencing identified a heterozygous RPS17 intronic variant of uncertain significance (VUS) that was not predicted to alter splicing by two prediction tools. No pathogenic mutations or VUSs were identified in RPL5, RPL11, RPL19, RPL26, RPL35a, RPS7, RPS10, RPS19, RPS24, or RPS26.
Corticosteroid therapy, which stimulates erythropoiesis in some RP or GATA1 gene–mutated DBA patients, did not improve the anemia in the current patient, and chronic RBC transfusions were continued. At 2 yr of age, he developed atopic dermatitis, anti-D and anti-E warm auto anti-RBC antibodies, and transfusion-related iron overload treated with subcutaneous deferoxamine. At 5 yr of age, he developed widespread molluscum contagiosum and laboratory signs of autoimmune neutropenia including absolute neutrophil counts (ANCs) fluctuating between 0 and 300/mm3 and bone marrow myeloid hyperplasia with neutrophil precursor arrest at the band stage. After multiple bouts of fever and neutropenia, granulocyte colony stimulating factor (G-CSF) was initiated, with an increase in ANC to 700–2000/mm3. Plasma immunoglobulin levels were in the low-normal range. Relevant laboratory data are summarized in Table 1.
Table 1.
Laboratory data
| Presentation | Age 2 | Age 5 | Age 6 | |
|---|---|---|---|---|
| Hgb (g/dl) | 1.5 | 11.5 (transfused) | 8 (transfusion trough) | 10.1 (transfused) |
| MCV (fl) | 91.8 | 82.8 (transfused) | 82.4 (transfusion trough) | 84.7 (transfused) |
| Reticulocyte percent | 0.50% | 0.21% (transfused) | 0.12% (transfusion trough) | 0.29% (transfused) |
| WBC (per mm3) | 12,900 | 6500 | 2900 | 4600 |
| ANC (per mm3) | 6800 | 1400 | 0 | 700 |
| Platelets (per mm3) | 364,000 | 343,000 | 219,000 | 178,000 |
| IgG (mg/dl) | 665 | 727 | ||
| IgM (mg/dl) | 100 | 42 | ||
| IgA (mg/dl) | 45 | 59 | ||
| RBC antibody screen | Negative | Positive | Positive | Positive |
| Neutrophil antibody | Positive | |||
| Ferritin (ng/mL) | 762 | 2008 | 2315 | 2828 |
| R2* Estimated liver iron content (mg Fe/gram dry weight) | 10.86 | 3.67 | 8.22 | |
| Plasma ADA2 activity | 0.4 mU activity/ml |
TECHNICAL ANALYSIS AND VARIANT INTERPRETATION
The patient and parents were tested by CLIA-approved commercial whole-exome sequencing (WES) with targeted analysis of 429 candidate genes associated with the Human Phenotype Ontology (Robinson et al. 2008) terms “Anemia (HP:0001903),” “Bone marrow hypocellularity (HP:0005528),” “Congenital neutropenia (HP:0005549),” and “Neutropenia (HP:0001875)” at 20× coverage depth or greater. We also performed whole-genome sequencing (WGS) under the research protocol “Investigation of the Genetics of Hematologic Diseases” (INSIGHT-HD, NCT 02720679).
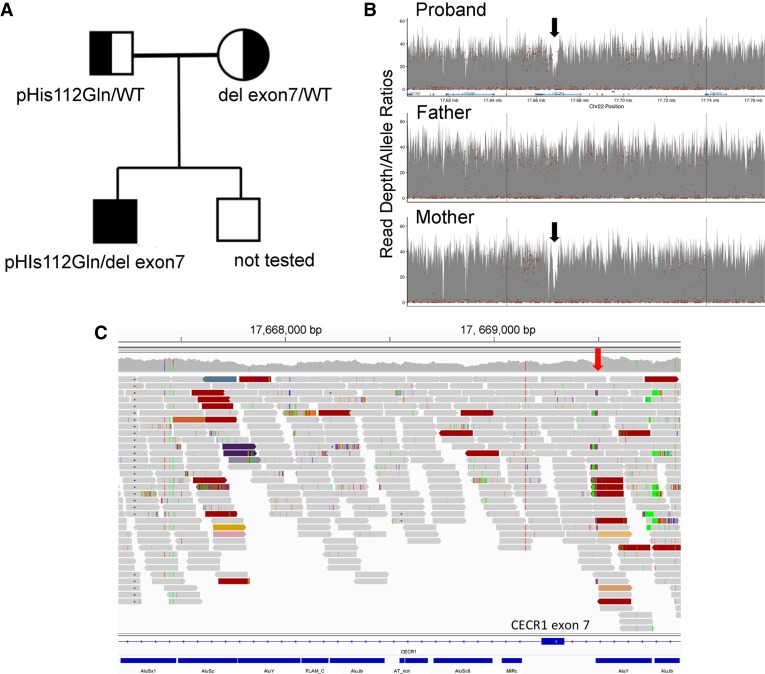
The WES identified a heterozygous paternally inherited mutation in the CECR1 gene (p.His112Gln), which encodes adenosine deaminase 2 (ADA2) (Fig. 1A; Table 2). Mutations in CECR1 lead to reduced levels of ADA2, a secreted extracellular regulator of adenosine signaling and modulator of cell proliferation and differentiation (Hashem et al. 2017a). This variant is present in the gnomAD population database at a frequency of 0.002% (5/245816 alleles) and was reported in a pediatric patient with early-onset polyarteritis nodosum and ADA2 deficiency (Navon Elkan et al. 2014). Because biallelic CECR1 mutations were recently reported to cause hypoplastic anemia (Uettwiller et al. 2016; Hashem et al. 2017a), we investigated the possibility of a second mutation in our patient, initially by investigating plasma ADA2 activity, which was found to be very low at 0.4 mU/ml (normal 13.9 ± 5.3). Review of WGS data confirmed the CECR1 missense mutation p.His112Gln in the patient and father. Visual analysis of read depth and mate–pair mapping information revealed compelling evidence for a CECR1 exon 7 deletion in the patient and mother (Fig. 1B; Table 2). Because the left deletion breakpoint occurred in a region of repetitive DNA (Fig. 1C), which can interfere with analytical algorithms for WGS, it was initially filtered away by a quality-control step designed to reduce false-positive rates. A similar deletion was reported previously in an ADA2-deficient family (Uettwiller et al. 2016).
Figure 1.
Congenital hypoplastic anemia caused by compound heterozygosity for a CECR1 missense mutation and exon 7 deletion. (A) Pedigree. (B) Plot of WGS coverage depth on Chromosome 22p of the proband suggests an ∼2-kb maternally inherited CECR1 deletion (black arrows). (C) Visualization of mapped reads of the proband (via the Integrated Genomics Viewer) in the region of the deletion event. The red arrow denotes the position of the right breakpoint. CECR1 exon 7 and Alu repeat sequences are shown at the bottom in blue. Colored reads indicate pairs with anomalous insert sizes or reads with a mate mapping to a repetitive sequence on another chromosome. Dark red reads indicate a deletion event. The left breakpoint is embedded in repetitive Alu-derived sequences and thus was identified by only two of the four calling algorithms used. Failure to precisely define the breakpoint locations caused this copy-number variant to be filtered away by a quality-control step in our standard analytical pipeline for WGS.
Table 2.
Genomic findings
| Gene | Genomic location | HGVS cDNA | HGVS protein | Zygosity | Parent of origin | Variant interpretation |
|---|---|---|---|---|---|---|
| CECR1 | Chr 22: 17207277 (GRCh38) Chr 22: 17688167 (GRCh37) |
NM_017424 c.336C>G NM_001282227 c.210C>G |
p.His112Gln | Heterozygous | Father | Pathogenic |
| CECR1 | Chr 22: CECR1 deletion, ∼2 kb spanning exon 7; breakpoints unknown |
NM_001282227 c.956-1113 del |
p.Asp319GlyfsTer6 | Heterozygous | Mother | Pathogenic |
Variant interpretation: ADA2 deficiency is an autosomal recessive condition caused by biallelic loss-of-function mutations in the CECR1 gene. The His112Gln variant was reported previously in a patient with ADA2 deficiency (Navon Elkan et al. 2014). The CECR1 exon 7 deletion causes a frameshift mutation with premature translational termination. The proband is a compound heterozygote for the paternal and maternal CECR1 mutant alleles and has minimal circulating ADA2 activity. Hence, both mutations are pathogenic.
Although WES is highly sensitive for capturing nucleotide substitutions, this approach does not capture all deletions. However, in light of the new findings, the commercial laboratory reevaluated the WES data, identified reduced read depth for CECR1 exon 7 in the patient and mother, and confirmed a heterozygous deletion via next-generation sequencing (NGS). Together, persistent investigation and multiple complementary laboratory approaches converged to the diagnosis of hypoplastic “DBA-like” anemia caused by compound heterozygous CECR1 mutations with ADA2 deficiency.
Germline biallelic CECR1 mutations with ADA2 deficiency causing recurrent fever, vasculopathy, and stroke were first described in 2014 (Navon Elkan et al. 2014; Zhou et al. 2014). Subsequent reports identified hematological manifestations including hypoplastic anemia, autoimmune cytopenias, and immunodeficiency in some patients (Uettwiller et al. 2016; Hashem et al. 2017b). The pathophysiology of ADA2 deficiency and reasons for the clinical variability of this syndrome are unknown. Treatment includes supportive care and immunosuppression (Caorsi et al. 2017; Hashem et al. 2017). A recent study treating 14 ADA2-deficient patients with allogeneic hematopoietic stem cell transplantation reported resolution of all disease-associated pathologies with no deaths (Hashem et al. 2017c).
SUMMARY
Approximately 70% of DBA cases are caused by autosomal dominant mutations in multiple RP genes. Congenital hypoplastic anemia also occurs less frequently with mutations in GATA1 (X-linked), TSR2 (X-linked), EPO (autosomal recessive), and CECR1 (autosomal recessive) (Hashem et al. 2017; Uettwiller et al. 2016; Da Costa et al. 2017; Kim et al. 2017). Obtaining a genetic diagnosis is important for counseling, therapeutic decisions, and reassurance to patients, families, and clinicians. The clinical manifestations of ADA2 deficiency are heterogenous and can include autoimmunity, immunodeficiency, vasculitis, stroke, and erythroid hypoplasia (Caorsi et al. 2017; Hashem et al. 2017b). Thus, the genetic diagnosis of biallelic CECR1 mutations with ADA2 deficiency explained our patient's severe eczema and immune cytopenias, which are atypical for RP-haploinsufficient DBA.
This case illustrates several important principles of genetic testing for rare blood disorders. First, mutations in different genes (characterized and uncharacterized) can produce similar phenotypes (i.e., locus heterogeneity). Our patient was initially assigned a clinical diagnosis of DBA, with no mutations detected in classical DBA genes. Subsequent testing identified biallelic mutations in CECR1 (Table 2), which was linked to hypoplastic anemia only recently. Second, genomic sequencing tests have limitations (Priest 2017). In this case, valid technical issues associated with WES and WGS interfered initially with detection of a 2-kb CECR1 deletion. Genome-wide array analysis such as comparative genome hybridization (CGH) or single-nucleotide polymorphism (SNP) array had not been performed in this patient. However, the commercially available array platforms would very likely miss the small intragenic one-exon deletion because of the low resolution that is inherent to such methods. Multiplex ligation–dependent probe amplification (MLPA), considered to be the gold standard for identification of small intragenic deletions, was not available for the CECR1 gene. Third, diagnostic WES or WGS frequently detect multiple VUSs in the same individual; pinpointing relevant disease alleles requires clinical expertise and consideration of data supporting variant pathogenicity (Richards et al. 2015). Finally, our understanding of inherited blood disorders and technologies to detect the underlying gene mutations is progressing rapidly. WGS is the most comprehensive approach, but it is more expensive and technically challenging than WES (Meienberg et al. 2016). Both methods typically interrogate a candidate gene panel, which may not include newly identified disease-associated genes. Thus it is important to periodically revisit phenotype-based genetic testing options, which are seldom all-inclusive and likely to evolve in parallel with new scientific discoveries. Five years after initial presentation, reevaluation of the progressive phenotype of our patient, combined with advances in genomics and gene discovery, provided a definitive genetic diagnosis.
Overall, this case illustrates the utility of genetic testing for rare blood disorders and the requirements for multidisciplinary expertise and persistence to optimize success in difficult cases. More specifically, ADA2 deficiency caused by biallelic CECR1 mutations should be considered for all cases of congenital hypoplastic anemia without RP gene haploinsufficiency.
ADDITIONAL INFORMATION
Data Deposition and Access
The genomic variants were submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000854421 (missense) and SCV000854422 (deletion). The raw sequencing data has been submitted to dbGaP (https://www.ncbi.nlm.nih.gov/gap), accession number pending.
Ethics Statement
We obtained parental written informed consent for participation in the St. Jude Children's Research Hospital Institutional Review Board (IRB) approved INSIGHT-HD research protocol.
Acknowledgments
The authors would like to thank all staff at St. Jude Children's Research Hospital for their support throughout the study, but especially Nidal Boulos, Teresa Carr, Neha Das Gupta, Nathan Gray, Melvanique Hale, Guolian Kang, Chimene Kesserwan, Amber Rose King, Janet Middlekauff, Jessica Uhrich, Bettina Utzelmann, and Kristine Woodward for their efforts to make INSIGHT-HD an ongoing success. We also acknowledge Evelyn Harris for her assistance in overseeing the logistics of ongoing teleconferences. Finally, we especially recognize the eager participation and time commitments made by the children and families who have enrolled in the INSIGHT-HD study.
Author Contributions
J.H.E. designed and wrote INSIGHT-HD and serves as the overall protocol principal investigator. D.C., M.J.W., and J.H.E. designed the research and wrote the first version of the manuscript, provided ongoing critical reviews, and edited the final manuscript. K.M.B., G.M.C., and M.W. analyzed the genomic data and wrote methods/results sections. M.B., S.L., and J.C. identified the proband and family, provided guidance on clinical course of the patient, and reviewed and edited the manuscript. M.H.’s laboratory performed ADA2 analyses and reviewed and edited the manuscript. All authors approved the final manuscript.
Funding
This project was sponsored by American Lebanese Syrian Associated Charities (ALSAC).
Competing Interest Statement
J.H.E. receives funding support from Pfizer and Eli Lilly and Co. and serves as a consultant for Daiichi Sankyo and Global Blood Therapeutics on projects not related to the one described here. The authors have declared no competing interest.
Referees
Neal Sondheimer
Anonymous
References
- Caorsi R, Penco F, Grossi A, Insalaco A, Omenetti A, Alessio M, Conti G, Marchetti F, Picco P, Tommasini A, et al. 2017. ADA2 deficiency (DADA2) as an unrecognised cause of early onset polyarteritis nodosa and stroke: a multicentre national study. Ann Rheum Dis 76: 1648–1656. [DOI] [PubMed] [Google Scholar]
- Da Costa L, O'Donohue MF, van Dooijeweert B, Albrecht K, Unal S, Ramenghi U, Leblanc T, Dianzani I, Tamary H, Bartels M, et al. 2017. Molecular approaches to diagnose Diamond–Blackfan anemia: The EuroDBA experience. Eur J Med Genet doi: 10.1016/j.ejmg.2017.10.017 [DOI] [PubMed] [Google Scholar]
- Hashem H, Egler R, Dalal J. 2017a. Refractory pure red cell aplasia manifesting as deficiency of adenosine deaminase 2. J Pediatr Hematol Oncol 39: e293–e296. [DOI] [PubMed] [Google Scholar]
- Hashem H, Kelly SJ, Ganson NJ, Hershfield MS. 2017b. Deficiency of adenosine deaminase 2 (DADA2), an inherited cause of polyarteritis nodosa and a mimic of other systemic rheumatologic disorders. Curr Rheumatol Rep 19: 70. [DOI] [PubMed] [Google Scholar]
- Hashem H, Kumar AR, Müller I, Babor F, Bredius R, Dalal J, Hsu AP, Holland SM, Hickstein DD, Jolles S, et al. 2017c. Hematopoietic stem cell transplantation rescues the hematological, immunological, and vascular phenotype in DADA2. Blood 130: 2682–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AR, Ulirsch JC, Wilmes S, Unal E, Moraga I, Karakukcu M, Yuan D, Kazerounian S, Abdulhay NJ, King DS, et al. 2017. Functional selectivity in cytokine signaling revealed through a pathogenic EPO mutation. Cell 168: 1053–1064 e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meienberg J, Bruggmann R, Oexle K, Matyas G. 2016. Clinical sequencing: is WGS the better WES? Hum Genet 135: 359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L, Khincha PP, Ellis SR, Giri N, Brodie S, Chandrasekharappa SC, Donovan FX, Zhou W, Hicks BD, Boland JF, et al. 2017. Novel and known ribosomal causes of Diamond–Blackfan anaemia identified through comprehensive genomic characterisation. J Med Genet 54: 417–425. [DOI] [PubMed] [Google Scholar]
- Navon Elkan P, Pierce SB, Segel R, Walsh T, Barash J, Padeh S, Zlotogorski A, Berkun Y, Press JJ, Mukamel M, et al. 2014. Mutant adenosine deaminase 2 in a polyarteritis nodosa vasculopathy. N Engl J Med 370: 921–931. [DOI] [PubMed] [Google Scholar]
- Priest JR. 2017. A primer to clinical genome sequencing. Curr Opin Pediatr 29: 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. 2008. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 83: 610–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uettwiller F, Sarrabay G, Rodero MP, Rice GI, Lagrue E, Marot Y, Deiva K, Touitou I, Crow YJ, Quartier P. 2016. ADA2 deficiency: case report of a new phenotype and novel mutation in two sisters. RMD Open 2: e000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yang D, Ombrello AK, Zavialov AV, Toro C, Zavialov AV, Stone DL, Chae JJ, Rosenzweig SD, Bishop K, et al. 2014. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med 370: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]