Abstract
Over the past decade, a focus on de novo mutations has rapidly accelerated gene discovery in autism spectrum disorder (ASD), intellectual disability (ID), and other neurodevelopmental disorders (NDDs). However, recent studies suggest that only a minority of cases are attributable to de novo mutations, and instead these disorders often result from an accumulation of various forms of genetic risk. Consequently, we adopted an inclusive approach to investigate the genetic risk contributing to a case of male monozygotic twins with ASD and ID. At the time of the study, the probands were 7 yr old and largely nonverbal. Medical records indicated a history of motor delays, sleep difficulties, and significant cognitive deficits. Through whole-genome sequencing of the probands and their parents, we uncovered elevated common polygenic risk, a coding de novo point mutation in CENPE, an ultra-rare homozygous regulatory variant in ANK3, inherited rare variants in NRXN3, and a maternally inherited X-linked deletion situated in a noncoding regulatory region between ZNF81 and ZNF182. Although each of these genes has been directly or indirectly associated with NDDs, evidence suggests that no single variant adequately explains the probands’ phenotype. Instead, we propose that the probands’ condition is due to the confluence of multiple rare variants in the context of a high-risk genetic background. This case emphasizes the multifactorial nature of genetic risk underlying most instances of NDDs and aligns with the “female protective model” of ASD.
Keywords: autism; intellectual disability, profound; language impairment; severe expressive language delay
INTRODUCTION
Autism spectrum disorder (ASD) is characterized by social communication deficits, repetitive behaviors, and restricted interests. In addition to these core features, ASD is associated with numerous comorbidities including intellectual disability (ID) and epilepsy. For decades, researchers have worked to identify causal genetic factors that lead to these neurodevelopmental disorders (NDDs), and much of this effort has been dedicated to isolating de novo mutations that make large contributions to the complex presentation of NDDs. This approach has proven highly fruitful to gene discovery, identifying more than 100 NDD risk genes shared across thousands of individuals (De Rubeis et al. 2014; Stessman et al. 2017). Nonetheless, despite the immense progress in this realm, recent studies suggest that de novo mutations explain the minority of NDD cases. A large-scale study of more than 2500 simplex families suggested that de novo mutations—including missense mutations, likely gene disrupting mutations, and copy-number variants—contribute to ∼30% of ASD cases (Iossifov et al. 2014). In the remaining cases, ASD is thought to result from a confluence of multiple risk alleles—an accumulation of various forms of genetic risk (Weiner et al. 2017). Thus, although de novo mutation studies lend themselves readily to robust statistical analyses and make invaluable contributions to large-scale gene discovery, most cases will not be explained by the presence of an interpretable de novo mutation. As such, we aim to exemplify an alternative approach to examining genetic risk in idiopathic cases of NDD that cannot be explained by single de novo mutations. In this study, we present a case of male identical twins who each received a diagnosis of ID and ASD. We performed whole-genome sequencing (WGS) of the probands and biological parents to thoroughly explore the genetic risk factors contributing to their condition.
RESULTS
Clinical Presentation and Family History
Twin Caucasian males, age 7, were referred to our genetic research study by the University of Iowa Child Psychiatry Clinic. The probands have two older sisters, both of whom are typically developing. Parents reported distant relatives with conditions including epilepsy, Asperger's syndrome, and language difficulties, but family history was generally unremarkable. At the probands’ time of birth, the mother was age 30 and father age 29. No pregnancy complications were reported before delivery. At 36 wk gestation, the probands were delivered by emergency Caesarean section because of nuchal cord. They initially required incubation, and remained in the NICU for breathing difficulties, jaundice, and low platelet count. After a few days, the probands were discharged.
During their first six months, the probands were described as irritable and “colicky.” Both exhibited sleeping and feeding difficulties, abnormal bowel movements, and delayed motor functions. At 30 mo, they were evaluated at a local autism center. Their expressive and receptive language skills were evaluated using the Preschool Language Scale (PLS-4), and their scores indicated that they each fell below the first percentile. They were also administered the Autism Diagnostic Observation Schedule (ADOS) and received a diagnosis of autism spectrum disorder.
Five months later, the probands received further psychological evaluation at a second mental health clinic. They were assessed for cognitive (Weschler Preschool and Primary Scale of Intelligence) and adaptive (Vineland Adaptive Behavior Scale) skills and were deemed below the first percentile in each. They were also administered the Child Behavior Checklist (CBCL), which indicated clinically significant levels of withdrawal, inattention, sleep problems, and internalizing/externalizing behaviors. The boys received diagnoses of autistic disorder, mild intellectual disability, and severe apraxia. At age 5, the probands were reassessed for intellectual disability and scored below the first percentile for both Intellectual Functioning (Weschler Preschool and Primary Scale of Intelligence) and Adaptive Skills (Vineland Adaptive Behavior Scales). In addition to their neurodevelopmental symptomatology, they have both experienced ongoing health concerns including eczema, skin infections, chronic ear infections, multiple allergies, and gross motor delays.
At the time of enrollment, the probands were largely nonverbal, although one twin had begun developing single words. Both utilized signs, vocalizations, and gestures for most of their communication. Inconsistent with their diagnosis of ASD, they exhibited very little sensory sensitivity and were generally quite social. They initiated and accepted physical contact, enjoyed interacting with both peers and adults, and rarely engaged in aggressive or self-harm behaviors.
Genomic Analyses
After enrollment in our Genetic Research Study of Neurodevelopmental Conditions (devGenes; IRB #201505743), saliva samples were obtained from the probands, mother, and father. DNA was extracted and sent to the University of Iowa Tissue Procurement Core for processing. WGS was performed at the Iowa Institute of Human Genetics—Genomics Division. Alignment and variant calling to the GRCh37 build of the human genome was carried out with the bcbio pipeline. See Methods for full details (a summary of the WGS statistics for each individual is provided in Table 1).
Table 1.
Whole-genome sequencing statistics
| Sample | Total reads | Mapped read | Mapping rate | Mean genome-wide coverage |
|---|---|---|---|---|
| Twin 1 | 945,349,752 | 832,773,579 | 88.0915848593 | 34.06 |
| Father | 656,563,978 | 638,368,673 | 97.2287079996 | 26.87 |
| Mother | 618,497,080 | 570,044,035 | 92.1660026269 | 22.81 |
| Twin 2 | 761,942,864 | 626,645,650 | 82.2431286659 | 25.14 |
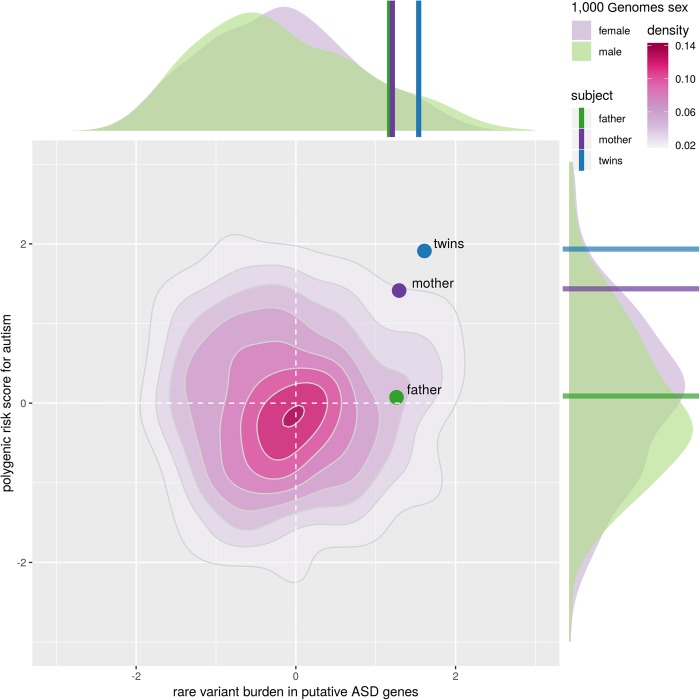
Samples were first assessed for polygenic risk: a summation of the association of common SNPs most strongly associated with ASD, as found by the Psychiatric Genomics Consortium (Cross-Disorder Group of the Psychiatric Genomics 2013). The family's polygenic risk scores (PGRs) were compared against those of the European population of the 1000 Genomes project, a cohort representing the distribution of risk in typically developing individuals (Genomes Project Consortium et al. 2015). PGR distributions were calculated separately for males and females to account for systematic differences between the sexes (two-sided KS test P = 0.0321). The probands’ PGR fell in the 98th percentile compared to males and 97th percentile compared to females (see x-axis of Fig. 1). Their mother's score was elevated, in the 93rd percentile (males) and 92nd percentile (females). The father's PGR was unexceptional, in the 60th percentile (males) and 49th percentile (females). To systematically quantify the family's rare genetic burden in a fashion similar to PGR, the ratio of damaging alleles in putative ASD-related genes to background genes was compared to that of 1000 Genomes Europeans (see Methods for details). No systematic difference between typically developing males and females was observed in 1000 Genomes (two-sided KS test P = 0.37). In this analysis, the probands fell in the 94th percentile of 1000 Genomes, whereas the father was in the 89th percentile and the mother in the 90th percentile. When considering the joint distribution of rare and common risk (Fig. 1), the probands fell well outside the expectation for typically developing individuals (empirical P < 0.01), suggesting that their condition is likely the joint effect of an accumulation of rare and common risk factors. Notably, this risk quadrant (high common variant risk, high rare variant risk) is significantly enriched for females (i.e., depleted for unaffected males) in the 1000 Genomes sample (OR = 1.68, P = 0.02).
Figure 1.
The unaffected mother and twins exhibit excessive rare-variant burden on a high PGR background. Rare-variant burden, shown on the x-axis, was estimated using an unaffected population of 506 individuals with European ancestors from the 1000 Genomes project. Because association with ASD is not broadly applicable for individual rare variants, only variants associated with happloinsufficient putative ASD genes were considered. PGRs, shown on the y-axis, were estimated through the association of ASD with common SNPs observed by the Psychiatric Genomics Consortium. These scores were then compared to the unaffected population from the 1000 Genomes project. Reported scores are normalized.
Individual rare variants were then examined for possible impact on the probands’ phenotype, with emphasis on inheritance pattern, coding changes, conservation, and predicted pathogenicity. The functional impact of rare coding variants was evaluated with PolyPhen (Flanagan et al. 2010), which estimates potential transcript-level effects of an amino acid substitution. Rare variants in evolutionarily conserved regions of the genome were identified via PhastCons score (Siepel et al. 2005). PhastCons estimates the probability that a variant lies within an evolutionarily constrained region using primate, placental mammal, and vertebrate conservation. Variant-level pathogenicity predictions were generated with CADD, a machine-learning-based score that estimates the deleteriousness of coding and noncoding human SNVs and indels (Kircher et al. 2014). As a gene-level complement to CADD, the probability of being loss-of-function intolerant (pLI) for genes impacted by rare variants was retrieved from the ExAC database (Lek et al. 2016). Notable results from these analyses are summarized in Table 2.
Table 2.
Variants of note
| Gene(s) affected | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVar ID | Maternal genotype | Paternal genotype | Twin genotype | Region | Scaled CADD | GnomAD allele frequency (European) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CENPE | 4 | NC_000004.11:g.104070056 G > C | NM_001813.1 p.Ala1263Gly | sub | Missense | — | −/− | −/− | −/+ | Coding (p.A1263G) | 14.74 | 0.00 |
| ANK3 | 10 | NC_000010.10:g.62493816 G > A | — | sub | — | rs112339619 | −/+ | −/+ | +/+ | Upstream | 36 | 0.04478 |
| NRXN3 | 14 | NC_000014.8:g.78951205 G > A | — | sub | — | rs117696080 | −/+ | −/− | −/+ | Intron | 23.8 | 0.01487 |
| NRXN3 | 14 | NC_000014.8:g.79664529 A > G | — | sub | — | rs12432103 | −/− | −/+ | +/− | Intron | 23.9 | 0.03190 |
| ZNF81/ZNF182 | X | NC_000023.10:g.47817751_47828450 del | — | del | — | — | −/+ | −/. | +/. | Upstream | — | — |
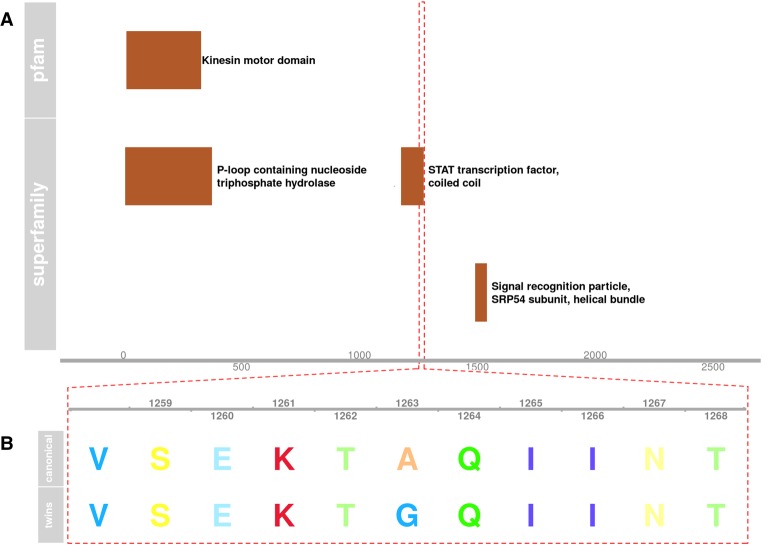
We discovered that the probands have a novel de novo missense mutation causing an alanine to glycine substitution in the central coiled-coil domain of Centromere Protein E (CENPE) (see Fig. 2). Two previously reported SNVs (rs144716013 and rs141488085) (Mirzaa et al. 2014) in this domain have been linked to rare autosomal recessive Microcephaly 13 (OMIM616051). CENPE has an elevated pLI score of 0.65, suggestive of haploinsufficiency. The PolyPhen (Flanagan et al. 2010) prediction for possible impact on the canonical transcript (ENST00000265148) is “Benign” (0.146) and “Possibly Damaging” (0.866) for a slightly shorter alternate transcript (ENST00000380026). We were not able to find any previously reported instance of this variation in humans, and the nucleotide position of this variant is conserved in most primates and some other mammals. The primate PhastCons (Siepel et al. 2005) for this genomic position is 0.81, whereas the placental mammal PhastCons is 0.31.
Figure 2.
(A) Diagram of superfamily and PFAM domains in Centromere Protein E (CENPE). (B) Amino acid sequence of the highlighted region in A, where the proband twins have a de novo nonsynonymous mutation.
We then identified a homozygous variant (rs112339619) in a regulatory region upstream of the known ASD risk gene ANK3 (pLI = 1.00) (Durak et al. 2015), which is responsible for maintenance of proper membrane organization in neurons (Kordeli et al. 1998). The identified variant is found at low frequency in the 1000 Genomes (Genomes Project Consortium et al. 2015) European populations (0.0258), in which it was found exclusively in heterozygotes. In Europeans from the larger gnomAD data set (Lek et al. 2016)—which includes some individuals from disease-specific studies, but none with ASD—this variant is slightly more frequent (0.0447) and is found in 24 homozygotes out of 7493 individuals. This position is highly conserved, with a primate PhastCons score of 0.96 and a vertebrate PhastCons of 0.98. rs112339619 also interrupts an ENCODE-defined (ENCODE Project Consortium 2012) binding site for RE1 silencing transcription factor (REST), which functions to repress transcription of neuronal genes in other tissue types (Bruce et al. 2009). It also intersects overlapping H3K4me3 and methyl-CpG peaks—marks of gene activation and repression, respectively—as found in gray matter (Maunakea et al. 2010).
We observed compound heterozygosity in the probands for rare intronic variants in conserved noncoding regions of the known ASD risk gene NRXN3 (pLI = 1.00) (Vaags et al. 2012). Both variants are found in European populations, in which the maternally inherited rs117696080 occurs at a frequency of 0.0148 (gnomAD) and the paternally inherited rs12432103 occurs at a frequency of 0.0319 (gnomAD). These variants are also located in regions of strong conservation: The locus of the maternal variant has a vertebrate PhastCons score of 1.00 and the paternal variant locus has a score of 0.99.
Finally, our analysis uncovered a novel, X-linked, ∼11-kb transmitted deletion between the zinc finger protein genes ZNF81 (pLI = 0.30) and ZNF182 (pLI = 0.86). Zinc finger proteins are the largest class of DNA binding proteins and play a critical role in regulating many biological processes. Both genes have been previously linked with intellectual disability, developmental delay, and ASD (Kleefstra et al. 2004; Alesi et al. 2012), but they are relatively understudied with unknown functional impact, and in the case of ZNF81, one unaffected male was documented with a truncating variant (Piton et al. 2013). ZNF182 is widely expressed in the brain, whereas ZNF81 shows lower expression, but data from the Genotype-Tissue Expression (GTEx) project reveals that both genes are preferentially expressed in the cerebellum (Carithers et al. 2015).
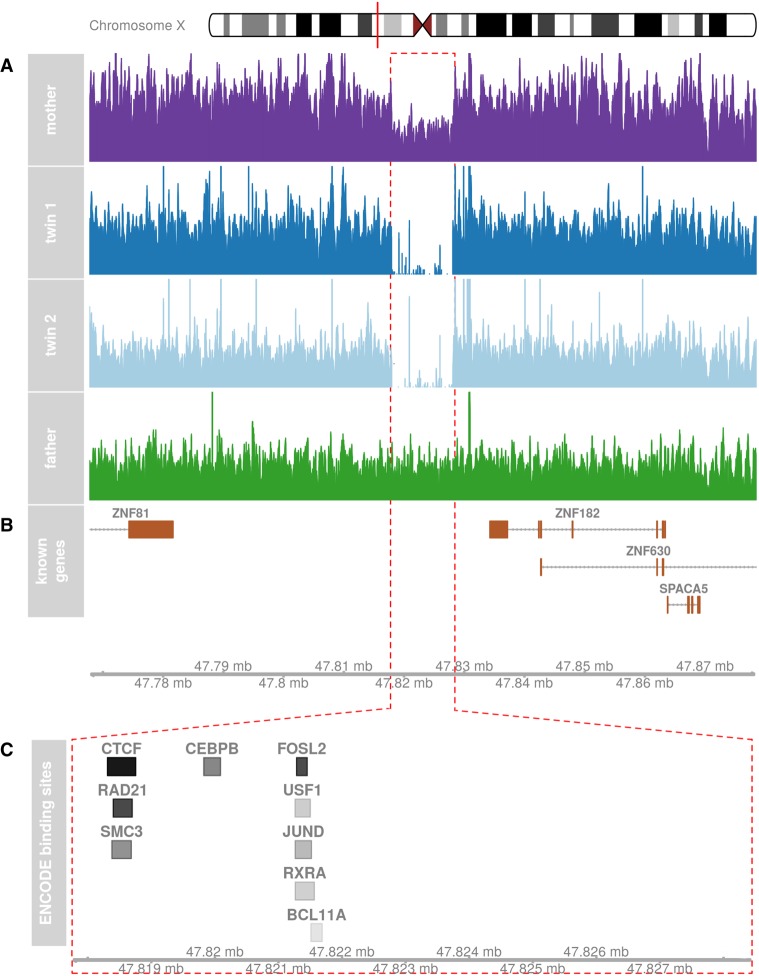
The probands’ deletion is downstream from both genes (which are transcribed on opposing strands) and eliminates regulatory content in the form of several clusters of ENCODE-defined transcription factor binding sites (Fig. 3). These sites include a cluster of CTCF, SMC3, and RAD21 binding sites, all of which are linked to the MECP2/Rett syndrome pathway (Liyanage and Rastegar 2014). JUND and FOSL2 binding sites form another cluster in this region. Both of these transcription factors are highly expressed in the brain (Fagerberg et al. 2014) and involved in forming the AP1 transcription factor complex (Huttlin et al. 2015), which responds to cytokines and growth factor signaling.
Figure 3.
(A) An ∼11-kb X-linked deletion inherited by the probands from their mother is evident in read coverage of the region. (B) The deletion is downstream from several zinc finger proteins (ZNF81 and ZNF182). (C) The deletion impacts as a cluster of ENCODE transcription factor ChIP-seq binding sites. A darker color indicates a higher score.
DISCUSSION
In this case study, we investigated the genetic burden carried by identical twin males with comorbid ASD and ID. Through WGS of both probands and their parents, we identified a de novo mutation, inherited rare variants, and elevated PGR that may collectively contribute to the boys’ condition. When aggregating rare and common risk factors, the boys showed a significant (empirical P < 0.01) joint increase of rare and common risk (Fig. 1).
Previous research has determined that most genetic liability for ASD is explained by common variants with additive genetic effects (Weiner et al. 2017). The extent of these effects can be quantified using a PGR score, which can be compared with the PGR scores of a control population—like 1000 Genomes—to assign a percentile ranking. Such individual-level PGR ranking is increasingly used as part of a clinical toolbox used to drive interventions (Torkamani et al. 2018). At the population level, PGR for autism has been positively correlated with educational attainment and other measures of cognitive ability, suggesting that at least some of these common variants may be under positive selection (Clarke et al. 2016). However, inheriting many common autism risk alleles may have an overall negative effect, as seen in our profoundly affected male probands. Interestingly, the probands’ unaffected mother also displayed elevated PGR (92nd percentile), which is consistent with the female protective model. ASD and ID both exhibit a striking sex bias, with males diagnosed at far higher rates than females (4:1 and 2:1 male-to-female ratios, respectively) (Fombonne 2009; Maulik et al. 2011). Studies examining the genetic basis of this sex difference have suggested that females with NDDs exhibit a higher mutational burden than males with NDDs, and unaffected mothers transmit deleterious mutations at higher rates than unaffected fathers (Jacquemont et al. 2014). Based on PGR alone, the present case appears to align with these findings, but without functional data we are not able to rule out alternative explanations for the unaffected mother's elevated PGR.
In addition to the elevated PGR, the entire family showed increased rare variant burden in putative ASD-related genes. None of these variants alone would be expected to lead to an ASD phenotype, but when aggregated, they represent an axis of risk that complements PGR (Fig. 1). These rare variants likely confer a broader range of risk than those used to calculate PGR, with some variants having large effects and others very little. To better characterize them, we identified several specific variants that previous literature supports as plausibly conferring risk for ASD.
The probands are compound heterozygotes for two possibly deleterious noncoding variants in the neurexin 3 gene (NRXN3), which has been previously linked to autism and other cognitive disorders (Vaags et al. 2012). NRXN3 is highly expressed in the brain and plays an essential role in the brain's ability to process information (Fagerberg et al. 2014). Through thousands of alternate transcripts encoded by NRXN3, the gene largely impacts cell adhesion and cell signaling (Ullrich et al. 1995). Given the impressive number of alternate transcripts encoded by NRXN3, the variants identified in the probands could conceivably impact one or several of the complicated splicing events needed to produce mature NRXN3 mRNA. Furthermore, the reported positions are extremely conserved throughout all vertebrates, indicating substantial evolutionary constraint. Taken together, these factors suggest the identified variants are likely deleterious and contribute to the probands’ condition.
The probands are also homozygous for a regulatory variant with a markedly high scaled CADD score of 36, which is upstream of the known autism risk gene ankyrin 3 (ANK3). This locus is also highly conserved throughout vertebrates and overlaps several important markers of gene regulation within the brain. This regulatory region exhibits signals of both gene activation and repression—H3K4me3 and methyl-CpG, respectively—indicating it may tightly control the expression of ANK3. In addition, the variant is located within a binding site for REST, a transcription factor that prevents improper expression of neuronal genes in other tissues. Disruption of the REST binding site could lead to nonspecific or overexpression of ANK3. In mice, overexpression of ANK3 has been shown to promote degradation of tyrosine-phosphorylated platelet-derived growth factor receptor (PDGFR) (Ignatiuk et al. 2006), which promotes the development and survival of both GABAergic and dopaminergic neurons (Smits et al. 1993; Othberg et al. 1995). PDGFR-knockout mice exhibit a variety of autism-like behaviors (Nguyen et al. 2011), including impaired spatial working memory, decreased social interaction, and compromised contextual memory. The variant identified in our probands could lead to a similar chain of events: Overexpression of ANK3 may lead to a reduction in PDGFR, thereby resulting in neuronal deficits that contribute to an ASD phenotype.
We also detected a de novo missense mutation of unknown significance in the gene CENPE. Although this specific mutation has not been previously identified, other missense variants in the same domain have been linked to a form of microcephalic primordial dwarfism (OMIM 616051) typified by absent speech, developmental delay, and gross motor deficits (Mirzaa et al. 2014). At the molecular level, CENPE is partly responsible for proper chromosome alignment in prometaphase and, thus, chromosome segregation. Mutations in several other genes vital to this cellular process (e.g., POGZ, CHAMP1) have been associated with ASD and ID, as well as other traits exhibited by our probands such as language impairment and recurrent infections (Deciphering Developmental Disorders Study 2015; Hempel et al. 2015; Stessman et al. 2016). The elevated pLI of CENPE (0.65) supports the notion that a single de novo mutation could be sufficient to contribute to the probands’ phenotype, as pLI > 0.5 indicates probable sensitivity to haploinsufficiency.
The largest novel variant we detected—an ∼11-kb deletion on Chromosome X—impacts ZNF81 and ZNF182. These genes have been associated with several large mutations tied to X-linked intellectual disability (Kleefstra et al. 2004), although the findings have not been replicated and have been challenged by studies utilizing large-scale human exome sequencing data (Piton et al. 2013). Specific research on these genes is needed to more precisely evaluate the impact of this mutation, but we provide speculation based on the preliminary information currently available. First, GTEx data indicates that the genes are preferentially expressed in the cerebellum, a brain region largely associated with coordination and motor functioning. As such, the deletion may contribute to the severe apraxia (Ackermann et al. 2007) and significant gross motor delays exhibited by the probands. Second, the deletion does not interrupt the coding regions of ZNF81 or ZNF182, but it potentially affects the transcription of both by eliminating overlapping binding sites for the transcription factors CTCF, SMC3, and RAD21. These transcription factors are responsible for regulating chromatin state (Ball et al. 2014) and each directly or indirectly interacts with MECP2. Altered expression of MECP2 has been tied to autism spectrum disorder (Samaco et al. 2004), as well as traits such as motor delays, disrupted sleep, and absent speech (Williamson and Christodoulou 2006), all of which were exhibited by the probands. Indeed, in GTEx (Carithers et al. 2015) expression of ZNF81 and ZNF182 is significantly correlated with that of MECP2 in the cerebellum (for both genes: r = 0.65 and P < 0.01). The locus deleted in the probands may therefore represent a downstream regulatory target of MECP2, but functional data is necessary to support this hypothesis, and a comprehensive investigation of the potential regulatory role for MECP2 of these genes will require focused experimental follow-up.
Notably, this maternally inherited deletion was transferred from a phenotypically unaffected mother, which leads us to consider two potential explanations: (a) the deletion is of zero consequence and does not contribute to the probands’ condition, or (b) the mother is buffered from any phenotypic effect because of the intact copy of this region on her other X chromosome (see Fig. 3). Interestingly, in addition to the mother's elevated PGR, this X-linked transmission is consistent with findings from several recent studies examining families with NDDs. Results suggest that the neurodevelopmental impact of large-effect genetic variants is compounded by secondary variants, and that these secondary variants are disproportionately maternally inherited (Girirajan et al. 2012; Jacquemont et al. 2014; Duyzend et al. 2016). Male offspring, lacking a second X chromosome and other elements of the “female protective effect” (Constantino et al. 2010, 2013; Constantino and Charman 2012; Gockley et al. 2015; Constantino 2016) are then more sensitive to genetic risk factors and more readily cross the threshold to disease as a result of these “multiple hits.” These findings may explain the complex clinical presentation of our probands, despite the absence of neurodevelopmental symptomatology in the probands’ mother (and two typically developing sisters). Our conclusions are limited by the absence of genetic data for the probands’ sisters, which might have provided further insight into the role of the female protective effect in this family.
Overall, the collective results of this study must be interpreted with consideration of a few important limitations. Although we did not identify a single mutation that appeared to unequivocally contribute to the probands’ condition and we uncovered multiple variants that likely contribute in a collective manner, a lack of functional data for these particular variants prevents us from concluding that any or all of those mutations are functionally significant. It is also possible that, despite our efforts to maximize sensitivity (see Methods section for further details), a pathogenic variant was missed because of sporadic low coverage, mapping problems, or other technical limitations. Finally, as with any case study, these findings cannot be assumed to generalize to outside cases of ASD, ID, or other NDDs. Nevertheless, idiopathic cases such as this allow for a more nuanced investigation of complex disorders like ASD and ID than large-scale genetic studies. In fact, such individual cases can illustrate exactly where current knowledge is strongest and where the field has room to improve. For example, the advantages of WGS over whole-exome sequencing (WES) have been illustrated in a variety of studies investigating the genetics of NDDs (Michaelson et al. 2012; Yuen et al. 2015; Stavropoulos et al. 2016; Turner et al. 2016; Werling et al. 2018), and the use of WGS allowed us to identify variants that would not have been found with WES or other targeted techniques. Notably, only one of the four identified variants—the missense mutation in CENPE—was found in a coding region, and there was little evidence to suggest that the probands’ phenotype could be solely attributed to such a variant. The use of WGS allowed us to provide a more detailed account of the probands’ genetic risk, and as the cost of WGS continues to decline, the advantages should continue to expand (Lacey et al. 2014). Until then, the interpretation of these findings is limited by the field's incomplete knowledge of NDD genetics. In this case, current knowledge allows us to identify this as an exceptional case of excess rare variants on a high-risk background but does not allow us to assign high confidence to individual genetic variants. Ultimately, this limitation highlights the importance of pursuing multiple methods of genetic research to deepen our understanding of NDDs.
In conclusion, previous genetic studies of autism spectrum disorder and intellectual disability have largely focused on the identification of causal de novo variants. Although this approach has been invaluable in uncovering specific genes involved in these disorders thanks to robust analysis and interpretation, de novo variants hold less clinical utility for the many cases that cannot be attributed to single mutations. We were confronted with this reality in the present case of monozygotic twins with idiopathic ASD and ID. We expected such a case to provide an optimal opportunity to identify a single causal variant; instead, we uncovered an example of the effects of accumulated genetic risk factors, many of which appear to have passed through an unaffected mother. Our results suggest that the probands’ phenotype is due to multiple rare variants in the context of a high-risk genetic background and a high-risk delivery (multiple births, preterm delivery, and Cesarean section). The study highlights our occasional tendency to overestimate the probability of identifying an obviously causal de novo mutation and reiterates the utility of considering multiple modes of genetic variation to identify the biological basis of NDDs. As more large-scale gene discovery studies adopt similarly integrative approaches to modeling the confluence of multiple forms of genetic risk, our appreciation of ASD as a solidly oligogenic phenomenon will grow, and fewer cases will be left indecipherable.
METHODS
Biological samples for genotyping were gathered from consenting families (devGenes; IRB #201505743). Both biological parents were invited to participate to facilitate completion of family-based association tests. DNA was obtained from buccal, or saliva, samples using Oragene kits (DNA Genotek Inc.). DNA extraction was carried out with prepIT (DNA Genotek Inc.). DNA quantification was performed using the Qubit 2.0 Fluorometer (Life Technologies Corporation), with quantities ranging from 100 to 800 ng/µL.
DNA samples were sheared on an E220 Focused-ultrasonicator (Covaris) to an average size of 400 bp. Sequencing libraries were generated with the Kapa Hyper Prep kit (Kapa Biosystems) following the manufacturer protocol. Prepared libraries were analyzed on the Fragment Analyzer (Advanced Analytical), and the four libraries were combined equally into a pool. The pool was size selected on the Blue Pippin (Sage Science) to a size range of 420–670 bp. The size selected pool was then sequenced on four lanes of the HiSeq4000 (Illumina) with 150-bp Paired End chemistry.
FASTQ files generated by the HiSeq4000 were processed with the bcbio-nextgen pipeline (v1.0.0a0-e1baf12). In brief, reads were mapped to the GRCh37 human reference genome with BWA-mem (v0.7.15). Variants were called as a pool with GATK (v3.5-0-g36282e4), Platypus (v0.8.1), FreeBayes (v1.0.2.29), and SAMtools (v1.3.1), producing squared off callsets. Filtering and quality assurance were performed on these variant calls using the default parameters of bcbio-nextgen. In brief, GATK variants were filtered by variant quality score recalibration; Platypus variants were filtered based on the goodness of fit of genotype calls, excessive region-based haplotype scores, root-mean-square mapping quality, variant quality and its ratio with read depth, low complexity sequence context, allele bias, region-based read quality, neighboring homopolymers, and strand bias; FreeBayes and SAMtools variants were filtered based on a combination of allele frequency, read depth, and quality with thresholds determined by bcbio. Table 3 provides further details.
Table 3.
Filtering procedure
| Variant caller | SAMtools | FreeBayes | GATK | Platypus defaults |
|---|---|---|---|---|
| Filter source | bcbio | Bcbio | GATK VQSR (tuned by bcbio) | Sequence context: variant is in low-complexity region |
| Alternative allele frequency > 0.5; read depth < 4; and variant quality < 20 | Alternative allele frequency > 0.5 (read depth < 4 | read depth < 13 and variant quality < 10) | Type = SNP and tranche level ≥ 99.97% | Haplotype score: excessive haplotypes in region | |
| Conditions | Read depth >13 and variant quality < 10 | Alternative allele frequency > 0.5; read depth < 4; and variant quality < 50 | Type = InDel and tranche level ≥ 98.0% | Mapping quality: RMS mapping quality across region |
| Alternate allele frequency < 0.5; read depth < 4; and variant quality < 50 | Alternative allele frequency > 0.5; read depth > 145; and variant quality < 290 | Variant quality < 20 Quality/depth ratio Strand bias Allele bias Goodness-of-fit test |
To improve sensitivity over individual variant callers, filtered variants from all callsets were combined into an ensemble callset in which every variant call was supported by at least two callers. This approach resulted in modest increases in the number of variants over those called by GATK and FreeBayes alone (+2% for each), whereas it produced a substantial increase over variants passing Platypus's stringent filtering (+15%) and a substantial decrease compared to the number of variants called by SAMtools (−12%). The specificity of this final callset is demonstrated by the 83% of variants that were supported by all four variant callers, whereas 11% were supported by three callers and 6% were supported by two callers. Variants were then annotated with VEP (v83) and GEMINI (v0.19.1) to assign genes association and functionality. Structural variants were called with ForestSV (v0.3.3).
The genome-wide coverage for our subjects varied from 22.8× to 34× (Table 1). Coverage over 30× would have been ideal, as previous work has estimated that the sensitivity of detecting CNVs > 1 kb reaches 99% at coverages of 30× (Yoon et al. 2009; Michaelson and Sebat 2012), whereas smaller indels may require up to 60× coverage (Fang et al. 2014) to identify with comparable confidence to SNVs. As such, we were not able to report indels in the present study. Nonetheless, it is estimated that coverage as low as 5× can detect CNVs > 1 kb with 80% sensitivity (Yoon et al. 2009), and heterozygous SNVs can be detected with ∼98% sensitivity for average coverages of 15× (Bentley et al. 2008) to 22× (Meynert et al. 2014).
Only variants found at an allele frequency of <5% in gnomAD Europeans, with a GATK-defined genotype quality greater than 20, and near genes with a previous association with ASD (i.e., SFARI Gene 3.0 category 1 and 2 genes), were considered for individual reporting. No de novo loss of function mutations met these criteria. The de novo missense mutation with greatest CADD score, the inherited homozygous variant (heterozygous in both parents) with highest CADD score, and two compound heterozygous variants (assigned by VEP to the same gene) with highest combined CADD scores were reported.
Polygenic risk scores were computed using PRSice (v1.25) with ASD GWAS summary statistics from the Psychiatric Genomics Consortium (03/25/15 version). The proband and parental genomes were combined with the 1000 Genomes European populations (which served as controls). SNPs were then clumped to reduce the impact of linkage disequilibrium. An optimal threshold on SNP inclusion was found at a P-value of 0.01, corresponding to the most ASD-associated polymorphisms (15,104 SNPs). The major histocompatibility complex region was excluded from the analysis.
The burden of rare and putatively damaging variants in the family and the typically developing population of 1000 Genomes Europeans was calculated by counting the number of rare variants (allele frequency < 0.05 in all of 1000 Genomes) with a GERP score ≥ 5. GERP (Davydov et al. 2010) is a base-level measure of evolutionary constraint based on substitution deficits (evidence of purifying selection), in which scores greater than zero represent greater conservation. GERP, rather than a substitution-specific pathogenicity score (like CADD), was used because CADD uses 1000 Genomes as training data for neutral mutations, leading all variants found in 1000 Genomes to have systematically lower scores. GERP was selected because conservation scores like it are the most important features used by CADD. To compensate for the reduced specificity of the GERP score, only variants in the region of putative ASD genes (Brueggeman et al. 2018) with pLI > 0.5 were counted. To correct for possible ascertainment bias between the family and 1000 Genomes, qualifying variants in a random set of size-matched, non-ASD related genes with pLI > 0.5 were also counted and the ratio of these counts were used to compare the family's rare-variant burden to that of 1000 Genomes Europeans.
ADDITIONAL INFORMATION
Data Deposition and Access
The sequencing presented in this study was not National Institutes of Health (NIH)-funded, and consequently NIH elected not to include it in dbGaP. Genotype data will be made available by the authors to qualified researchers who are able to demonstrate proper human subjects research authorization. The variants were deposited in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000853274–SCV000853278.
Ethics Statement
Written consent for the probands, mother, and father to participate in the Genetic Research Study of Neurodevelopmental Conditions (devGenes; IRB #201505743) was obtained from the mother and father. Because of the probands’ age and level of intellectual functioning, verbal assent was not deemed necessary by the University of Iowa IRB.
Author Contributions
J.M., B.M., and J.K. conceived and planned the case study. B.M. carried out the data collection and processing, with assistance from J.K. T.K. and K.V. conducted the analyses and created the figures. B.M. and T.K. took the lead in writing the manuscript, with critical feedback and revisions from J.M.
Funding
The authors are grateful for the financial support of the National Institute of Mental Health (R01MH105527) and the National Institute on Deafness and Other Communication Disorders (R01DC 014489).
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Ackermann H, Mathiak K, Riecker A. 2007. The contribution of the cerebellum to speech production and speech perception: clinical and functional imaging data. Cerebellum 6: 202–213. [DOI] [PubMed] [Google Scholar]
- Alesi V, Bertoli M, Barrano G, Torres B, Pusceddu S, Pastorino M, Perria C, Nardone AM, Novelli A, Serra G. 2012. 335.4 kb microduplication in chromosome band Xp11.2p11.3 associated with developmental delay, growth retardation, autistic disorder and dysmorphic features. Gene 505: 384–387. [DOI] [PubMed] [Google Scholar]
- Ball AR Jr, Chen YY, Yokomori K. 2014. Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies. Biochim Biophys Acta 1839: 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AW, López-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, Koch CM, Langford CF, Dunham I, Andrews RM, et al. 2009. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res 19: 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman L, Koomar T, Michaelson J. 2018. Forecasting autism gene discovery with machine learning and genome-scale data. bioRxiv 10.1101/370601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. 2015. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank 13: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T-K, Lupton MK, Fernandez-Pujals AM, Starr J, Davies G, Cox S, Pattie A, Liewald DC, Hall LS, MacIntyre DJ, et al. 2016. Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol Psychiatry 21: 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN. 2016. Data from the Baby Siblings Research Consortium confirm and specify the nature of the female protective effect in autism: a commentary on Messinger et al. Mol Autism 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Charman T. 2012. Gender bias, female resilience, and the sex ratio in autism. J Am Acad Child Adolesc Psychiatry 51: 756–758. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. 2010. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry 167: 1349–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Todorov A, Hilton C, Law P, Zhang Y, Molloy E, Fitzgerald R, Geschwind D. 2013. Autism recurrence in half siblings: strong support for genetic mechanisms of transmission in ASD. Mol Psychiatry 18: 137–138. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381: 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study. 2015. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519: 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O, de Anda FC, Singh KK, Leussis MP, Petryshen TL, Sklar P, Tsai LH. 2015. Ankyrin-G regulates neurogenesis and Wnt signaling by altering the subcellular localization of β-catenin. Mol Psychiatry 20: 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyzend MH, Nuttle X, Coe BP, Baker C, Nickerson DA, Bernier R, Eichler EE. 2016. Maternal modifiers and parent-of-origin bias of the autism-associated 16p11.2 CNV. Am J Hum Genet 98: 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Wu Y, Narzisi G, O'Rawe JA, Barrón LT, Rosenbaum J, Ronemus M, Iossifov I, Schatz MC, Lyon GJ. 2014. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med 6: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SE, Patch AM, Ellard S. 2010. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genet Test Mol Biomarkers 14: 533–537. [DOI] [PubMed] [Google Scholar]
- Fombonne E. 2009. Epidemiology of pervasive developmental disorders. Pediatr Res 65: 591–598. [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, et al. 2012. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med 367: 1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gockley J, Willsey AJ, Dong S, Dougherty JD, Constantino JN, Sanders SJ. 2015. The female protective effect in autism spectrum disorder is not mediated by a single genetic locus. Mol Autism 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel M, Cremer K, Ockeloen CW, Lichtenbelt KD, Herkert JC, Denecke J, Haack TB, Zink AM, Becker J, Wohlleber E, et al. 2015. De novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet 97: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. 2015. The BioPlex network: a systematic exploration of the human interactome. Cell 162: 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiuk A, Quickfall JP, Hawrysh AD, Chamberlain MD, Anderson DH. 2006. The smaller isoforms of ankyrin 3 bind to the p85 subunit of phosphatidylinositol 3′-kinase and enhance platelet-derived growth factor receptor down-regulation. J Biol Chem 281: 5956–5964. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, Beckmann JS, Rosenfeld JA, Eichler EE. 2014. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet 94: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Yntema HG, Oudakker AR, Banning MJ, Kalscheuer VM, Chelly J, Moraine C, Ropers HH, Fryns JP, Janssen IM, et al. 2004. Zinc finger 81 (ZNF81) mutations associated with X-linked mental retardation. J Med Genet 41: 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Ludosky MA, Deprette C, Frappier T, Cartaud J. 1998. AnkyrinG is associated with the postsynaptic membrane and the sarcoplasmic reticulum in the skeletal muscle fiber. J Cell Sci 111(Pt 15): 2197–2207. [DOI] [PubMed] [Google Scholar]
- Lacey S, Chung JY, Lin H. 2014. A comparison of whole genome sequencing with exome sequencing for family-based association studies. BMC Proc 8: S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VR, Rastegar M. 2014. Rett syndrome and MeCP2. Neuromolecular Med 16: 231–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. 2011. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil 32: 419–436. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynert AM, Ansari M, Fitzpatrick DR, Taylor MS. 2014. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics 15: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Sebat J. 2012. forestSV: structural variant discovery through statistical learning. Nat Methods 9: 819–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, et al. 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa GM, Vitre B, Carpenter G, Abramowicz I, Gleeson JG, Paciorkowski AR, Cleveland DW, Dobyns WB, O'Driscoll M. 2014. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum Genet 133: 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PT, Nakamura T, Hori E, Urakawa S, Uwano T, Zhao J, Li R, Bac ND, Hamashima T, Ishii Y, et al. 2011. Cognitive and socio-emotional deficits in platelet-derived growth factor receptor-β gene knockout mice. PLoS One 6: e18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othberg A, Odin P, Ballagi A, Ahgren A, Funa K, Lindvall O. 1995. Specific effects of platelet derived growth factor (PDGF) on fetal rat and human dopaminergic neurons in vitro. Exp Brain Res 105: 111–122. [DOI] [PubMed] [Google Scholar]
- Piton A, Redin C, Mandel JL. 2013. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet 93: 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Nagarajan RP, Braunschweig D, Lasalle JM. 2004. Multiple pathways regulate MeCP2 expression in normal brain development and exhibit defects in autism-spectrum disorders. Hum Mol Genet 13: 629–639. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits A, Ballagi AE, Funa K. 1993. PDGF-BB exerts trophic activity on cultured GABA interneurons from the newborn rat cerebellum. Eur J Neurosci 5: 986–994. [DOI] [PubMed] [Google Scholar]
- Stavropoulos DJ, Merico D, Jobling R, Bowdin S, Monfared N, Thiruvahindrapuram B, Nalpathamkalam T, Pellecchia G, Yuen RKC, Szego MJ, et al. 2016. Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine. NPJ Genom Med 1: 15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, Wang T, Hoekzema K, Vives L, Vogel I, et al. 2016. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet 98: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman HA, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, Kvarnung M, Gerdts J, Trinh S, Cosemans N, et al. 2017. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet 49: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Wineinger NE, Topol EJ. 2018. The personal and clinical utility of polygenic risk scores. Nat Rev Genet 19: 581–590. [DOI] [PubMed] [Google Scholar]
- Turner TN, Hormozdiari F, Duyzend MH, McClymont SA, Hook PW, Iossifov I, Raja A, Baker C, Hoekzema K, Stessman HA, et al. 2016. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet 98: 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Sudhof TC. 1995. Cartography of neurexins more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14: 497–507. [DOI] [PubMed] [Google Scholar]
- Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, Ogilvie C, Ahn JW, Drmic I, Senman L, et al. 2012. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet 90: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, et al. 2017. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49: 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Brand H, An JY, Stone MR, Zhu L, Glessner JT, Collins RL, Dong S, Layer RM, Markenscoff-Papadimitriou E, et al. 2018. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 50: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SL, Christodoulou J. 2006. Rett syndrome: new clinical and molecular insights. Eur J Hum Genet 14: 896–903. [DOI] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res 19: 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y, et al. 2015. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 21: 185–191. [DOI] [PubMed] [Google Scholar]