Abstract
X-linked agammaglobulinemia (XLA, OMIM#300300) is a rare monogenic primary immunodeficiency caused by mutations in the Bruton tyrosine kinase (BTK) gene. XLA is characterized by insufficient immunoglobulin levels and susceptibility to life-threatening bacterial infections. We report on a patient that presented with ecthyma gangrenosum and septicemia. Rapid trio whole-genome sequencing (rWGS) revealed an apparently de novo hemizygous pathogenic variant (c.726dupT; p.Ile243TyrfsTer15) in the BTK gene. Metagenomic analysis of rWGS sequences that did not align to the human genome revealed 770 aligned to the Pseudomonas aeruginosa PAO1 genome. The patient was diagnosed with XLA and pseudomonal sepsis.
Keywords: congenital neutropenia, immune dysregulation, sepsis
CASE PRESENTATION
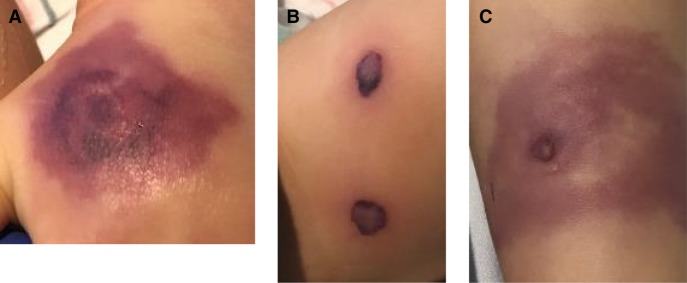
A 19-mo-old twin Hispanic male was admitted to the hospital with fever (38.5°C) and compensated septic shock. Physical examination revealed approximately one dozen deep, indurated purpuric plaques with necrotic changes on the extremities and buttocks (Fig. 1). The patient's twin brother had died of septic shock, purpura fulminans, and disseminated intravascular coagulation in an outside hospital 1 d prior, after a 2-d hospitalization for similar symptoms. The twin brothers were born at 36-wk gestation and had not previously been hospitalized, but the mother reported that for the past 8 mo they had been frequently ill with respiratory tract infections requiring multiple courses of antibiotics. At the time of admission, the patient's weight was 12 kg, which fell into the 70th percentile for age-matched male controls. There was no family history of immunodeficiency.
Figure 1.
Skin lesions in the affected patient showing (A) left hand, (B) left thigh, and (C) right lower leg pseudomonal ecthyma gangrenosum.
Laboratory evaluation was significant for a white blood cell count of 2800 L−1 (reference 6000-14000) with 50% monocytes (2%–12%), 32% lymphocytes (30%–60%), 10% atypical lymphocytes (0%–10%), 2% segmented neutrophils (45%–70%), and 7% band neutrophils (0%–10%), C-reactive protein (CRP) level of 30.10 mg/dl (0.00–0.99), an erythrocyte sedimentation rate of 5 mm/h (0–10), procalcitonin level of 49.04 ng/ml (<2.0), prothrombin time (PT) of 34.7 sec (11.4–14), and an international normalized ratio (INR) of 3.4 (0.9–1.2). Immunoglobulin levels were IgG < 135 mg/dl (413–1112), IgA = 32 mg/dl (21–117), and IgM = 19 mg/dl (30–146).
The patient was started on vancomycin, ceftriaxone, doxycycline, and gentamicin, as the identity of the causative organism for this presumed community-acquired, life-threatening infection was not initially known. Cultures from the patient's blood were sterile. Filgrastim (G-CSF) was also administered subcutaneously at a dose of 5 mcg/kg daily for an absolute neutrophil count of 252 µl in the setting of overwhelming infection. Within the first 24 h of admission, the patient required intubation for respiratory failure as well as vasopressor therapy with multiple medications for fluid-refractory hypotension and poor perfusion. Because of concerns about hypercoagulability, intravenous immunoglobulin (IVIG) was initially held, but as the patient's condition worsened, a dose of 0.4 mg/kg of IVIG was given, which increased the patient's IgG level to 646 mg/dl.
A tissue culture obtained from a 5-mm punch biopsy of a cutaneous lesion grew Pseudomonas aeruginosa on the third hospital day. At that point, meropenem was added to the regimen and other antibiotics were eventually stopped. The diagnosis of pseudomonal ecthyma gangrenosum in this patient and the presumptive identical diagnosis in his deceased twin brother were highly suspicious for a primary immune deficiency, but the combination of neutropenia and low IgG and IgM levels did not conclusively point to one diagnosis. A lymphocyte enumeration panel was sent and resulted a CD19 absolute count of <20 cells/µl (830–1880) the following day, consistent with absent B cells. Given the patient's critically ill state and the recent death of his twin, rapid trio whole-genome sequencing (rWGS) was undertaken simultaneously on the patient and his parents. In just more than 72 h, on the fourth hospital day, an apparently de novo hemizygous pathogenic variant (c.726dupT; p.Ile243TyrfsTer15; Table 1) in the BTK gene was detected and the patient was diagnosed with X-linked agammaglobulinemia (XLA, OMIM#300300). As a result, filgrastim injections were discontinued and IVIG was administered to maintain IgG levels of >800 mg/dl. On the same day (hospital day 4), verbal communication from the hospital where the patient's brother expired confirmed a positive blood culture for pseudomonas in the deceased twin. The patient's clinical status and level of cardiorespiratory support slowly improved on anti-pseudomonal antibiotics and IVIG therapy. He required regular debridement of the gangrenous lesions by plastic surgery and eventually underwent skin grafting. He was discharged from the hospital after a total of 5 mo.
Table 1.
Genomic findings
| Gene | Genomic location | HGVS cDNA | HGVS protein | Zygosity | Parent of origin | Variant interpretation |
|---|---|---|---|---|---|---|
| BTK | NC_000023.10: Chr X:100615605 (GRCh37/hg19) | NM_000061.2: c.726dupT |
NP_000052.1 p.Ile243TyrfsTer15 |
Hemizygous | De novo | Pathogenic |
TECHNICAL ANALYSIS AND METHODS
The patient was enrolled in an IRB-approved research study. Blood was drawn immediately following consent for trio rWGS. The WGS testing was performed in a CAP/CLIA environment. DNA was subsequently extracted and sequenced on a HiSeq 4000 (Illumina). Rapid alignment and nucleotide variant calling was performed using the Dragen (Edico Genome) hardware and software (Miller et al. 2015). Proband and parental samples were sequenced to a mean coverage of 40×. Variants were annotated and analyzed in Opal Clinical (Fabric Genomics) (Coonrod et al. 2013). Initially, variants were filtered to retain those with allele frequencies of <1% in the Exome Variant Server, 1000 Genomes Samples, and Exome Aggregation Consortium database (http://evs.gs.washington.edu/EVS/2016; Karczewski et al. 2017). A gene panel was built in Phenolyzer (Yang et al. 2015) using Human Phenotype Ontology (HPO) (Köhler et al. 2016). This panel included 1098 genes related to the following HPO terms: immunodeficiency (HP:0002721), neutropenia (HP:0001875), and sepsis (HP:0100806). Variants were further filtered to retain those mapping to these 1098 genes yielding 1068 proband calls (465 homozygous variants, 509 heterozygous inherited variants, and 15 heterozygous de novo variants). Manual curation revealed one variant as pathogenic by ACMG guidelines (Richards et al. 2015). Specifically, the criteria invoked to score this variant as pathogenic included PVS1 (null variant in a gene where loss of function is a known mechanism of disease), PS2 (de novo variant), and PM2 (variant is absent from controls). This variant was confirmed by Sanger sequencing. The diagnosis was made by rWGS in 4 d.
VARIANT INTERPRETATION
This patient was found to be hemizygous for an apparently de novo known pathogenic frameshift variant (c.726dupT; p.Ile243TyrfsTer15) in the Bruton tyrosine kinase (BTK) gene. Pathogenic variants in BTK have been implicated in XLA (OMIM#300300).
The de novo p.Ile243TyrfsTer15 pathogenic variant has been previously reported in a single individual with XLA (Holinski-Feder et al. 1998). p.Ile243TyrfsTer15 is absent from the ExAC and gnomAD population databases. The deletion causes a frameshift starting with codon Isoleucine 243, changes this amino acid to a tyrosine residue, and creates a premature stop codon at position 15 of the new reading frame. This pathogenic variant is predicted to cause loss of normal protein function through protein truncation or nonsense-mediated decay of the mRNA transcript.
METAGENOMICS
Per institutional protocol, samples were processed via a standard clinically validated workflow, in addition to passing a battery of quality control (QC) scripts (Farnaes et al. 2018). Using the reference human genome hg37dh, the QC metric of percent aligned reads passed at 98.8% and did not have a statistically significant deviation from median of past samples (N = 100). Given the pseudomonas sepsis phenotype, a metagenomics exploration was performed: After isolating the unaligned portion of reads, the most frequent, randomly sampled 500 reads were checked against the NCBI NR database, and significant similarity to P. aeruginosa genome was noted, whereas similarity to other pathogenic or nonpathogenic genomes was not found. The random sampling was drawn from the 500 most abundant reads (including duplicates). Then, all unaligned reads were aligned to reference P. aeruginosa PAO1 genome and 770 paired reads (654 unique; duplication rate 14%) were aligned. The qualities of the PAO1-aligned reads were not significantly different from those that aligned to the reference human genome (mean Q in read 1 = 36.28 and in read 2 = 36.04, aligned to PAO1; mean Q in read 1 = 36.09 and in read 2 = 35.66, aligned to hg19). For negative controls, unaligned portions of the patient's parental genomic DNA as well as genomic DNA from five random, unrelated persons were aligned to the reference PAO1 genome and 0–3 paired reads aligned in each (establishing the level of random alignments). Furthermore, the pattern of spatial distribution of the 770 reads from the proband's DNA sample to PAO1 genome was nearly uniform over the full length of the PAO1 genome (6.2 Mb, 25 ± 10 reads in each 200 kb window, i.e., ∼1 read/8 kb), resulting in the conclusion of the bona fide low-titer presence of P. aeruginosa in the proband's DNA sample (Fig. 2).
Figure 2.

Uniformly random spatial distribution of 770 reads from the proband's DNA sample (in which the y-axis represents the number of reads) aligned to the PAO1 genome (in which the x-axis is position Mb of the PAO1 genome; total size = 6.2 Mb).
DISCUSSION
Inherited defects of the immune system have an incidence of nearly 1 in 2000 children (Bonilla et al. 2015). More than 300 primary immunodeficiencies (PIDs) are currently known, with the list rapidly expanding (Bonilla et al. 2015; Picard et al. 2015). Between 2013 and 2015 alone, 34 new gene defects for PID were identified (Picard et al. 2015). The severity of infection in affected patients encompasses a spectrum ranging from recurrent respiratory illnesses to life-threatening systemic disease. Current consensus guidelines advocate for a stepwise approach to identifying a suspected PID (Bonilla et al. 2015). However, identifying specific immunodeficiencies in this manner can take weeks to months and delay in diagnosis is significantly associated with increased mortality in these patients (Joshi et al. 2009). More recently, whole-exome sequencing has started to be explored as a high-throughput approach to diagnosing patients with suspected PIDs, with a diagnostic rate of up to 40% (Stray-Pedersen et al. 2017). rWGS is emerging as a technology capable of making a specific genetic diagnosis in time to effect changes in acute clinical management. In our patient, the diagnosis of XLA was highly likely after the lymphocyte enumeration panel demonstrated absent B cells, and the next traditional step in the absence of rWGS would be to sequence the suspected gene (in this case, BTK), which takes several weeks to result. rWGS was able to make the diagnosis of XLA within 74.5 h.
XLA is a primary humoral immunodeficiency typically characterized by severe hypogammaglobinemia and recurrent bacterial infections (Smith and Berglöf 2016). It is caused by mutations in BTK, which encodes Bruton tyrosine kinase (Btk) (Smith and Berglöf 2016). Btk is a signal transduction molecule essential to B-cell lineage development, and its loss impairs the progression of pre-B cells to mature lymphocytes (Ochs and Smith 1996). The lack of mature B lymphocytes results in insufficient immunoglobulin levels, and affected patients are unable to mount an appropriate antibody response to infection. Affected persons have normal levels of IgG at birth because of transplacental passage of maternal immunoglobulin, but become susceptible to bacterial and enteroviral infections as these levels gradually wane during the first year of life (Ochs and Smith 1996). The diagnosis of XLA is suspected when laboratory investigations demonstrate absent B cells and a reduction in all classes of immunoglobulins, and it is confirmed by identifying a pathogenic variant in BTK (Ochs and Smith 1996; Smith and Berglöf 2016).
The bacterial infections responsible for causing severe disease in patients with XLA are most frequently the pyogenic organisms Streptococcus pneumoniae and Hemophilus influenzae (Zenone and Souillet 1996; Winkelstein et al. 2006). Although uncommon, in addition to our patient there have been case reports of patients presenting with P. aeruginosa septicemia and ecthyma gangrenosum that were subsequently diagnosed with XLA (Nussinovitch et al. 1991; Zenone and Souillet 1996).
The diagnosis of XLA by rWGS explained the patient's susceptibility to pseudomonal sepsis, indicated the appropriate management to be regular IVIG infusions, and may have prevented additional life-threatening infections. Retrospectively, it also provided an explanation for the death of his twin brother. The patient will be closely followed by immunology for the duration of his lifetime. In addition to monthly IVIG infusions, the patient will have to avoid live vaccinations and his treating physicians should have a low threshold for initiating antibiotic therapy when he is ill.
The metagenomic analysis identification of 770 P. aeruginosa genome sequences confirmed the presence of the bacterial DNA in the patient's bloodstream (DNAemia) and the clinical diagnosis of severe sepsis. Blood cultures in our patient, while receiving broad spectrum antibiotics, never grew Pseudomonas, and the bacterium was identified by culture of a skin biopsy of one of the gangrenous lesions. Patients with bacterial sepsis infrequently have skin lesions available for biopsy—in the absence of that sample for this patient, nucleic acid analysis would potentially have been the only acute method for identifying the responsible bacterial agent. This raises the possibility that rWGS could potentially have utility in identification of disease-causing organisms, especially in cases of culture-negative sepsis, and warrants further exploration.
SUMMARY
We report on a case of XLA presenting as pseudomonal ecthyma gangrenosum and severe sepsis that was diagnosed expeditiously by rWGS. rWGS identified a de novo frameshift variant (c.726dupT; p.Ile243TyrfsTer15) in BTK, the Bruton tyrosine kinase gene on Chromosome X. Thus, the proband was determined to be hemizygous for the monogenic disorder XLA. Additionally, metagenomic analysis of rWGS sequences that did not align to the human genome were found to align to the P. aeruginosa PAO1 genome.
ADDITIONAL INFORMATION
Data Deposition and Access
The variant was submitted to ClinVar (http://www.ncbi.b\nih.nlm.gov/clinvar) and can be found under accession number SCV000809043.
Ethics Statement
Informed and signed consent forms were obtained for all sequenced individuals in this study. The project is approved by the Institutional Review Board of the University of California at San Diego under protocol #160468 and has received nonsignificant risk status in a pre-Investigational Device Exemption submission to the Food and Drug Administration.
Author Contributions
E.S. prepared the manuscript and performed the phenotyping; L.F., S.F.K., and J.B. supervised and prepared the manuscript; S.B., M.B., and M.T. performed the variant interpretation and supervised the analysis; and S.L. and H.M.W. performed the clinical implementation. All authors contributed to the reviewing of the final version.
Funding
Supported by Rady Children's Hospital, National Institute of Child Health and Human Development, and National Human Genome Research Institute (grant U19HD077693).
Referees
Dustin Baldridge
Jim Connelly
Eric W. Klee
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Bonilla FA, Khan DA, Ballas ZK, Chinen J, Frank MM, Hsu JT, Keller M, Kobrynski LJ, Komarow HD, Mazer B. 2015. Practice parameter for the diagnosis and management of primary immunodeficiency. J Allergy Clin Immunol 136: 1186–1205. [DOI] [PubMed] [Google Scholar]
- Coonrod EM, Margraf RL, Russell A, Voelkerding KV, Reese MG. 2013. Clinical analysis of genome next-generation sequencing data using the Omicia platform. Expert Rev Mol Diagn 13: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, Cakici J, Bensen W, Kaplan R, Kronick R, et al. 2018. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinski-Feder E, Weiss M, Brandau O, Jedele KB, Nore B, Bäckesjö CM, Vihinen M, Hubbard SR, Belohradsky BH, Smith CI, et al. 1998. Mutation screening of the BTK gene in 56 families with X-linked agammaglobulinemia (XLA): 47 unique mutations without correlation to clinical course. Pediatrics 101: 276–284. [DOI] [PubMed] [Google Scholar]
- Joshi AY, Iyer VN, Hagan JB, St. Sauver JL, Boyce TG. 2009. Incidence and temporal trends of primary immunodeficiency: a population-based cohort study. Mayo Clin Proc 84: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Weisburd B, Thomas B, Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha KE, Cummings BB, et al. 2017. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res 45: D840–D845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, Baynam G, Bello SM, Boerkoel CF, Boycott KM, et al. 2016. The Human Phenotype Ontology in 2017. Nucleic Acids Res 45: D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, et al. 2015. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med 7: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinovitch M, Frydman M, Cohen HA, Varsano I. 1991. Congenital agammaglobulinemia presenting with ecthyma gangrenosum. Acta Paediatr Scand 80: 732–734. [DOI] [PubMed] [Google Scholar]
- Ochs HD, Smith CE. 1996. X-linked agammaglobulinemia a clinical and molecular analysis. Medicine 75: 287–299. [DOI] [PubMed] [Google Scholar]
- Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, Cunningham-Rundles C, Etzioni A, Holland SM, Klein C, et al. 2015. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for primary immunodeficiency. J Clin Immunol 35: 696–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Berglöf A. 2016. X-linked agammaglobulinemia In GeneReviews® [Internet] (ed. Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A). University of Washington, Seattle: https://www.ncbi.nlm.nih.gov/books/NBK1453 [PubMed] [Google Scholar]
- Stray-Pedersen A, Sorte HS, Samarakoon P, Gambin T, Chinn IK, Coban Akdemir ZH, Erichsen HC, Forbes LR, Gu S, Yuan B, et al. 2017. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol 139: 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelstein JA, Marino MC, Lederman HM, Jones SM, Sullivan K, Burks AW, Conley ME, Cunningham-Rundles C, Ochs HD. 2006. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine 85: 193–202. [DOI] [PubMed] [Google Scholar]
- Yang H, Robinson PN, Wang K. 2015. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods 12: 841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenone T, Souillet G. 1996. X-linked agammaglobulinemia presenting as Pseudomonas aeruginosa septicemia. Scand J Infect Dis 28: 417–418. [DOI] [PubMed] [Google Scholar]