Abstract
Oncolytic viruses are designed for cancer treatment. Cell-virus interactions are key determinants for successful viral replication. Therefore, the extensive reprogramming of gene expression that occurs in tumor cells might create a hurdle for viral propagation. We used a replication-based approach of a microRNA (miRNA) adenoviral library encoding up to 243 human miRNAs as a bioselection strategy to identify miRNAs that facilitate adenoviral oncolytic activity in pancreatic ductal adenocarcinoma. We identify two miRNAs, miR-99b and miR-485, that function as enhancers of adenoviral oncolysis by improving the intra- and extracellular yield of mature virions. An increased adenoviral activity is the consequence of enhanced E1A and late viral protein expression, which is probably mediated by the downregulation of the transcriptional repressors ELF4, MDM2, and KLF8, which we identify as miR-99b or miR-485 target genes. Arming the oncolytic adenovirus ICOVIR15 with miR-99b or miR-485 enhances its fitness and its antitumoral activity. Our results demonstrate the potential of this strategy to improve oncolytic adenovirus potency, and they highlight miR-99b and miR-485 as sensitizers of adenoviral replication.
Keywords: oncolytic adenovirus, miRNAs, pancreatic cancer
Oncolytic adenoviruses are a promising alternative therapy for refractory cancers, including pancreatic cancer; however, more potent viruses are needed. This paper describes an approach to identify oncolytic adenoviruses with enhanced features. It is based on the construction of a library encoding up to 243 miRNAs, followed by a bioselection strategy to determine miRNAs that enhance adenoviral replication. We identified miR-99b and miR-485 as oncolytic adenoviral sensitizers, and we show the mechanism of enhanced activity. The new viruses display improved anticancer efficacy in xenograft models. This approach could be applied to other oncolytic viruses, expanding the interest in the oncolytic virus community.
Introduction
Viruses have evolved a number of strategies to hijack cellular resources, forcing them to synthesize new virions for a successful viral propagation.1, 2 These strategies have been acquired as a consequence of an adaptive process in specific cellular contexts. While the use of viruses in cancer treatments has long been studied,3 tumor cells present a number of genetic alterations that elicit massive reprogramming of cellular gene expression, creating a novel context in which viruses have not adapted to replicate.4 The inherent complexity of tumors makes the oncolytic viral activity highly dependent on the specific characteristics of tumor cells. In fact, the differential expression of certain genes in cancer cells can negatively impact viral replication.5, 6, 7 Nevertheless, molecular mechanisms explaining viral replication heterogeneity in cancer cells remain to be fully appraised.8
MicroRNAs (miRNAs) are post-transcriptional negative regulators of gene expression that modulate a variety of cellular physiological processes. miRNAs also play important roles in host-virus interactions.9 Viral infections can modulate the cellular miRNome with changes induced either by the virus, to promote a proviral environment, or by the antiviral cellular response, to limit the viral life cycle.10, 11, 12 Although the mechanisms of miRNA-mediated regulation of viral infection have not been completely clarified, it is clear that host miRNAs can regulate viral infections.
In the context of tumorigenesis, many miRNAs with essential roles in cancer-associated pathways are dysregulated.13 Participation of miRNAs has been demonstrated in central cancer processes, such as cell proliferation and death, cell differentiation, and maintenance of stem cell potency.14 Aberrant expression of miRNAs has been well documented for pancreatic ductal adenocarcinoma (PDAC), and specific miRNA signatures have been associated with diagnosis, staging, progression, prognosis, and treatment response.15, 16 Hence, when viruses are used as oncolytic agents, they face an already perturbed miRNome in cancer cells. These miRNA profile changes may not necessarily favor viral replication and propagation within the tumor; indeed, the downregulation of certain miRNAs could induce the overexpression of viral-limiting factors.
Adenoviruses (Ads) are non-enveloped icosahedral viruses with double-stranded DNA genomes. The viral genome is sub-divided into regions termed early and late according to when transcription activation initiates. Early genes remodel the intracellular environment to prepare the cell for viral replication and activate the expression of other viral genes. Late genes primarily constitute viral structural proteins. Adenoviruses, and particularly adenovirus serotype 5 (Ad5), are engineered as oncolytic agents for therapeutics against different cancer types, including PDAC.17, 18 Upon adenoviral infection, cellular miRNA profiles suffer profound changes that influence viral replication.10, 11, 19 For this reason, dysregulated miRNA profiles in cancer cells may generate an inadequate environment for adenoviral activity. Recently, Hodzic and coworkers11 identified a group of miRNAs that sensitize cells to Ad5-induced cell death. However, several of these miRNAs triggered premature cell death that interfered with the adenoviral cycle, and only miR-26b was found to be an enhancer of Ad5 propagation. Intriguingly, and contrary to what was expected, miR-26b was highly expressed in PC-3 cells, and Ad5 infection only caused a slight reduction of its expression, thus confirming the complexity of miRNA-virus interactions.11
Here we describe the identification of human miRNAs that favor adenoviral activity in pancreatic cancer, using a replication-based bioselection strategy. We identified miR-99b and miR-485 as enhancers of the adenovirally mediated oncolysis in PDAC, whereby they facilitate the formation of mature virions by means of negatively regulating the expression of specific transcription factors. Arming the ICOVIR15 oncolytic adenovirus with miR-99b or miR-485 resulted in new viruses that greatly improved the antitumor efficacy. We believe that this approach could be used to bypass the complexity of the host-virus interactions, as miRNA sequences are codified directly within the adenoviral genome, giving rise to a single therapeutic agent.
Results
In Vitro Bioselection of the miRNA Adenoviral Library AdwtE miRLib in PDAC Cell Lines
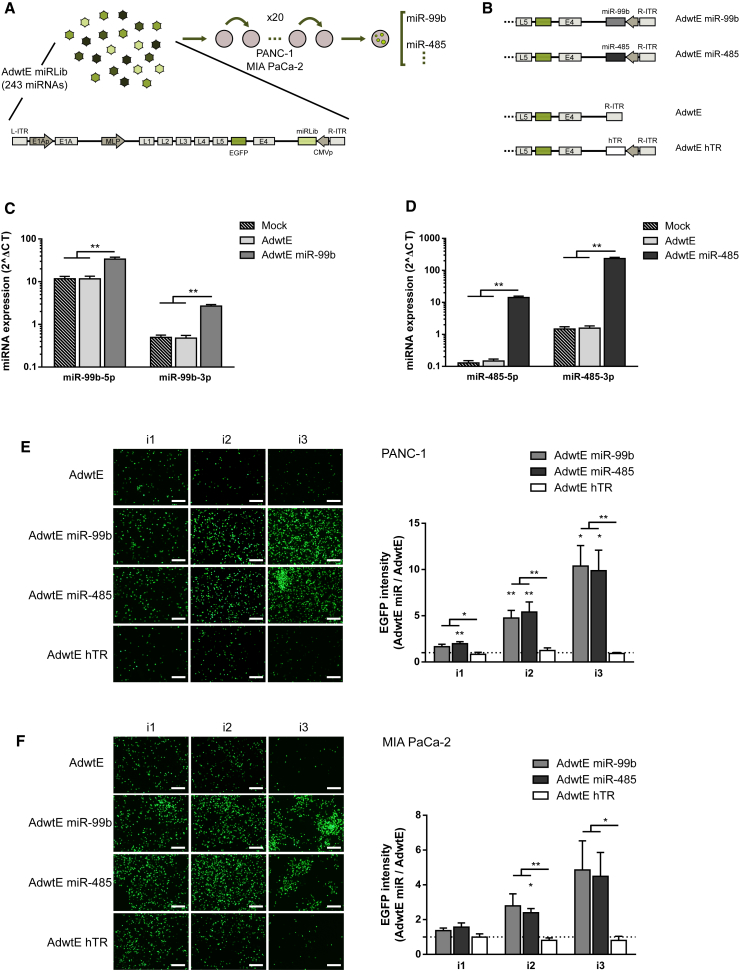
To identify miRNAs that enhance adenoviral activity, we generated a miRNA adenoviral library (AdwtE miRLib). The AdwtE miRLib consisted of a pool of adenoviruses coding for up to 243 different human primary miRNA (pri-miRNA) genomic sequences under the control of the cytomegalovirus (CMV) promoter. These sequences were introduced next to the right inverted terminal repeat (R-ITR) of a wild-type adenovirus 5 expressing the EGFP gene under the control of the major later promoter (MLP) (AdwtE) (Figure 1A). The AdwtE miRLib was subjected to 20 rounds of bioselection in PANC-1 and MIA PaCa-2 PDAC cell lines. After this, adenoviral clones were isolated by the plaque assay, and the miRNAs present were identified by Sanger sequencing (Figure 1A). Although several miRNAs were detected to be enriched following PANC-1 and MIA PaCa-2 bioselection, adenoviruses encoding miR-99b and miR-485 were enriched in the two cell lines (Figure 1A; Table S1). In MIA PaCa-2 cells, clones coding for miR-99b were highly enriched with respect to the total number of clones encoding for a given miRNA (87.9%), whereas miR-485 was less represented (9.1%). In PANC-1 cells, the most abundant miRNA-encoding represented clone was miR-485 (28.6%), with miR-99b encoded in fewer clones (7.1%).
Figure 1.
miR-99b and miR-485 Identified through a Bioselection Strategy as Enhancers of the Adenoviral Replication
(A) Schematic representation of the miRNA adenoviral library bioselection procedure. 243 human miRNAs under the control of a CMV promoter were cloned into the adenoviral genome (between the E4 gene and R-ITR) to generate a library (AdwtE miRLib). The library was bioselected through a replication-based strategy in PANC-1 and MIA PaCa-2 cell lines (see the Materials and Methods for a detailed description). As viruses encoding miR-99b and miR-485 were bioselected in both cell lines, they were chosen for further validation. (B) Schematic representation of the candidate viruses, AdwtE miR-99b and AdwtE miR-485, and the control viruses, AdwtE and AdwtE hTR. (C and D) Expression of miR-99b (5p and 3p) (C) and miR-485 (5p and 3p) (D) in PANC-1 cells at 24 hr PI with mock or 10 IFU/cell AdwtE, AdwtE miR-99b, or AdwtE miR-485. (E and F) AdwtE miR-99b and AdwtE miR-485 activity. PANC-1 (E) and MIA PaCa-2 (F) cells were infected (at 1 IFU/cell or 5 IFU/cell, respectively) with AdwtE, AdwtE miR-99b, AdwtE miR-485, or AdwtE hTR. At 48 hr PI, virus-containing supernatants were collected and used to infect new cells (10% for PANC-1 and 50% for MIA PaCa-2). The procedure was repeated for three sequential steps. EGFP expression was analyzed and quantified at the end of each infection. The dashed line represents AdwtE values. Representative images are shown. Original magnification 4×; scale bar, 200 μm. Data are shown as mean ± SEM for at least five independent biological replicates. Significance was assessed using a two-tailed Mann-Whitney test (C and D) and by comparison to AdwtE-infected cells using a one-sample t test and to AdwtE hTR infected cells using a two-tailed Mann-Whitney test (E and F). *p < 0.05, **p < 0.01.
Next, we took advantage of the miRNome profile of 11 PDAC and 3 normal pancreatic tissues, performed by next-generation sequencing (NGS) (Illumina Genome Analyzer), in the context of a previous study focused on the identification of miRNA biomarkers for early disease detection.15 Analysis of miR-99b and miR-485 in this NGS dataset revealed that both miRNAs’ 3′ arms (3p), miR-99b-3p and miR-485-3p, were significantly downregulated in PDAC tumors (Figure S1A). Using The Cancer Genome Atlas (TCGA) dataset (https://tcga-data.nci.nih.gov/docs/publications/tcga/?), we classified patients into groups with high or low expression for each miRNA, and we evaluated relative survival. Interestingly, low miRNA expression of both miRNAs correlated with poor clinical outcome (Figures S1B and S1C). These results suggest that miR-99b and miR-485 may play a role in PDAC tumorigenesis.
Together, the data suggest that miR-99b and miR-485 may be potential regulators of the host response to adenoviral activity. Thus, we considered AdwtE miR-99b and AdwtE miR-485 to be good candidates for further characterization.
Viruses Encoding miR-99b or miR-485 Have Increased Activity in PDAC
Adenoviral clones encoding either miR-99b or miR-485 were selected, expanded, and purified to generate AdwtE miR-99b and AdwtE miR-485. Two control viruses were also generated: the parental virus (AdwtE) and a similar-sized virus of AdwtE miR encoding the human telomerase RNA (hTR) (AdwtE hTR) (Figure 1B).
To verify that candidate miRNAs were expressed from AdwtE miR-99b and AdwtE miR-485 viruses, PANC-1 cells were infected with the corresponding viruses; after 24 hr, specific expression of the corresponding miRNAs was analyzed. We determined that both the 3′ arm (3p) and the 5′ arm (5p) of the miRNA chains were overexpressed, indicating that candidate miRNAs were correctly processed from the viral genome (Figures 1C and 1D).
Next, we examined AdwtE miR-99b and AdwtE miR-485 viral activities in PANC-1 and MIA PaCa-2 cells in comparison to the control viruses (AdwtE and AdwtE hTR). For this, cells were exposed to three consecutive rounds of infection, and EGFP fluorescence levels were evaluated at the end of each infection. Cells infected with the two AdwtE miR viruses displayed higher EGFP levels after the first round, which increased throughout the infection rounds, reaching levels 10-fold higher than AdwtE in PANC-1 cells and 4-fold higher than AdwtE in MIA PaCa-2 cells (Figures 1E and 1F). Similar results were obtained in CP15-Luc, NP-18, Capan-2, HPAF-II, and Hs766T PDAC cells (Figure S2A).
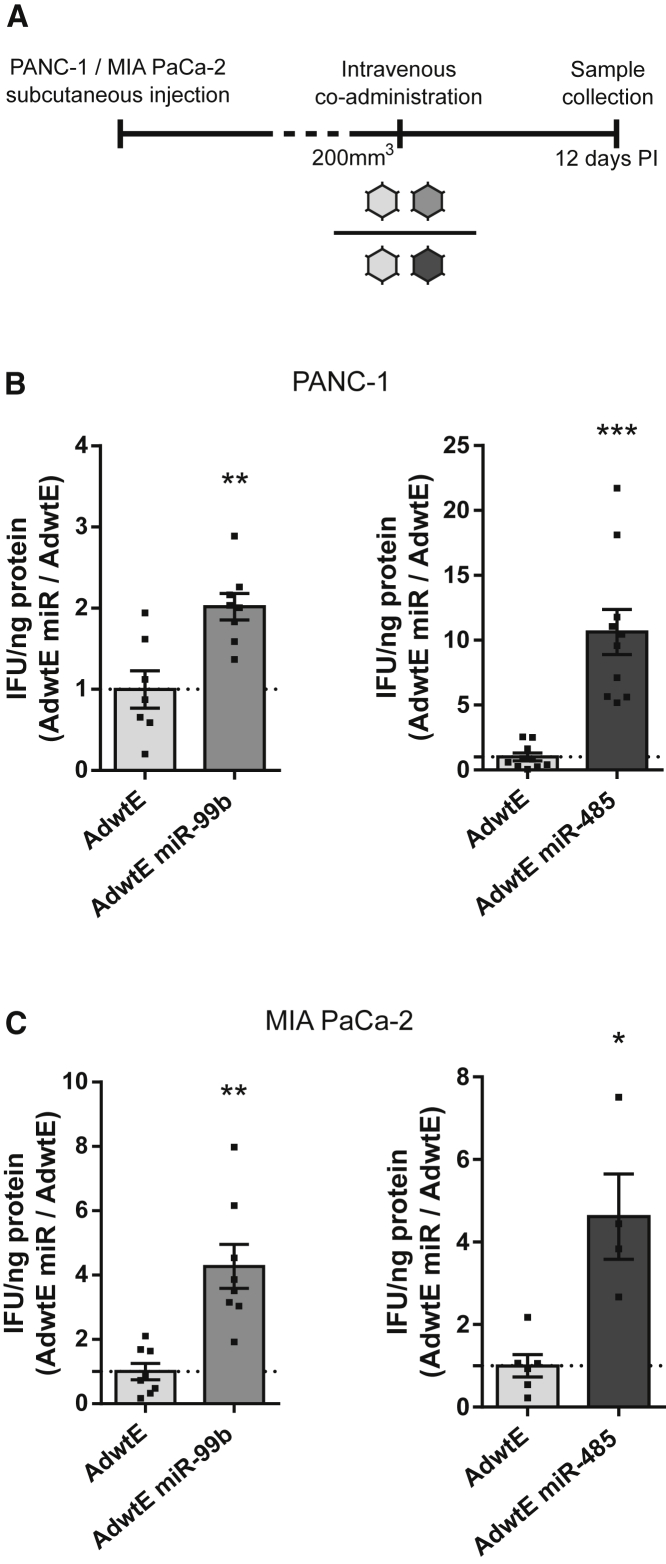
To assess if the enhanced in vitro activity of AdwtE miR viruses could also be recapitulated in vivo, we performed an in vivo competition assay. Animals with subcutaneous PANC-1 or MIA PaCa-2 tumors in the lateral flanks received a mixture of 1:1 AdwtE:AdwtE miR-99b or AdwtE:AdwtE miR-485 through the lateral tail vein. After 12 days, the amount of infective particles of each virus present in the tumors was quantified (Figure 2A). Adenoviruses encoding either of the miRNA candidates were more abundant in both PANC-1 and MIA PaCa-2 tumors (Figures 2B and 2C). In PANC-1 tumors, AdwtE miR-485 was up to 10-fold superior to AdwtE, while AdwtE miR-99b replicated only 2-fold more than AdwtE (Figure 2B). These results coincide with the bioselection approach in which AdwtE miR-485 was the most represented clone in PANC-1 cells (Table S1). In MIA PaCa-2 tumors, miR-99b and miR-485 similarly improved AdwtE fitness by 4-fold (Figure 2C). These results suggest that miR-99b- and miR-485-encoding viruses may trigger a more potent antitumoral activity than the parental unmodified virus.
Figure 2.
AdwtE miR-99b and AdwtE miR-485 Viruses Have Viral Activity Superior to the Parental Virus AdwtE in an In Vivo Context
(A) Scheme of the in vivo competition assay. Mice bearing subcutaneous PANC-1 and MIA PaCa-2 tumors were intravenously administered a 1:1 mixture of AdwtE:AdwtE miR-99b or AdwtE:AdwtE miR-485 (2 × 1010 vp/animal). Tumors were recovered at 12 days PI and the infective viral particles of each virus were quantified. (B and C) Infective viral particle quantification at the end of the competition assay in (B) PANC-1 tumors treated with AdwtE:AdwtE miR-99b (n = 7) or AdwtE:AdwtE miR-485 (n = 10) or in (C) MIA PaCa-2 tumors treated with AdwtE:AdwtE miR-99b (n = 8) or AdwtE:AdwtE miR-485 (n = 4). Data are shown as mean ± SEM. Significance was assessed by comparison to AdwtE infective particles present in tumors using a two-tailed Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-99b and miR-485 Expression Enhances Infective Viral Particle Formation, Viral Release, and In Vitro Cytotoxicity
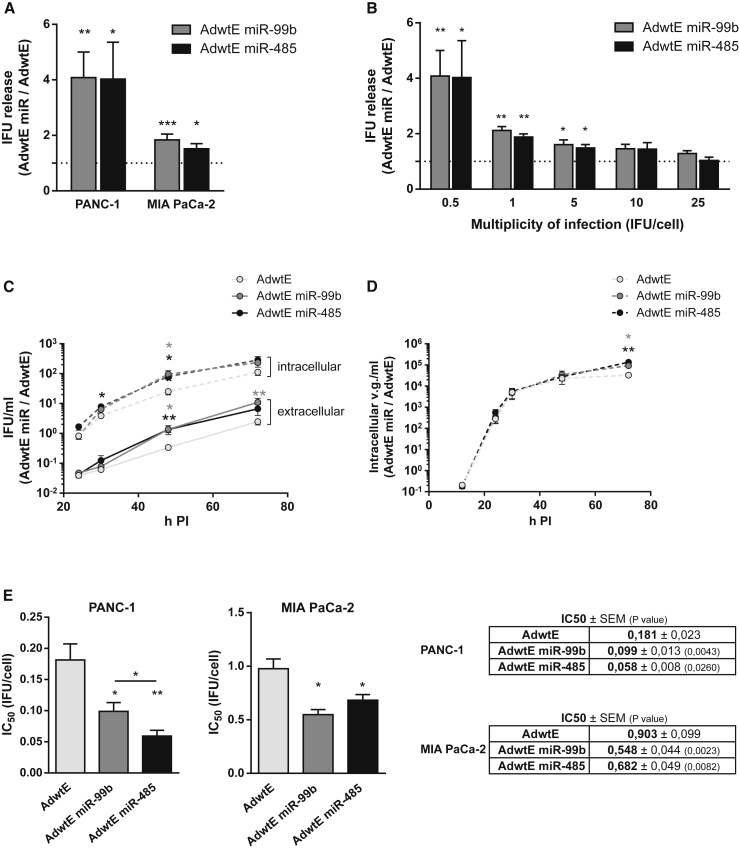
To evaluate the effects of miR-99b and miR-485 on adenoviral replication, PANC-1 and MIA PaCa-2 cells were infected with AdwtE miR-99b and AdwtE miR-485 at a low viral dose; after one replication cycle, the concentration of infective viral particles present in the supernatant was determined, as stated and at 4 hr after infection to ensure equal viral entry between viruses. Indeed, both miRNAs led to higher numbers of released infective particles compared to AdwtE (Figure 3A). This effect was dose dependent. Differences between AdwtE miR viruses and AdwtE diminished at higher infecting doses (Figure 3B), suggesting a saturation effect, where all the viruses already reached the maximum effects.
Figure 3.
miR-99b- and miR-485-Encoding Adenoviruses Display Higher Viral Yield, Correlating with Increased In Vitro Oncolytic Activity
(A) Relative infective viral particle release. PANC-1 and MIA PaCa-2 cells were seeded in triplicate and infected (at 0.5 IFU/cell or 5 IFU/cell, respectively) with AdwtE, AdwtE miR-99b, or AdwtE miR-485. At 48 hr PI, supernatants were collected and titrated by viral infectious units. The dashed line represents AdwtE values. (B) Dose dependence effect on infective viral particles released in PANC-1 cells. Cells were seeded in triplicate and infected with AdwtE, AdwtE miR-99b, or AdwtE miR-485 at a range of doses. At 48 hr PI, supernatants were collected and titrated by viral infectious units. The dashed line represents AdwtE values. (C) Time course of intra- and extracellular infective viral particles in PANC-1 cells. Cells were seeded in triplicate and infected with AdwtE, AdwtE miR-99b, or AdwtE miR-485 at a dose of 0.5 IFU/cell. Cells and supernatants were collected at 24, 30, 48, and 72 hr PI and titrated by viral infectious units. (D) Time course of total intracellular viral genome content in PANC-1 cells. Cells were seeded in triplicate and infected with AdwtE, AdwtE miR-99b, or AdwtE miR-485 at a dose of 0.5 IFU/cell. Cells were collected at 24, 30, 48, and 72 hr PI, and the intracellular viral genomes were quantified by qPCR. (E) In vitro oncolytic activity in PANC-1 and MIA PaCa-2 cells. Cells were seeded in triplicate and infected with a dose range of AdwtE, AdwtE miR-99b, or AdwtE miR-485. Cell viability was measured 7 days PI by (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) assay and IC50 values were determined. Data are shown as mean ± SEM for at least five independent biological replicates. Significance was assessed by comparison to AdwtE-infected cells using a two-tailed Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001.
To confirm that the increase in released infective particles was specific for miR-99b and miR-485 activity, PANC-1 cells were transfected with miRNA expression plasmids (miRVec-99b or miRVec-485) or a control plasmid (miRVec hTR), followed by AdwtE infection. The release of AdwtE infective particles was increased in the presence of either miRNA (Figure S3A). Furthermore, infection of PANC-1 cells with AdwtE miR-759, a miRNA that remains stable during adenovirus infection,10, 11 resulted in a viral release similar to that of the control viruses AdwtE and AdwtE hTR (Figure S3B), indicating that the observed effects of candidate miRNAs were specific and not due to universal miRNA effects, such as a block in the miRNA biogenesis pathway.
We next investigated whether the increase in the number of infective particles in the supernatants was due to an enhanced viral release or to an increase in the number of intracellular particles. PANC-1 cells were infected with AdwtE, AdwtE miR-99b, or AdwtE miR-485, and the amounts of infective viral particles present in the intracellular and extracellular compartments were measured at the indicated time points. Infection with AdwtE miR-99b or AdwtE miR-485 led to a faster release of infective viral particles than did AdwtE. This higher release correlated with a more efficient intracellular content of AdwtE miR infective particles (Figure 3C). In contrast, quantification of intracellular viral genomes showed no differences between viruses at early time points. An increase in the number of intracellular viral genomes was observed at 72 hr post-infection (PI), probably due to the internalization of viral genomes in a second infection cycle (Figure 3D).
The increase in the viral release of AdwtE miR-99b and AdwtE miR-485 led to an enhanced cytotoxic activity, with AdwtE miR-485 leading to a 3-fold decrease in the IC50 values in PANC-1 cells and to a 2-fold decrease in MIA PaCa-2 cells. Similarly, AdwtE miR-99b promoted a 2-fold decrease in the IC50 in both cell lines (Figure 3E). AdwtE miR-485 and AdwtE miR-99b also had significantly reduced IC50 values in CP15-Luc, NP-18, Capan-2, HPAF-II, and Hs766T PDAC cells (Figure S2B). Altogether, these results establish that miR-99b and miR-485 increase the adenoviral fitness through an enhancement in the intracellular content and release of mature virions, thereby conferring increased cytotoxicity to miR candidate-encoding viruses.
miR-99b and miR-485 Increase the Expression of Viral Genes
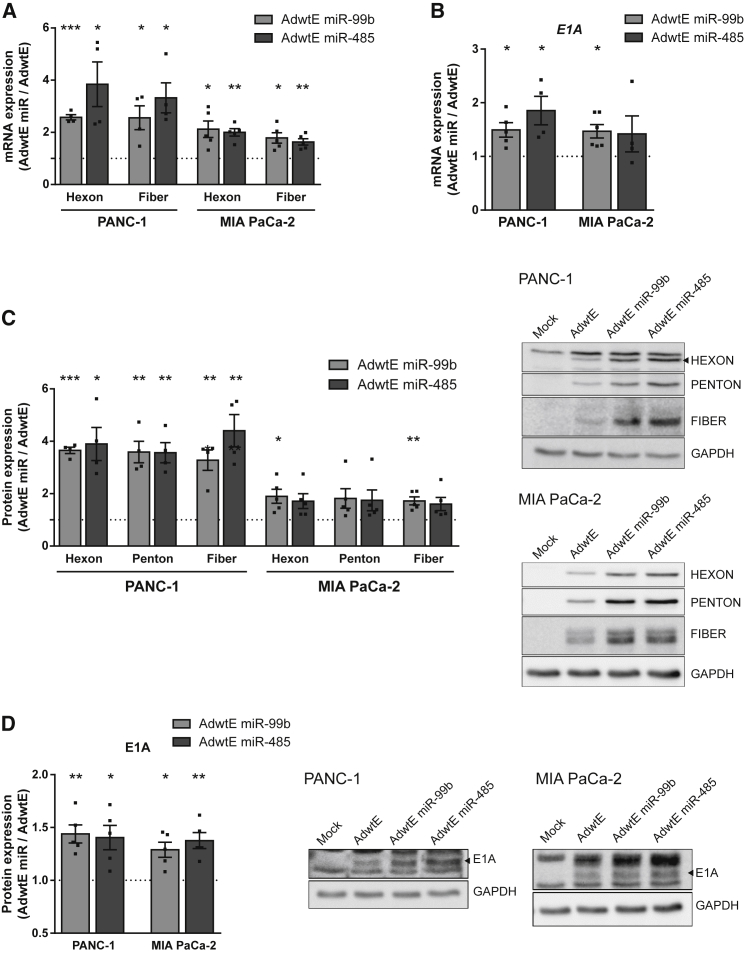
To further characterize AdwtE miR-99b and AdwtE miR-485, we analyzed the mRNA levels of late (hexon and fiber) and immediate early (E1A) genes at 24 or 8 hr PI, respectively, in PANC-1 and MIA PaCa-2 cells. Increased mRNA levels of the three genes were observed in AdwtE miR-infected cells (Figures 4A and 4B). Consistent with these results, higher protein contents of hexon, penton, fiber, and E1A were also detected (Figures 4C and 4D). These results suggest that miR-99b and miR-485 may modulate cellular pathways, affecting both early and late transcription of viral genes.
Figure 4.
The Increase in AdwtE miR-99b and AdwtE miR-485 Infective Particles Is Facilitated by Higher Expression Levels of Viral Genes
(A and B) qPCR analysis of hexon, fiber (24 hr PI) (A), and E1A (8 hr PI) (B) mRNA levels in PANC-1 and MIA PaCa-2 cells infected (at 1 and 5 IFU/cell, respectively) with AdwtE, AdwtE miR-99b, or AdwtE miR-485. mRNA values are given relative to cellular GAPDH in each replicate. The dashed line represents AdwtE values. (C and D) Quantification of hexon, penton, fiber (24 hr PI) (C), and E1A (8 hr PI) (D) protein levels in PANC-1 and MIA PaCa-2 cells infected (at 1 and 5 IFU/cell, respectively) with AdwtE, AdwtE miR-99b, or AdwtE miR-485. Representative western blot images are shown. Quantification of signal was normalized to cellular GAPDH in each replicate. The dashed line represents AdwtE values. Data are shown as mean ± SEM for at least four independent biological replicates. Significance was assessed by comparison to AdwtE-infected cells using a one-sample t test. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-99b and miR-485 Inhibit the Expression of Genes Related to Transcription Regulation
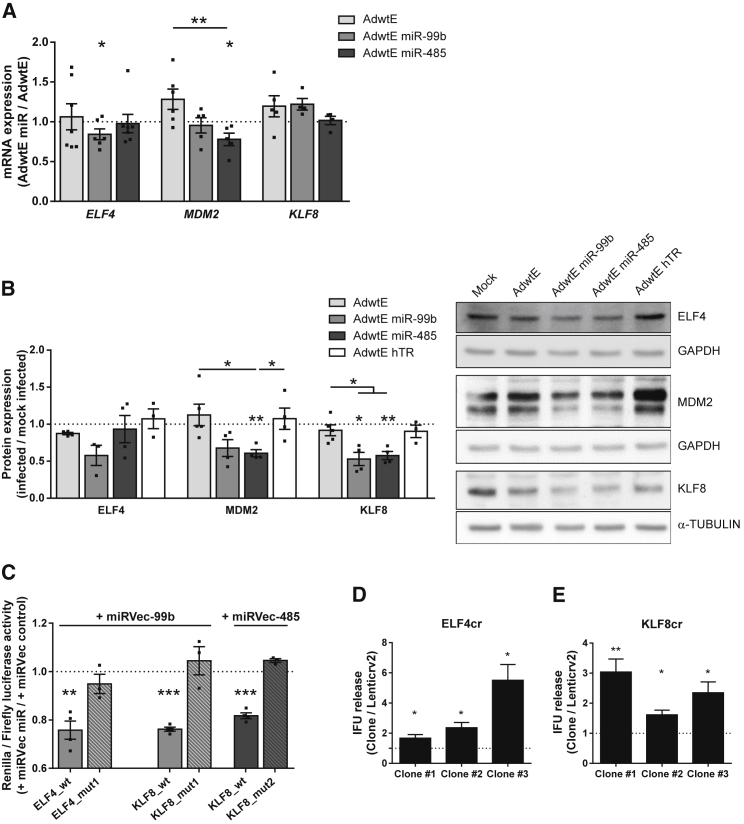
We next performed a bioinformatics study to elucidate the mechanisms by which miR-99b and miR-485 regulate the expression of viral genes (Supplemental Materials and Methods; Figure S4A). The list of potential target genes for miR-99b and miR-485, generated by different miRwalk prediction algorithms, was classified by the biological process subontology of gene ontology (GO) terms. One of the enriched GO terms that was common to both miRNAs and interesting in light of the experimental data was “regulation of gene expression.” Among the genes belonging to this term, we selected those with better prediction scores as targets for miR-99b and miR-485 (Table S2), which we could infer from published data also possessed some activity modulating viral infections or the adenoviral cycle. Thus, we selected the transcriptional regulators E74-like ETS transcription factor 4 (ELF4) and Kruppel-like factor 8 (KLF8), both of which have predicted target sites for miR-99b and miR-485, and MDM2 proto-oncogene (MDM2), a miR-485-validated target gene20 (Figures S4B and S5).
We first investigated whether ELF4, MDM2, and KLF8 gene expression was regulated by the candidate miRNAs. PANC-1 cells were mock infected or infected with control or AdwtE miR viruses, and both mRNA and protein levels were analyzed. Reduction in the ELF4 mRNA and protein levels was observed only in AdwtE miR-99b-infected cells (Figures 5A and 5B). Thus, although ELF4 had a miR-485 prediction site (Figure S4B), we could not validate any effect in our experimental setting. MDM2 expression was significantly reduced in cells infected with either AdwtE miR-485 or AdwtE miR-99b (Figures 5A and 5B). The reduced expression observed after AdwtE miR-99b was unexpected, as MDM2 is not a direct target of miR-99b (Figure S4B). However, MDM2 downregulation could be mediated by limited transcription due to the decrease in its transcriptional regulator ELF4 or by other indirect mechanisms.21 KLF8 expression was significantly reduced in cells infected with either of the miR candidate viruses, as predicted, although it could be detected only at the protein level (Figures 5A and 5B).
Figure 5.
miR-99b and miR-485 Downregulate Genes Involved in Transcriptional Regulation
(A) qPCR analysis of ELF4, MDM2, and KLF8 mRNA levels in PANC-1 cells at 24 hr PI with mock or 10 IFU/cell AdwtE, AdwtE miR-99b, or AdwtE miR-485. mRNA values are expressed relative to cellular GAPDH in each replicate. The dashed line represents mock-infected values. (B) Quantification of ELF4, MDM2, and KLF8 mRNA levels in PANC-1 cells at 24 hr PI with mock or 10 IFU/cell AdwtE, AdwtE miR-99b, AdwtE miR-485, or AdwtE hTR. Representative western blot images are shown. Quantification of signal was normalized to cellular GAPDH for each replicate. The dashed line represents mock-infected values. (C) Validation of the miR-99b target site at ELF4 3′ UTR and miR-99b and miR-485 target sites at KLF8 3′ UTR. HEK293T cells were co-transfected with a psiCHECK-2 reporter plasmid, containing wild-type (ELF4_wt, KLF8_wt) or mutated ELF4 or KLF8 3′ UTRs (ELF4_mut1, KLF8_mut1, and KLF8_mut2) at the 3′ end of the renilla gene, and the corresponding miRNA expression plasmid (miRVec control, miRVec-99b, or miRVec-485). Renilla and luciferase activities were evaluated 72 hr post-transfection. (D and E) Relative infective viral particle release in control PANC-1 cells (Lenticrv2) and three CRISPR/Cas9-modified PANC-1 clones for ELF4 (D) and KLF8 (E). Cells were seeded in triplicate and infected with AdwtE (0.5 IFU/cell). At 48 hr PI, supernatants were collected and titrated by viral infectious units. The dashed line represents PANC-1 Lenticrv2 cells values. For (A) and (B), data are shown as mean ± SEM for at least three independent biological replicates. Significance was assessed by comparison to mock-infected cells using a one-sample t test and to infected cells using a two-tailed Mann-Whitney test. For (C), data are shown as mean ± SEM for at least three independent biological replicates. Significance was assessed by comparison to miRVec control-transfected cells using a one-sample t test. For (D), data are shown as mean ± SEM for at least five independent biological replicates. Significance was assessed by comparison to PANC-1 Lenticrv2 cells using a one-sample t test. *p < 0.05, **p < 0.01, ***p < 0.001.
MDM2 is a known miR-485 target.20 To validate the miR-99b target site for ELF4 and the miR-99b and miR-485 target sites for KLF8, we performed reporter assays. A fragment of the ELF4 or KLF8 3′ UTR, which contains predicted wild-type or mutated miR-binding sites (Figure S5), was cloned into a dual luciferase reporter construct and co-transfected into HEK293T cells with miRVec-99b or miRVec-485 expression plasmids. As shown in Figure 5C, Renilla activity was significantly decreased for ELF4 wild-type 3′ UTR with miR-99b and for KLF8 wild-type 3′ UTR with miR-99b and miR-485. These effects were not observed when miR-binding sites were mutated (Figure 5C; Figure S5). These data confirmed the regulation of ELF4 by miR-99b and of KLF8 by both miR-99b and miR-485.
These results suggest that miR-99b and/or miR-485 modulation of ELF4 and KLF8 could affect adenoviral replication. To test this, we generated ELF4 and KLF8 knockdown PANC-1 cells and infected them with AdwtE. Indeed, AdwtE-infected cells released higher levels of infective particles in all clones with reduced expression of ELF4 or KLF8 (Figures 5D and 5E; Figure S6), thus validating our hypothesis.
miR-99b and miR-485 Confer a Superior In Vitro and In Vivo Activity to the Oncoselective Adenovirus ICOVIR15
We next evaluated whether the improved adenoviral fitness triggered by miR-99b and miR-485 translates into enhanced antitumor activity. To this end, we armed ICOVIR15 with the candidate miRNAs. ICOVIR15 is an Ad5 derivative oncolytic adenovirus that keeps intact all the virus functions, with modifications in the E1A and fiber genes. The E1A region contains a 24-bp deletion and incorporates E2F-responsive elements to redirect E1A transcription toward deregulation of the Rb/p16 pathway. The fiber contains an RGD motif inserted in the HI-loop region.22
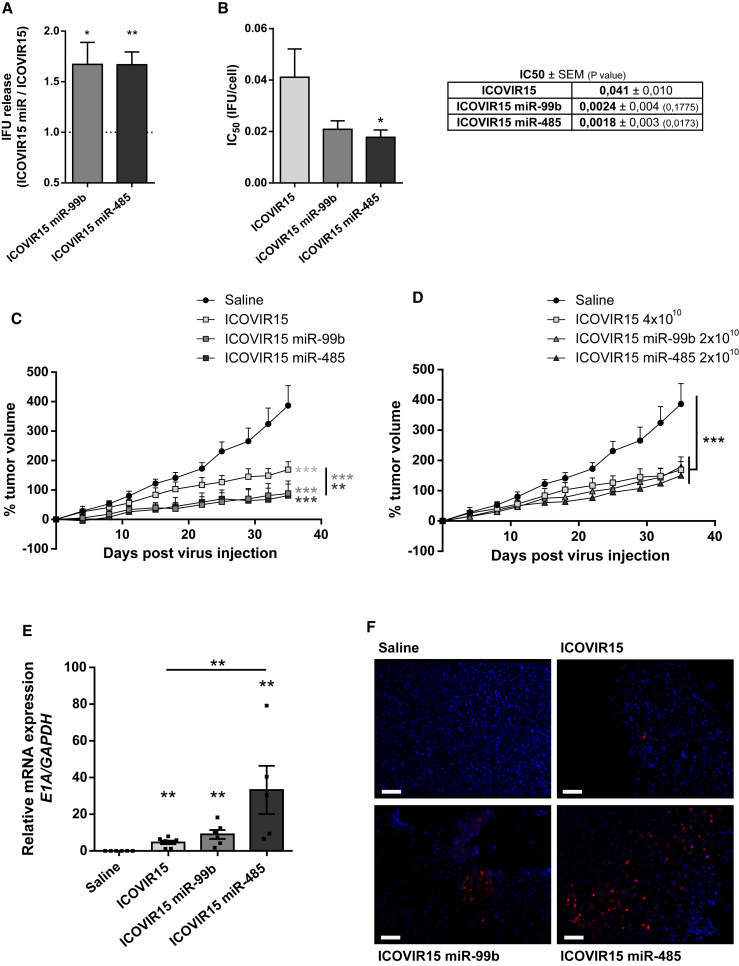
miR-99b and miR-485 human genomic sequences were incorporated into an ICOVIR15 genome under a CMV promoter next to the R-ITR. First, we tested the in vitro activity of these miR candidate-encoding viruses. In line with previous results with AdwtE miR viruses, ICOVIR15 miR-99b and ICOVIR15 miR-485 displayed a significant increase in infective particles released from PANC-1 cells, and they had a 2-fold increase in cytotoxicity as compared to the parental virus ICOVIR15 (Figures 6A and 6B). Similar data were obtained from MIA PaCa-2 cells (Figure S7).
Figure 6.
miR-99b and miR-485 Confer Increased Oncolytic Activity and Enhanced Antitumoral Response to ICOVIR15
(A) Relative infective viral particle release. PANC-1 cells were seeded in triplicate and infected with ICOVIR15, ICOVIR15 miR-99b, or ICOVIR15 miR-485 (0.5 IFU/cell). At 48 hr PI, supernatants were collected and titrated by viral infectious units. The dashed line represents ICOVIR15 values. (B) In vitro oncolytic activity in PANC-1 cells. Cells were seeded in triplicate and infected with a dose range of ICOVIR15, ICOVIR15 miR-99b, or ICOVIR15 miR-485. Cell viability was measured at 7 days PI by MTT assay and IC50 values were determined. (C and D) Mice bearing subcutaneous PANC-1 tumors were intravenously administered with saline solution; 4 × 1010 vp/animal of ICOVIR15, ICOVIR15 miR-99b, or ICOVIR15 miR-485 (C); or 4 × 1010 vp/animal of ICOVIR15, 2 × 1010 vp/animal of ICOVIR15 miR-99b, or 2 × 1010 vp/animal of ICOVIR15 miR-485 (D) (n ≥ 7 tumors/treatment group). Samples were collected 35 days after treatment. Follow-up of PANC-1 tumor volumes is represented as percentage of growth. *p < 0.05, **p < 0.01, ***p < 0.001. (E) qPCR analysis of E1A mRNA levels in PANC-1-treated tumors. mRNA values are expressed as relative to GAPDH in each replicate. (F) E1A immunofluorescence of PANC-1-treated tumors (blue, DAPI; red, E1A). Representative images are shown; scale bar, 100 μm. Data are shown as mean ± SEM for at least five independent biological replicates. Significance was assessed using a two-tailed Mann-Whitney test. *p < 0.05, **p < 0.01, ***p < 0.001.
To evaluate the antitumor effects of ICOVIR15 miR viruses, mice with subcutaneous PANC-1 tumors were treated with a systemic dose of saline solution, ICOVIR15, ICOVIR15 miR-99b, or ICOVIR15 miR-485 at 4 × 1010 viral particles (vp)/animal. Treatment with ICOVIR15 miR viruses strongly inhibited tumor growth that was consistently higher than inhibition observed using the parental ICOVIR15 (Figure 6C). Interestingly, ICOVIR15 miR viruses administered at 2 × 1010 vp/animal displayed the same antitumoral activity as ICOVIR15 administered at the double dose (4 × 1010 vp/animal) (Figure 6D). The presence of replicating active adenovirus in tumors was determined by analyzing E1A expression by qRT-PCR and by immunostaining at the end of the experiment. Tumors treated with ICOVIR15 miR viruses, and especially with ICOVIR15 miR-485, displayed increased E1A mRNA levels (Figure 6E). Larger areas of adenovirus replication were also observed in the tumors that were treated with ICOVIR15 miR viruses (Figure 6F). These data strongly suggest that miR-99b and miR-485 confer enhanced potency to the ICOVIR15 oncolytic adenovirus through increased adenoviral release.
Discussion
Oncolytic adenoviruses are therapeutics under clinical development. Nonetheless, although they display good safety profiles, their oncolytic activity does not fully eliminate tumors,17 highlighting the need for novel adenoviral designs with increased potency. miRNAs have been shown to play a role in the progression of the adenoviral cycle.9, 10, 11 However, it remains unclear how adenoviral replication is affected by the extensively deregulated miRNA profile of tumor cells. We hypothesized that, by expressing specific miRNAs, it would be feasible to re-establish the levels of key cellular genes relevant for productive viral infection, leading to improved adenoviral oncolysis.
Adenoviruses have the potential of being armed with sequences of interest that provide additional functions. We performed an approach based on the generation of an adenoviral miRNA library encoding 243 human miRNAs, followed by a replication-based bioselection strategy that identified miR-99b and miR-485 as enhancers of the adenoviral activity in pancreatic cancer cells. A similar strategy has been previously used in the context of an in vivo alpha virus infection to identify interactions between the virus and host factors impacting viral replication.23 However, the designs of these approaches were substantially different, since they were not expressing specific miRNAs but hairpin sequences designed to target unique murine open reading frames. Our strategy is also distinct from that of a previous work studying miRNA effects on adenovirus propagation, in which the modulation of miRNA content was dissociated from adenoviral infection as miRNAs were introduced into cells 1 day before adenoviral infection.11 Here, by incorporating the miRNAs directly into the adenoviral genome, we were able to couple their expression with the infection, forcing cancer cells to parse out those factors that are important for the adenoviral replication cycle. The temporal differences in cellular miRNA expression, and probably the tumor model of pancreatic versus prostate cancer, may at least partially explain the identification of different miRNAs regulating adenoviral activity.
Our data show that miR-99b and miR-485 improved the intracellular formation and led to the earlier release of infective virions, leading to enhanced lytic effects. Changes in the expression of these two miRNAs in response to viral infections have been documented for other viruses. During an influenza virus infection, miR-485 expression increases in order to reduce RIG-1 levels and to diminish the induction of downstream antiviral proteins.24 On the other hand, miR-99 family members facilitate hepatitis B virus (HBV) replication in hepatoma cells by increasing autophagic activity and by promoting HBV protein production.25 Thus, miR-99b and miR-485 seem to have a proviral function that supports their bioselection during our screening strategy.
We observed that the infection of pancreatic cancer cells with AdwtE miR-99b and AdwtE miR-485 was associated with higher expression of viral genes encoding structural proteins (namely, hexon, penton, and fiber) and the E1A gene, both at the RNA and protein levels, without changes in the content of total viral genomes. This suggests that the miR-99b and miR-485 effects were probably related to the transcriptional activation of viral genes. This induction was superior in the late genes, probably because the increase in E1A expression facilitates the transactivation of late genes through its binding to the MLP.26
Through bioinformatics analysis, we identified potential miRNA target genes for both miR-99b and miR-485 belonging to the transcription factor family of proteins. In the current study, ELF4 was validated as a target gene of miR-99b and KLF8 as that of both miR-99b and miR-485. MDM2 has already been described as a validated target for miR-485. Infection of pancreatic cancer cells with AdwtE miR-99b and AdwtE miR-485 resulted in decreased expression of these three proteins, in line with their regulation by miR-99b and/or miR-485. Interestingly, MDM2 is an adenoviral limiting factor.7, 20, 27 Importantly, we show that genetic downregulation of ELF4 or KLF8 resulted in enhanced release of adenoviral particles, pointing to a role for them in adenoviral activity modulation. KLF8 is a CACCC (GT-box)-binding dual-transcription factor, and its binding sites have been identified at or next to the promoters of different adenoviral genes, including two sites at the left ITR (L-ITR) and two in the MLP. In this study, we could validate that KLF8 reduces transcription from the adenoviral L-ITR/E1A promoter and MLP. Promoter reporter assays showed higher transcriptional activity in cells with downregulated KLF8 than in cells expressing KLF8 (Figure S8). These results support the view that, through the recruitment of the C-terminal-binding protein (CtBP), KLF8 represses the expression of these genes.28 Indeed, CtBP2 has been previously identified as an adenoviral limiting factor.29, 30
Furthermore, KLF8 recruits p300 and PCAF co-activators to promoters.31 To promote productive virus infection, E1A removes CtBP2 from repressed promoters, sequesters p300 from transcriptional active regions, and redirects it together with Rb to selected host genes, thereby repressing their expression.29, 32 In those cases, high levels of KLF8 might compete with E1A and lead to reduced adenoviral yields. ELF4 is an ETS transcription factor that becomes activated during the antiviral response and in cells with oncogenic Ras.33 ELF4 promotes MDM2 expression, which could explain, at least in part, why MDM2 is downregulated in AdwtE miR-99b-infected cells even though it is not a target for this miRNA.21 The effects of ELF4 on the adenoviral activity might be explained by its regulation of MDM2. In fact, different hypotheses have been proposed to explain MDM2 antiviral activity. MDM2 might send early adenoviral proteins (E1A and E1B-55K) or cellular proteins required for a productive viral cycle (PP2A) to proteasomal degradation,27, 34 but it can also induce the expression of CtBP235 or act as a transcriptional repressor by interacting with the 34K subunit of TFIIE.36 Thus, we propose that the downregulation of KLF8, ELF4, and MDM2 through miR-99b or miR-485 expression promotes the expression of the adenoviral late genes and E1A, which in turn facilitates the adenoviral life cycle.
The expression of miR-99b and miR-485 may also provide pancreatic cancer with tumor suppressor activities. Interestingly, we observed that miR-99b and miR-485 were poorly expressed in PDAC samples and that their reduced expression correlated with poorer survival rates of patients. Thus, the enhanced antitumor activity of ICOVIR15 miR viruses might be the result of miR-99b and miR-485 contributing to improve the adenoviral fitness and their tumor-suppressive effects.
Our work was restricted to pancreatic cancer; however, other authors have identified a contribution of miR-99b and miR-485 in the progression of different neoplasias. Low miR-99b expression has been associated with advanced stages of glioma and with the presence of lymphatic metastasis in cervical cancer.37, 38 A metastatic inhibitor role has been proposed for miR-485 in lung adenocarcinoma and breast cancer.39, 40 Also, low miR-485 expression has been found to correlate with poor prognosis in gastric cancer.41 Moreover, high expression levels of either KLF8 or ELF4 have been associated with pro-tumorigenic processes, suggesting that their downregulation may also be beneficial for controlling tumor growth.42, 43, 44 Altogether, these findings indicate that the expression of miR-99b and miR-485 from the adenoviral genome produces a significant antitumor effect as a consequence of the enhanced adenoviral activity, with a potential contribution of their tumor suppressor activity. The similarities in miR-99b and/or miR-485 alterations between cancer cells of different origins allow us to speculate that the improved antitumoral response of ICOVIR15-miR99b and ICOVIR15-miR-485 observed in PDAC applies to other solid tumors, which would make them very attractive anticancer agents.
Materials and Methods
Cell Lines
PANC-1, MIA PaCa-2, Capan-2, HPAF-II, Hs766T, A549, HEK293, and HEK293T cell lines were obtained from the American Type Culture Collection (ATCC). Luciferase-expressing cells (CP15-Luc) were established by transducing the parental cell line CP15 (derived from CP15 tumors) with a recombinant retrovirus pLHCluc.45 NP-18 cells were established and provided by Dr. Capellà.46 Cells were cultured in DMEM (Gibco-BRL) supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (100 μg/mL), and streptomycin (100 μg/mL) (Gibco-BRL), and they were maintained in a humidified atmosphere of 5% CO2 at 37°C. Interspecies contamination and cell morphology were evaluated by microscopic observation. Cell lines obtained from the ATCC were immediately expanded and frozen. Every 2 months cells were plated again from the original batch. Cells were not authenticated by the authors. Interspecies contamination was tested by PCR routinely.
Expression and Reporter Plasmids
miRVec plasmids were obtained from NKI human miRNA library (miRLib) bacteria cells grown in Luria-Bertani broth (LB) containing ampicillin (100 μg/mL). The miRLib library was generated by Voorhoeve et al.47 and commercialized by Source BioScience LifeSciences. psiCHECK-2_ELF4_wt, psiCHECK-2_ELF4_mut1, psiCHECK-2_KLF8_wt, psiCHECK-2_KLF8_mut1, and psiCHECK-2_KLF8_mut2 were generated by cloning the corresponding 3′ UTR fragments (genomic DNA: ELF4, 45,233–45654 bp; KLF8, 53,454–54,679 bp) into psiCHECK-2 vector. The 3′ UTR fragments contained XhoI and NcoI restriction sites at their 5′ and 3′ ends, respectively, and they were purchased as G-Blocks (Integrated DNA Technologies). G-blocks were digested with XhoI and NcoI restriction enzymes (Roche) and introduced into the psiCHECK-2 vector, digested with the same restriction enzymes. Plasmid constructions were tested by colony PCR and validated by Sanger sequencing using the primer set 1 (Table S3).
Adenovirus Generation and Titration
pAdwtE hTR, pAdwtE miR-759, pICOVIR15 miR-99b, and pICOVIR15 miR-485 were generated by an adapted recombineering protocol, based on homologous recombination in bacteria using a positive-negative selection screen with the RpsLNeo cassette.48, 49 Primer sets used for the recombination step contain long tails homologous to the insertion point at the adenoviral genome (Table S3). The RpsLNeo cassette was amplified by PCR from the pJetRpsL plasmid (with primer set 2), and it was introduced by homologous recombination downstream of the adenoviral fiber gene at the adenoviral genome (pAdwt). The EGFP gene was then amplified (with primer set 3). The RpsLNeo cassette was replaced with the EGFP amplicon by homologous recombination, generating the adenoviral genome pAdwtE. The RpsLNeo cassette was then amplified by PCR from the pJetRpsL plasmid (with primer set 4), and it was introduced by homologous recombination 3 bp upstream of the R-ITR in the reverse strand of the corresponding adenoviral genome (pAdwtE or pICOVIR15).
Sequences encoding for hTR, miR-759, miR-99b, or miR-485, as well as the CMV promoter regulating their expression, were amplified from the corresponding miRVec plasmid by PCR (with primer set 5). The RpsLNeo cassette was replaced by each one of the amplicons by homologous recombination. pAdwtE miRLib was generated by replacing the RpsLNeo cassette with a mixture of 243 amplicons amplified from miRVec plasmids by PCR (with primer set 5). Plasmid constructions were verified by PCR (with primer sets 6 and 7), EcoRI enzymatic digestion, and Sanger sequencing. The pAdwtE, pAdwtE hTR, pAdwtE miR-759, pICOVIR15, pICOVIR15 miR-99b, and pICOVIR15 miR-485 plasmids were transfected into HEK293 cells to obtain a first round of viral particles. Viruses were propagated in A549 cells and purified by cesium chloride density gradient centrifugation according to standard techniques.50 Adenoviral concentrations were calculated based on optical density (vp/mL) or on viral infectious units (IFU/mL), as previously described.51
AdwtE miRLib Bioselection and Identification of Isolated Clones
pAdwtE miRLib plasmids were transfected in HEK293 cells to obtain a first viral homogenate and titrated based on viral IFU, as previously described.51 PANC-1 and MIA PaCa-2 cells were infected with 1 IFU/cell and 5 IFU/cell of the viral homogenate, respectively. When cytopathic effect (CPE) was observed, 10% of the supernatant from PANC-1 and 30%–70% from MIA PaCa-2 were used to infect new cells; 20 rounds of infection were carried out. Individual adenoviral clones were isolated by plaque assay in PANC-1 cells (66 clones) or in A549 cells (45 clones) (for MIA PaCa-2 bioselected viruses).52 Plaque assays from MIA PaCa-2 bioselected clones was carried out in A549 cells since MIA PaCa-2 cells do not grow well at high confluency, a characteristic that is recommended for plaque assay. The largest isolated clones were selected and tested for miRNA identification. Encoded miRNAs were identified by Sanger sequencing using the primer set 7 (Table S3) in the purified viral genomes.
Quantification of Viral Genomes and Infective Viral Particles
Cellular DNA and viral DNA were obtained from cellular extracts using UltraClean BloodSpin DNA Isolation Kit. Adenoviral genomes were analyzed by qPCR on a ViiA7 System (Applied Biosystems), using SYBER Green I Master plus mix (Roche) and the primer set 4 (Table S4). The adenoviral copy number was quantified with a standard curve of adenovirus DNA dilutions (using 102–107 copies) in a background of genomic DNA. Viral DNA was expressed relative to the cellular DNA content, determined by qPCR using albumin 12 intron primers (primer set 5; Table S4) and a standard curve of genomic DNA and corrected by the number of cells per well. Infective viral particles were determined by viral DNA quantification in A549 at 4 hr PI.
miRNA qRT-PCR
Total RNA was obtained from cell cultures using miRNeasy Mini RNA Extraction Kit (QIAGEN). About 10 ng total RNA was reverse transcribed using Applied Biosystems TaqMan MicroRNA Reverse Transcription Kit and stem-loop primers (Thermo Fisher Scientific), according to the manufacturers’ instructions. Applied Biosystems TaqMan Universal PCR Master Mix No AmpErase UNG, qPCR probes (Thermo Fisher Scientific), and 1.5 μL RT reaction were used for the qPCR amplification reaction, performed as indicated by the manufacturer. Expression data were normalized to small nucleolar RNA U6 expression (RNU6B). The following stem-loop primers and qPCR probes were purchased from Applied Biosystems TaqMan MicroRNA Assay (Thermo Fisher Scientific): RNU6B (001093), hsa-miR-99b* (002196), hsa-miR-99b (000436), hsa-miR-485-3p (001277), and hsa-miR-485-5p (001036). Reactions were performed on a ViiA7 System.
In Vivo Competition Assay
Subcutaneous tumors were generated in 8-week-old male athymic nude Foxn 1nu nu/nu mice (ENVIGO) by injecting 2 × 106 PANC-1 or MIA PaCa-2 cells embedded in a ratio 1:1 of Matrigel (BD Biosciences) into each flank. Tumors were measured twice per week, and their volumes were calculated using the formula . Mice were randomly assigned to different groups of treatment. Once tumor volume reached a median of 200 mm3, mice were intravenously administered a 1:1 mixture of AdwtE:AdwtE miR-99b or AdwtE:AdwtE miR-485 (2 × 1010 vp/animal). Animals were sacrificed 12 days after virus administration and tumors were collected. To determine the number of infective particles of each virus, tumors were mechanically homogenized in PBS 1×, followed by three freeze-thaw cycles. Cell debris was eliminated by centrifugation, and a fraction of the supernatant was titrated (as above) using specific primer sets for each virus (Table S2). The results were corrected by the sample protein content, as determined by the BCA Protein Assay Kit (Thermo Fisher Scientific). Animal procedures met the guidelines of European Community Directive 86/609/EEC and were approved by the Local Ethical Committee.
Antitumoral Efficacy Study
PANC-1 subcutaneous tumors were established as described above. Mice were randomly assigned to different groups of treatment, and they were intravenously administered with a single dose of saline solution or virus (2 × 1010 or 4 × 1010 vp/animal) when tumor volumes reached a median of 100 mm3. Tumors were measured twice per week. Animals were euthanized 40 days after virus administration and tumors were collected.
Statistical Analysis
Experimental data are represented by the mean ± SEM of at least three independent experiments. Statistical analysis was performed on GraphPad Prism version (v.)6.01. Statistical differences were evaluated using a 2-tailed non-parametric Mann-Whitney test or a one-sample t test; p < 0.05 was taken as the level of significance. The in vivo tumor growth statistical analysis was evaluated using R v.2.14.1 software with a linear mixed-effect model using the Ime4 package. Statistical differences were evaluated using a multiple comparison of means by Tukey contrasts; p < 0.05 was taken as the level of significance.
Author Contributions
M.R.-R. designed and performed the experiments and contributed to manuscript writing. G.R. contributed to the CRISPR-Cas9 experiments and cytotoxicity studies. M.A.M. contributed to target validation and nanoparticle experiments. M.G. provided miRNA NGS data of PDAC and healthy tissue. R.A. provided some reagents and expertise and contributed to manuscript writing. C.F. coordinated the study and wrote the manuscript.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We thank Sagetis Biotech for kindly providing oligopeptide-modified poly(β-amino ester)s (OM-pBAE) polymers. This work was developed at the Centro Esther Koplowitz, Barcelona, Spain. M.R.-R. and G.R. are recipients of an FPI predoctoral contract (BES-2012-053726 and BES-2015-071612) from MINECO, Spain. This work was supported by grants to C.F. from the Spanish Ministry of Economia y Competitividad (BIO2014-57716-C2-R and BIO2017-89754-C2-2R), with partial support from the Generalitat de Catalunya (SGR14/248, SGR17/861, and 2014/LLAVOR0061). CIBERER and CIBEREHD are an initiative of the ISCIII. The C.F. group was partially financed by the Instituto de Salud Carlos III (IIS10/00014) and co-financed by Fondo Europeo de Desarrollo Regional (FEDER); the C.F. group also acknowledges the support of COST Action BM1204 EUPancreas and the Spanish Adenovirus Network (AdenoNet, BIO2015-68990-REDT). We also acknowledge the support of CERCA Programme/Generalitat de Catalunya. This work has been developed in the context of ADVANCE(CAT) with the support of ACCIÓ (Catalonia Trade & Investment; Generalitat de Catalunya) and the European community under the Catalonian European Regional Development Fund operational program 2014–2020.
Footnotes
Supplemental Information includes eight figures, five tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.09.016.
Supplemental Information
References
- 1.Goodwin C.M., Xu S., Munger J. Stealing the Keys to the Kitchen: Viral Manipulation of the Host Cell Metabolic Network. Trends Microbiol. 2015;23:789–798. doi: 10.1016/j.tim.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller D.L., Myers C.L., Rickards B., Coller H.A., Flint S.J. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 2007;8:R58. doi: 10.1186/gb-2007-8-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell S.J., Peng K.-W., Bell J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012;30:658–670. doi: 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorer D.E., Holtrup F., Fellenberg K., Kaufmann J.K., Engelhardt S., Hoheisel J.D., Nettelbeck D.M. Replication and virus-induced transcriptome of HAdV-5 in normal host cells versus cancer cells–differences of relevance for adenoviral oncolysis. PLoS One. 2011;6:e27934. doi: 10.1371/journal.pone.0027934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monsurrò V., Beghelli S., Wang R., Barbi S., Coin S., Di Pasquale G., Bersani S., Castellucci M., Sorio C., Eleuteri S. Anti-viral state segregates two molecular phenotypes of pancreatic adenocarcinoma: potential relevance for adenoviral gene therapy. J. Transl. Med. 2010;8:10. doi: 10.1186/1479-5876-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moerdyk-Schauwecker M., Shah N.R., Murphy A.M., Hastie E., Mukherjee P., Grdzelishvili V.Z. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: role of type I interferon signaling. Virology. 2013;436:221–234. doi: 10.1016/j.virol.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Beusechem V.W., van den Doel P.B., Gerritsen W.R. Conditionally replicative adenovirus expressing degradation-resistant p53 for enhanced oncolysis of human cancer cells overexpressing murine double minute 2. Mol. Cancer Ther. 2005;4:1013–1018. doi: 10.1158/1535-7163.MCT-05-0010. [DOI] [PubMed] [Google Scholar]
- 8.Mahoney D.J., Stojdl D.F. A call to arms: using RNAi screening to improve oncolytic viral therapy. Cytokine Growth Factor Rev. 2010;21:161–167. doi: 10.1016/j.cytogfr.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Bofill-De Ros X., Rovira-Rigau M., Fillat C. Implications of MicroRNAs in Oncolytic Virotherapy. Front. Oncol. 2017;7:142. doi: 10.3389/fonc.2017.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H., Chen M., Tellgren-Roth C., Pettersson U. Fluctuating expression of microRNAs in adenovirus infected cells. Virology. 2015;478:99–111. doi: 10.1016/j.virol.2015.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Hodzic J., Sie D., Vermeulen A., van Beusechem V.W. Functional screening identifies human miRNAs that modulate adenovirus propagation in prostate cancer cells. Hum. Gene Ther. 2017;28:766–780. doi: 10.1089/hum.2016.143. [DOI] [PubMed] [Google Scholar]
- 12.Lagos D., Pollara G., Henderson S., Gratrix F., Fabani M., Milne R.S.B., Gotch F., Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 13.Lujambio A., Lowe S.W. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vila-Navarro E., Vila-Casadesús M., Moreira L., Duran-Sanchon S., Sinha R., Ginés À., Fernández-Esparrach G., Miquel R., Cuatrecasas M., Castells A. MicroRNAs for Detection of Pancreatic Neoplasia: Biomarker Discovery by Next-generation Sequencing and Validation in 2 Independent Cohorts. Ann. Surg. 2017;265:1226–1234. doi: 10.1097/SLA.0000000000001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giovannetti E., Funel N., Peters G.J., Del Chiaro M., Erozenci L.A., Vasile E., Leon L.G., Pollina L.E., Groen A., Falcone A. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 17.Rosewell Shaw A., Suzuki M. Recent advances in oncolytic adenovirus therapies for cancer. Curr. Opin. Virol. 2016;21:9–15. doi: 10.1016/j.coviro.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva E., Navarro P., Rovira-Rigau M., Sibilio A., Méndez R., Fillat C. Translational reprogramming in tumour cells can generate oncoselectivity in viral therapies. Nat. Commun. 2017;8:14833. doi: 10.1038/ncomms14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi Y., Tu J., Cui L., Guo X., Shi Z., Li S., Shi W., Shan Y., Ge Y., Shan J. High-throughput sequencing of microRNAs in adenovirus type 3 infected human laryngeal epithelial cells. J. Biomed. Biotechnol. 2010;2010:915980. doi: 10.1155/2010/915980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratovitski E.A. Phospho-ΔNp63α/microRNA network modulates epigenetic regulatory enzymes in squamous cell carcinomas. Cell Cycle. 2014;13:749–761. doi: 10.4161/cc.27676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sashida G., Liu Y., Elf S., Miyata Y., Ohyashiki K., Izumi M., Menendez S., Nimer S.D. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol. Cell. Biol. 2009;29:3687–3699. doi: 10.1128/MCB.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas J.J., Guedan S., Searle P.F., Martinez-Quintanilla J., Gil-Hoyos R., Alcayaga-Miranda F., Cascallo M., Alemany R. Minimal RB-responsive E1A promoter modification to attain potency, selectivity, and transgene-arming capacity in oncolytic adenoviruses. Mol. Ther. 2010;18:1960–1971. doi: 10.1038/mt.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varble A., Benitez A.A., Schmid S., Sachs D., Shim J.V., Rodriguez-Barrueco R., Panis M., Crumiller M., Silva J.M., Sachidanandam R., tenOever B.R. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe. 2013;14:346–356. doi: 10.1016/j.chom.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Ingle H., Kumar S., Raut A.A., Mishra A., Kulkarni D.D., Kameyama T., Takaoka A., Akira S., Kumar H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8:ra126. doi: 10.1126/scisignal.aab3183. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y., Deng W., Pang J., Kemper T., Hu J., Yin J., Zhang J., Lu M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell. Microbiol. 2017;19:e12709. doi: 10.1111/cmi.12709. [DOI] [PubMed] [Google Scholar]
- 26.Royds J.A., Hibma M., Dix B.R., Hananeia L., Russell I.A., Wiles A., Wynford-Thomas D., Braithwaite A.W. p53 promotes adenoviral replication and increases late viral gene expression. Oncogene. 2006;25:1509–1520. doi: 10.1038/sj.onc.1209185. [DOI] [PubMed] [Google Scholar]
- 27.Yang H., Zheng Z., Zhao L.Y., Li Q., Liao D. Downregulation of Mdm2 and Mdm4 enhances viral gene expression during adenovirus infection. Cell Cycle. 2012;11:582–593. doi: 10.4161/cc.11.3.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Vliet J., Turner J., Crossley M. Human Krüppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grand R.J.A., Baker C., Barral P.M., Bruton R.K., Parkhill J., Szestak T., Gallimore P.H. The significance of the CtBP – AdE1A interaction during viral infection and transformation. In: Chinnadurai G., editor. GtBP Family Proteins. Springer; 2007. pp. 44–60. [Google Scholar]
- 30.Subramanian T., Zhao L.J., Chinnadurai G. Interaction of CtBP with adenovirus E1A suppresses immortalization of primary epithelial cells and enhances virus replication during productive infection. Virology. 2013;443:313–320. doi: 10.1016/j.virol.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urvalek A.M., Wang X., Lu H., Zhao J. KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription. Cell Cycle. 2010;9:601–611. doi: 10.4161/cc.9.3.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrari R., Gou D., Jawdekar G., Johnson S.A., Nava M., Su T., Yousef A.F., Zemke N.R., Pellegrini M., Kurdistani S.K., Berk A.J. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe. 2014;16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suico M.A., Shuto T., Kai H. Roles and regulations of the ETS transcription factor ELF4/MEF. J. Mol. Cell Biol. 2017;9:168–177. doi: 10.1093/jmcb/mjw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riley M.F., Lozano G. The Many Faces of MDM2 Binding Partners. Genes Cancer. 2012;3:226–239. doi: 10.1177/1947601912455322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Barrios O., Győrffy B., Fernández-Aceñero M.J., Sánchez-Tilló E., Sánchez-Moral L., Siles L., Esteve-Arenys A., Roué G., Casal J.I., Darling D.S. ZEB1-induced tumourigenesis requires senescence inhibition via activation of DKK1/mutant p53/Mdm2/CtBP and repression of macroH2A1. Gut. 2017;66:666–682. doi: 10.1136/gutjnl-2015-310838. [DOI] [PubMed] [Google Scholar]
- 36.Thut C.J., Goodrich J.A., Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M., Guo Y., Wu J., Chen F., Dai Z., Fan S., Li P., Song T. Roles of microRNA-99 family in human glioma. OncoTargets Ther. 2016;9:3613–3619. doi: 10.2147/OTT.S99363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Chang L., Li Z., Gao Q., Cai D., Tian Y., Zeng L., Li M. miR-99a and -99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Med. Oncol. 2014;31:934. doi: 10.1007/s12032-014-0934-3. [DOI] [PubMed] [Google Scholar]
- 39.Mou X., Liu S. MiR-485 inhibits metastasis and EMT of lung adenocarcinoma by targeting Flot2. Biochem. Biophys. Res. Commun. 2016;477:521–526. doi: 10.1016/j.bbrc.2016.04.043. [DOI] [PubMed] [Google Scholar]
- 40.Lou C., Xiao M., Cheng S., Lu X., Jia S., Ren Y., Li Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016;7:e2159. doi: 10.1038/cddis.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jing L.-L., Mo X.-M. Reduced miR-485-5p expression predicts poor prognosis in patients with gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2016;20:1516–1520. [PubMed] [Google Scholar]
- 42.Yi X., Li Y., Zai H., Long X., Li W. KLF8 knockdown triggered growth inhibition and induced cell phase arrest in human pancreatic cancer cells. Gene. 2016;585:22–27. doi: 10.1016/j.gene.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 43.Wei Y., Chen G., You L., Zhao Y. Krüpel-like factor 8 is a potential prognostic factor for pancreatic cancer. Chin. Med. J. (Engl.) 2014;127:856–859. [PubMed] [Google Scholar]
- 44.Yi X., Zai H., Long X., Wang X., Li W., Li Y. Krüppel-like factor 8 induces epithelial-to-mesenchymal transition and promotes invasion of pancreatic cancer cells through transcriptional activation of four and a half LIM-only protein 2. Oncol. Lett. 2017;14:4883–4889. doi: 10.3892/ol.2017.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-Torras S., Vidal-Pla A., Miquel R., Almendro V., Fernández-Cruz L., Navarro S., Maurel J., Carbó N., Gascón P., Mazo A. Characterization of human pancreatic orthotopic tumor xenografts suitable for drug screening. Cell Oncol. (Dordr.) 2011;34:511–521. doi: 10.1007/s13402-011-0049-1. [DOI] [PubMed] [Google Scholar]
- 46.Villanueva A., García C., Paules A.B., Vicente M., Megías M., Reyes G., de Villalonga P., Agell N., Lluís F., Bachs O., Capellá G. Disruption of the antiproliferative TGF-β signaling pathways in human pancreatic cancer cells. Oncogene. 1998;17:1969–1978. doi: 10.1038/sj.onc.1202118. [DOI] [PubMed] [Google Scholar]
- 47.Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J.M., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv. Exp. Med. Biol. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 48.Puig-Saus C., Rojas L.A., Laborda E., Figueras A., Alba R., Fillat C., Alemany R. iRGD tumor-penetrating peptide-modified oncolytic adenovirus shows enhanced tumor transduction, intratumoral dissemination and antitumor efficacy. Gene Ther. 2014;21:767–774. doi: 10.1038/gt.2014.52. [DOI] [PubMed] [Google Scholar]
- 49.Stanton R.J., McSharry B.P., Armstrong M., Tomasec P., Wilkinson G.W.G. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques. 2008;45:659–662, 664–668. doi: 10.2144/000112993. [DOI] [PubMed] [Google Scholar]
- 50.Graham F.L., Prevec L. Methods for construction of adenovirus vectors. Mol. Biotechnol. 1995;3:207–220. doi: 10.1007/BF02789331. [DOI] [PubMed] [Google Scholar]
- 51.Villanueva E., Martí-Solano M., Fillat C. Codon optimization of the adenoviral fiber negatively impacts structural protein expression and viral fitness. Sci. Rep. 2016;6:27546. doi: 10.1038/srep27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puig-Saus C., Gros A., Alemany R., Cascalló M. Adenovirus i-leader truncation bioselected against cancer-associated fibroblasts to overcome tumor stromal barriers. Mol. Ther. 2012;20:54–62. doi: 10.1038/mt.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.