Abstract
Editing the β-globin locus in hematopoietic stem cells is an alternative therapeutic approach for gene therapy of β-thalassemia and sickle cell disease. Using the CRISPR/Cas9 system, we genetically modified human hematopoietic stem and progenitor cells (HSPCs) to mimic the large rearrangements in the β-globin locus associated with hereditary persistence of fetal hemoglobin (HPFH), a condition that mitigates the clinical phenotype of patients with β-hemoglobinopathies. We optimized and compared the efficiency of plasmid-, lentiviral vector (LV)-, RNA-, and ribonucleoprotein complex (RNP)-based methods to deliver the CRISPR/Cas9 system into HSPCs. Plasmid delivery of Cas9 and gRNA pairs targeting two HPFH-like regions led to high frequency of genomic rearrangements and HbF reactivation in erythroblasts derived from sorted, Cas9+ HSPCs but was associated with significant cell toxicity. RNA-mediated delivery of CRISPR/Cas9 was similarly toxic but much less efficient in editing the β-globin locus. Transduction of HSPCs by LVs expressing Cas9 and gRNA pairs was robust and minimally toxic but resulted in poor genome-editing efficiency. Ribonucleoprotein (RNP)-based delivery of CRISPR/Cas9 exhibited a good balance between cytotoxicity and efficiency of genomic rearrangements as compared to the other delivery systems and resulted in HbF upregulation in erythroblasts derived from unselected edited HSPCs.
Keywords: genome editing, CRISPR/Cas9 delivery, β-hemoglobinopathies
Lattanzi et al. compared plasmid-, LV-, RNA- and ribonucleoprotein (RNP)-mediated CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells (HSPCs) to generate large genomic rearrangements in the β-globin locus. The RNP-based protocol allowed selection-free CRISPR/Cas9 delivery associated with minimal cytotoxicity and resulted in fetal hemoglobin upregulation in erythroblasts derived from edited HSPCs.
Introduction
Ex vivo genome editing in human hematopoietic stem and progenitor cells (HSPCs) is a promising therapeutic option for many hematopoietic inherited and acquired diseases.1, 2, 3 β-thalassemias and sickle cell disease (SCD) are caused by reduced or abnormal production of hemoglobin β-chain, resulting in anemia, organ damage, and reduced life expectancy.4, 5, 6 Given their prevalence and severity, β-hemoglobinopathies represent prototypic diseases for genome-editing-based therapeutic strategies. We and others have previously reported a genome-editing strategy aimed at reproducing naturally occurring large deletions in the β-globin locus,7, 8, 9, 10 associated with hereditary persistence of fetal hemoglobin (HPFH), a benign condition, which results in the increased expression of the fetal γ-globin genes and amelioration of both β-thalassemic and SCD clinical phenotypes. Genome editing was obtained by the CRISPR/Cas9 system, an RNA-guided nuclease targeting genomic sequences with 20-bp complementary to CRISPR guide RNA (gRNA).11, 12 Cas9-induced double-strand breaks (DSBs) are mostly resolved through two endogenous eukaryotic DNA-repair mechanisms: non-homologous end joining (NHEJ) and homology-directed repair (HDR). Due to its flexibility and multiplex editing capability, the CRISPR/Cas9 system can be harnessed to simultaneously target different DNA sequences and generate large genomic rearrangements (e.g., deletions, inversions, duplications, and translocations). Optimizing delivery and efficiency of the CRISPR/Cas9 complex for genome editing of primary human HSPCs is still object of intense investigation. Both viral13, 14, 15, 16 and non-viral delivery methods have been described,17 with different balance in terms of DSB efficiency, off-target cleavage, and toxicity.17, 18, 19 While NHEJ-mediated gene disruption and HDR-mediated gene correction are relatively well established,3, 20, 21, 22, 23, 24, 25 generation of large genomic rearrangements in HSPCs in clinically relevant conditions is far from being optimized.8, 10, 26, 27
In a proof-of-principle study, we previously showed by plasmid-based delivery of CRISPR/Cas9 that deletions in the γ-β intergenic region in the β-globin locus lead to potentially therapeutic elevation of fetal hemoglobin (HbF) in erythroblasts differentiated from genome-edited HSPCs.10 Here, we compare plasmid-, lentiviral vector (LV)-, RNA-, and ribonucleoprotein complex (RNP)-mediated CRISPR/Cas9 delivery to mobilized peripheral blood CD34+ HSPCs, with the aim of optimizing experimental conditions that maximize genome-editing efficiency and minimize cell toxicity. Both plasmid- and RNP-based delivery of the system resulted in efficient generation of deletions and inversions in the β-globin locus and reactivation of HbF synthesis. Importantly, an optimized RNP-based protocol allowed DNA- and selection-free delivery of CRISPR/Cas9 associated with minimal cytotoxicity.
Results
Design and Testing of gRNAs for Targeted Genomic Rearrangements in the β-Globin Locus
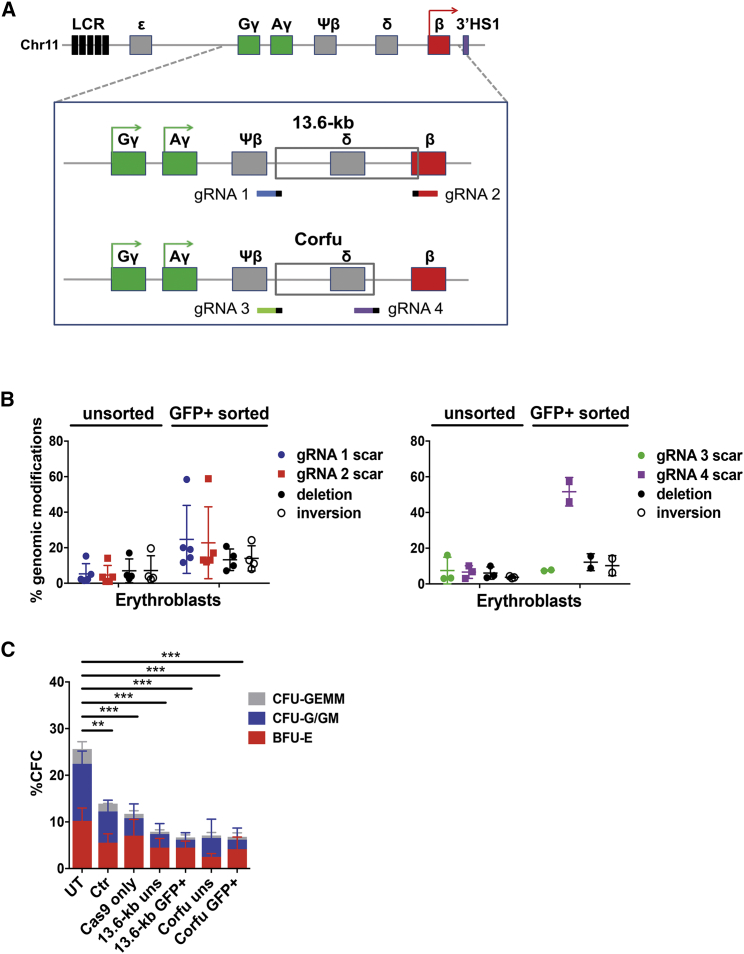
In this study, we used CRISPR/Cas9 to generate two large HPFH-like genomic deletions in the β-globin locus, which we previously showed to reactivate HbF synthesis in human adult erythroblasts:10 (1) the 7.2-kb “Corfu” deletion of γ-δ intergenic region (NG_000007.3:g.57237_64443del7207), associated with elevated HbF levels in β-thalassemic patients,28 and (2) a 13.6-kb deletion including the γ-δ intergenic region and extending to the first intron of the β-globin gene, similar to the “Sicilian” 12.9-kb HPFH-5 deletion29 (Figure 1A).
Figure 1.
Plasmid Delivery of the CRISPR/Cas9 System in HSPCs to Generate Genomic Rearrangements in the β-Globin Locus
(A) CRISPR/Cas9-based strategy targeting the 13.6-kb and the Corfu regions. gRNA target regions are indicated in different colors together with Protospacer-adjacent motif (PAM, small black bar). LCR, locus control region; ε, HBE gene; Gγ, HBG2 gene; Aγ, HBG1 gene; Ψβ, HBBP1 pseudogene; δ, HBD gene; β, HBB gene. (B) Genome-editing efficiency in erythroblasts derived from unsorted and GFP+ sorted adult mobilized HSPCs. Error bars denote SD. (C) Frequency of CFC in unsorted (uns) and GFP+-sorted HSPCs. Untreated (UT), mock-transfected (Mock), and cells transfected only with the Cas9-GFP plasmid (Cas9_only) served as controls (n = 2–4 donors with at least two replicates/donor); **p < 0.01, ***p < 0.001 (two-way ANOVA plus Tukey’s multiple comparison test). The percentage of CFU-G/GMs were significantly decreased in Cas9_only (p < 0.05), 13.6-kb_uns (p < 0.05), Corfu_GFP+ (p < 0.05), and 13.6-kb_GFP+ (p < 0.01) samples compared to UT. Error bars denote SD.
We first designed several ad hoc gRNA pairs, targeting the 5′ and 3′ ends of the Corfu and 13.6-kb regions (Table S1) and selected those with minimal predicted off-target activity. To increase gRNA binding specificity, we shortened their DNA targeting sequence to 18–19 bp, as previously reported.30 To evaluate DNA cleavage efficiency, the selected gRNAs were delivered individually or as gRNA pairs by plasmid transfection into K562 human erythroleukemia cells stably expressing Cas9. Single gRNAs showed up to ∼80% of on-target cleavage activity, measured as indel (insertion or deletion) frequency (Figure S1A). Paired gRNAs generated different genome modifications: (1) the expected deletions, with a frequency of 20.0% ± 7.7% and 8.1% ± 1.5% (mean ± SD) for the 13.6-kb and the Corfu regions, respectively; (2) inversion of the targeted regions with a respective frequency of 14.2% ± 3.5% and 4.7% ± 1.3%; (3) “scarring,” i.e., indel mutations without deletion or inversion of the target regions due to single gRNA cleavage or asynchronous cleavage of both gRNAs, in up to ∼70% of the remaining loci (Figures S1B and S1C).
Induction of Genomic Rearrangements of the β-Globin Locus in Human HSPCs by Plasmid-Mediated Delivery of CRISPR/Cas9
To test the efficiency of the selected gRNA pairs in primary human adult G-CSF (granulocyte-colony stimulating factor)-mobilized HSPCs, we electroporated the cells with three plasmids encoding, respectively, a Cas9-GFP fusion protein, gRNA 1 and gRNA 2 targeting the 13.6-kb region in different experimental conditions. Cell washing soon after transfection and treatment with Z-Vad-FMK, a pan-caspase inhibitor that prevents apoptosis,31 improved transfection rate (Figures S1D and S1E), cell viability (Figure S1F), and genome-editing efficiency particularly at the highest gRNA plasmid doses of 1.6 and 3.2 μg (Figure S1G).
Under these optimized conditions, we then electroporated adult HSPCs with plasmids carrying the Cas9-GFP and gRNAs targeting either the 13.6-kb or the Corfu region. GFP+ cells were fluorescence-activated cell sorting (FACS)-sorted to enrich for edited HSPCs. Unsorted populations and GFP+-sorted cells were then differentiated in liquid culture toward the erythroid lineage for 13 days or plated in clonogenic cultures (colony forming cell [CFC] assay) allowing the growth of multipotent, erythroid, and granulomonocytic progenitors (CFU-GEMMs, BFU-Es, and CFU-G/GMs). In mature erythroblasts derived from GFP+ HSPCs (day 13), we achieved a deletion/inversion frequency of up to 21%/24% and 16%/14% for the 13.6-kb and the Corfu regions, respectively (mean ± SD, 13.2% ± 6.1%/14.1% ± 7.1% and 12.2% ± 4.7%/10.3% ± 5.6%, respectively; Figure 1B), whereas scarring activity of single gRNAs ranged from 10% to 60% (Figure 1B). We observed a linear correlation between the enrichment in Cas9-GFP+ HSPCs and the fold increase in genome editing efficiency in mature erythroblasts, with scarring events more enriched than deletion/inversion (Figure S1H). Genome-editing efficiency did not substantially change between early and late time points of erythroid differentiation (data not shown). Compared to untreated HSPCs, the plating efficiency of total hematopoietic progenitors was significantly reduced in mock-transfected cells and further decreased upon transfection of Cas9-GFP ± gRNA plasmids (Figure 1C). The proportion of CFU-G/GMs was significantly reduced compared to BFU-Es in most of the plasmid-electroporated samples (Figure 1C), suggesting a higher sensitivity to DNA transfection of granulomonocytic versus erythroid progenitors. PCR analysis performed in pools of BFU-Es and CFU-G/GMs derived from FACS-sorted HSPCs showed similar genome-editing efficiencies compared to mature erythroblasts differentiated in liquid culture: the proportion of deleted/inverted alleles were 10.9% ± 3.8%/27.2% ± 27.4% in BFU-Es and 9.7% ± 2.0%/6.6% ± 8.8% in CFU-G/GMs for the 13.6-kb region, and 8.4%/10% in BFU-Es and 22.6% ± 19.2%/7.7% ± 1.0% in CFU-G/GMs for the Corfu region (Figure S1I). In unsorted populations, however, editing efficiency was generally lower in CFU-G/GMs compared to BFU-Es (Figure S1I). In BFU-Es, GFP enrichment tended to be correlated with genome-editing efficiency, with scarring events more enriched than deletion/inversion, as observed in mature erythroblasts (Figure S1H). CFU-G/GM populations showed a poor correlation, with no trend toward the enrichment of specific genomic modifications (Figure S1H).
We then evaluated HbF expression in erythroblasts derived from GFP+-edited and control adult HSPCs. We observed a 1.8-fold increase in γ-globin and a concomitant decrease in β-globin mRNA levels by disrupting the 13.6-kb region, compared to control samples (Figure 2A). A milder effect on γ-globin expression was observed by targeting the Corfu region (1.4-fold increase; Figure 2A). FACS analysis showed an increase in the proportion of F cells (erythroid cells expressing HbF) up to ∼70% and ∼50% for the 13.6-kb- and Corfu-edited samples, respectively (representative samples in Figure 2B). Overall, the increase in the percent of F cells was positively correlated with the frequency of deletion/inversion of the target regions (Figure 2C). Finally, reversed phase (RP)-HPLC confirmed a more robust increase in γ-globin content and the reduction of β-globin chains when targeting the 13.6-kb region compared to the Corfu region (Figures 2D and 2E).
Figure 2.
Reactivation of Fetal Hemoglobin upon Plasmid Delivery of the CRISPR/Cas9 System
(A) β- and γ-globin mRNA quantification in erythroblasts derived from edited adult HSPCs and control (Ctr) samples (unsorted and GFP+-sorted HSPCs transfected only with Cas9-GFP plasmid and GFP− HSPCs from samples transfected with Cas9-GFP and gRNA-expressing plasmids) (n = 3–6). *p < 0.05, **p < 0.01 (two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD. (B) FACS analysis of F cells derived from a control sample (Ctr, Cas9 only) and 13.6-kb (deletion/inversion 15.3/13.0%)- and Corfu (deletion/inversion 15.5/14.2%)-edited adult HSPCs. The percentage of HbF+ cells and the geometric mean fluorescence intensity (in brackets) are indicated. (C) Correlation between frequency of genomic rearrangements (deletion + inversion) and fold change in the percentage of F cells in edited erythroblasts compared to control samples. (D) RP-HPLC quantification of β-like globin chains in erythroblasts derived from edited adult HSPCs and in control samples (Ctr). Normalized β-like globin expression (right) and relative abundance of β-like chains (left) are reported (n = 1–8). ****p < 0.0001 (two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD. (E) Chromatograms showing peaks corresponding to α- and β-like-globins. AγT, AγT γ-globin chain variant. The ratio α/non-α chains (in brackets) was similar in edited and control samples.
Development of LVs to Deliver Cas9 and gRNA Pairs in HSPCs
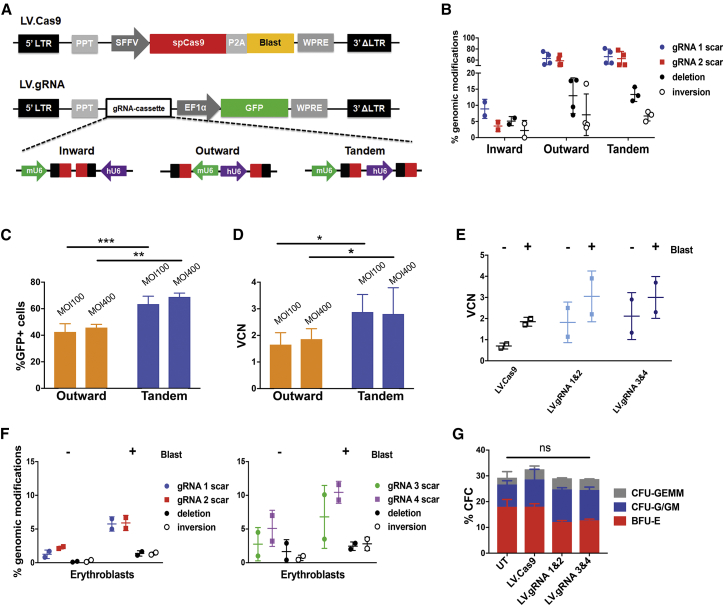
To reduce the toxicity associated with electroporation, we developed LVs to deliver Cas9 and gRNA pairs in HSPCs. We designed an LV expressing Cas9 and a selectable drug-resistance gene (blasticidin) under the control of the spleen focus-forming virus (SFFV) promoter (LV.Cas9; Figure 3A). In parallel, we introduced dual gRNA expression cassettes in an LV carrying a GFP gene under the control of the elongation factor 1-alpha (EF1α) short promoter (LV.gRNA; Figure 3A). To minimize sequence homology that may potentially trigger intramolecular recombination, we used the human and murine U6 promoters to drive the expression of gRNAs 1 and 2, respectively. We tested three different orientations of the gRNA expression cassettes within the LV, i.e., inward, tandem, and outward (Figure 3A). In HCT116 cells, the tandem and outward configurations yielded viral titers (transducing units [TU]/mL) comparable to a control vector containing only the GFP expressing cassette, whereas inward configuration caused a reduction of the LV titer (Figure S2A). Vector infectivity (TU/ng p24) was however comparable for the three LVs (Figure S2B). We then co-transduced K562 cells with LV.Cas9 and LV.gRNAs at equivalent MOI. All LV.gRNAs were able to transduce >80% of the cells (Figure S2C), although the LV carrying gRNAs in the inward configuration showed the lowest transduction efficiency (Figure S2C), vector copy number (VCN) (Figure S2D), and gRNA expression (Figure S2E) and generated negligible genomic modifications (Figure 3B). Conversely, tandem and outward configurations allowed higher VCN and gRNA expression levels (Figures S2D and S2E), resulting in higher deletion, inversion, and scarring frequencies (Figure 3B). Outward and tandem configurations were finally tested in primary cord-blood-derived HSPCs transduced with LV.Cas9. The tandem configuration yielded the best results in terms of transduction efficiency and VCN (Figures 3C and 3D) and was selected for further experiments.
Figure 3.
LVs Expressing Cas9 and gRNA Pairs Targeting the 13.6-kb and Corfu Regions
(A) LV.Cas9 and LV.gRNA vectors. SpCas9, Streptococcus pyogenes Cas9; SFFV, spleen focus forming virus promoter; P2A, porcine 2A self-cleaving peptide; Blast, blasticidin resistance gene; EF1α, human elongation factor 1-alpha promoter; LTR, long terminal repeat; PPT, polypurine tract; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; mU6, murine U6 promoter. hU6, human U6 promoter. gRNA protospacers and scaffolds are indicated with black and red boxes, respectively. (B) Frequency of deletion, inversion, and scarring events in LV-transduced K562. Error bars denote SD. (C and D) Transduction efficiency of LV.Outward and LV.Tandem in cord-blood-derived HSPCs (n = 4–5). Percentage of GFP+ cells (C) and VCN (D) are indicated. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired t test and Kolmogorov-Smirnov test). Error bars denote SD. (E) VCN analysis in adult mobilized HSPCs transduced with LV.Cas9 alone or in combination with LV.gRNA 1 and 2 and LV.gRNA 3 and 4 in tandem configuration. Error bars denote SD. (F) Genome-editing efficiency in erythroblasts derived from LV-transduced adult HSPCs. Error bars denote SD. (G) Frequency of CFC in adult HSPCs transduced with LV.Cas9 alone or in combination with LV.gRNA 1 and 2 and LV.gRNA 3 and 4. Untreated (UT) cells were used as control (n = 2–3 donors with at least two replicates/donor). ns, not significant (two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD.
To test the genome-editing efficiency induced by LVs, we transduced primary adult HSPCs with LV.Cas9, followed after 24 hr by a second round of transduction with LV.gRNA 1 and 2 or LV.gRNA 3 and 4. In erythroid liquid cultures, genome-editing efficiency increased over time in a fraction of the samples (Figure S2F). To enrich for Cas9-expressing cells, HSPCs were expanded in erythroid liquid culture in the presence of blasticidin, resulting in a total VCN of ∼1.8 in cells transduced with LV.Cas9 and ∼3.0 in cells transduced with both LV.Cas9 and LV.gRNAs (Figure 3E). Blasticidin selection led to an overall increase in genome-editing efficiency that was correlated with the fold increase in VCN, with no trend toward the enrichment of specific genome-editing events (Figures S2G and 3F). Despite the selection and high transduction efficiency, we observed low levels of deletion/inversion (up to 1.7%/1.5% and 3%/3.5% for 13.6-kb and Corfu regions, respectively) and scarring (<8% and <12% for gRNAs 1 and 2 and gRNAs 3 and 4, respectively) events in mature erythroblasts (day 13) (Figure 3F). A CFC assay showed no evidence of cytotoxicity in unselected transduced hematopoietic progenitors compared to mock-transduced control (Figure 3G). We were unable to perform a CFC assay in the presence of blasticidin due to heavy cell toxicity (data not shown).
Optimization of RNA-Mediated CRISPR/Cas9 Delivery in HSPCs
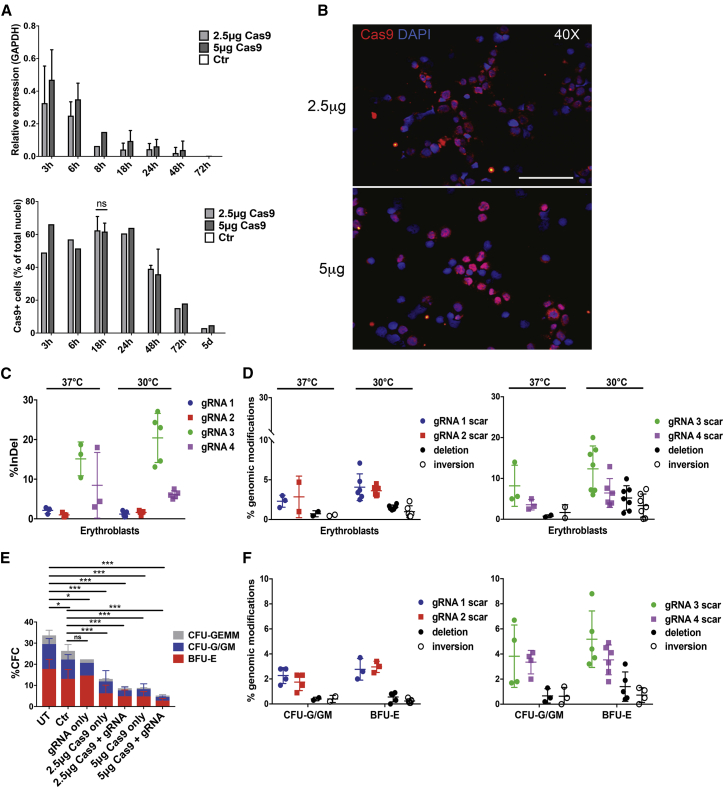
Non-viral, plasmid-free delivery of CRISPR/Cas9 can be achieved by electroporation of Cas-9 mRNA and individual gRNA molecules. We first electroporated cord-blood-derived HSPCs with 2 μg of a synthetic GFP mRNA bearing nucleotide modifications (5-methyl-cytidine triphosphate [CTP] and pseudo-uridine triphosphate [UTP]) that stabilize RNA, enhance translation, and decrease innate immunity responses32 using an Amaxa Nucleofector 4D. Afterward, cells were either cultured at 37°C or subjected to a transient cold shock (30°C for 24 hr), which is known to increase nuclease activity by probably decreasing mRNA and protein turnover.33, 34 We tested 16 different Amaxa programs and eventually selected the CA137, which yielded the highest proportion of GFP+ cells and mean fluorescence intensity (MFI) with modest effects on cell viability (Figure S3). The transient cold shock increased GFP expression without impairing cell viability (Figure S3). We then transfected cord-blood-derived HSPCs with two different amounts of synthetic Cas9 mRNA (2.5 and 5 μg) using the CA137 program and evaluated Cas9 mRNA and protein levels at different time points. Cas9 mRNA was detected at high levels 3 hr after electroporation and rapidly decreased over time (Figure 4A). Immunofluorescence analysis showed that Cas9 protein accumulated in up to ∼60% of HSPCs 3 hr after electroporation and substantially decreased at 72 hr (Figures 4A, 4B, and S4). Both Cas9 mRNA levels and protein immunofluorescence signals tended to be higher in cells transfected with 5 μg of RNA at early time points (Figures 4A, 4B, and S4). We then co-electroporated adult HSPCs with 5 μg of Cas9 mRNA together with synthetic gRNA pairs chemically modified at both termini with 2′-O-methyl-3′ phosphorothioate, a modification that increases resistance to base hydrolysis.20 Electroporated HSPCs were either kept at 37°C or shocked at 30°C and subsequently differentiated into mature erythroblasts. Incubation at 30°C tended to enhance the overall genome-editing frequency. The gRNAs delivered individually displayed a variable cleavage activity, with gRNA 3 and 4 showing the highest indel frequency (Figure 4C). Delivery of gRNA pairs resulted in up to 2.0%/2.3% and 9.9%/7.4% of deletions/inversions for gRNAs 1 and 2 and gRNA 3 and 4, respectively, in mature erythroblasts (day 13), with relatively low scarring activity at each gRNA target site (Figure 4D). A CFC assay revealed a significant reduction in the number of hematopoietic progenitors after electroporation proportional to the amount of transfected RNA (Figure 4E). The overall amount of genomic modifications in pools of erythroid and granulomonocytic colonies was lower compared to that obtained in erythroid liquid cultures, with deletion/inversion frequencies of <2% (Figures 4D and 4F).
Figure 4.
RNA-Mediated CRISPR/Cas9 Delivery to Target 13.6-kb and Corfu Regions
(A) Time-course analysis of Cas9 mRNA and protein expression after RNA electroporation of cord blood-derived HSPCs by qRT-PCR and immunofluorescence (IF). ns, not significant (unpaired t test; n = 3 for Cas9+ cells at 18 hr; n = 1–2 for the other time points and qRT-PCR analysis). Error bars denote SD. (B) IF representative pictures of Cas9+ cells acquired 24 hr after electroporation of cord-blood-derived HSPCs using 2.5 and 5 μg Cas9 mRNA. Original magnification, 40×. Scale bar, 200 μm. (C) Cleavage efficiency of gRNAs individually delivered in adult HSPCs, as determined in HSPC-derived erythroblasts. Error bars denote SD. (D) Genome-editing efficiency in erythroblasts derived from adult HSPCs transfected with Cas9 and gRNA pairs. The frequency of total scarring and deletion but not inversion events was significantly higher at 30°C (p < 0.05; two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD. (E) Percentage of CFC in adult HSPCs electroporated with gRNA alone (gRNA only) and different amounts of Cas9 mRNA alone (Cas9 only) or in combination with gRNAs (Cas9 + gRNA). Untreated (UT) and mock-transfected (Ctr) HSPCs were used as controls (n = 2–8 donors with at least two replicates/donor). *p < 0.05, ***p < 0.001 (two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD. (F) Genome-editing frequency in CFU-G/GMs and BFU-Es was lower compared to erythroblasts (p < 0.01; two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD.
Delivery of Cas9 RNPs Leads to High Frequency of Genomic Rearrangements in HSPCs with Minimal Cytotoxicity
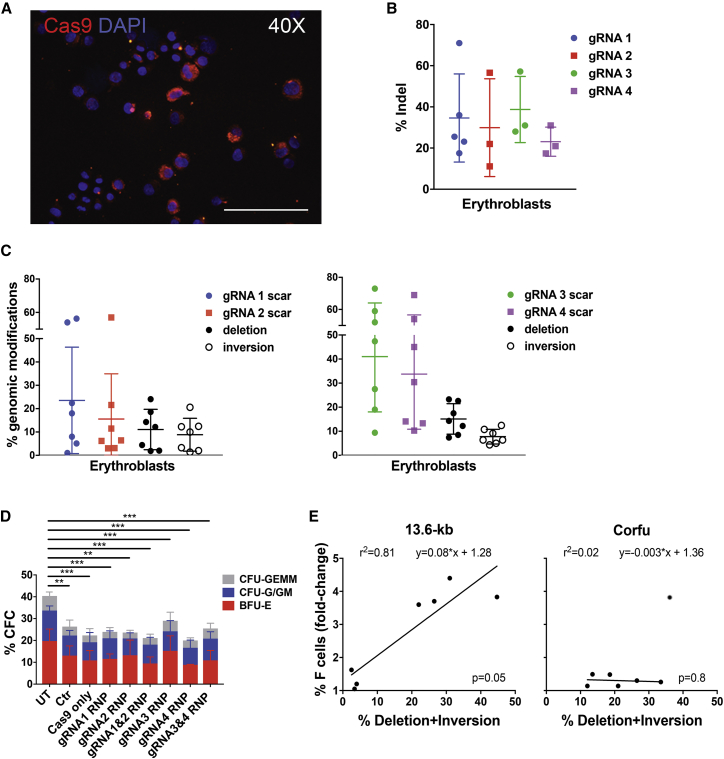
As an alternative to RNA-mediated delivery, we tested delivery of Cas9 and gRNAs as RNP complexes. Cas9 protein and gRNA were assembled at a 1:2.5 molar ratio and electroporated in cord-blood-derived HSPCs using the CA137 program. Cas9 protein was detected by immunofluorescence in >80% of HSPCs as early as 6 hr post-transfection (Figure 5A) and persisted in ∼15% of HSPC-derived erythroblasts after 13 days of culture (Figures S5A and S5B). Electroporation of Cas9 assembled with single gRNAs in adult HSPCs led to an indel frequency of ∼30% (Figure 5B), which did not increase upon transient cold shock (Figure S5C). By delivering RNPs containing gRNA pairs, we achieved up to 24%/20% and 23%/11% of deletion/inversion frequency by targeting the 13.6-kb and the Corfu region, respectively, in HSPC-derived mature erythroblasts (day 13) (11.1% ± 8.6%/8.9% ± 7.0% and 15.1% ± 6.3%/7.8% ± 3.1%, respectively; Figure 5C). Scarring activity was variable, ranging between 5% and 70% (Figure 5C). Genome-editing frequency was stable between day 3 and day 13 post-transfection (Figure S5D). Notably, the CFC assay showed no impairment in the clonogenic potential of HSPCs electroporated with Cas9 alone or with Cas9 RNPs as compared to mock-transfected cells, giving rise to similar numbers of multipotent, erythroid, and granulomonocytic colonies (Figure 5D). These data indicate that delivery of Cas9 RNPs adds no toxicity to the electroporation procedure, a significant improvement with respect to plasmid- or RNA-mediated delivery. The frequency of deletion/inversion and scarring activity in pooled granulomonocytic and erythroid progenitors were comparable to those observed in mature erythroblasts in liquid culture (Figure S5E). We could observe a linear correlation between the achieved level of genome editing and the increase in F cells in 13.6-kb-, but not in Corfu-edited adult erythroid populations (Figure 5E). These data are in line with the results obtained in mature erythroblasts derived from plasmid-transfected HSPCs, where disruption of the 13.6-kb region led to a more pronounced and consistent HbF upregulation compared to targeting of the Corfu region (Figure 2).
Figure 5.
CRISPR/Cas9-Induced Therapeutic Rearrangements in HSPCs by RNP-Mediated Delivery
(A) IF representative picture of Cas9+ cord blood-derived HSPCs 6 hr after RNP electroporation. Original magnification, 40×. Scale bar, 200 μm. (B) Cleavage efficiency of individual gRNAs in adult HSPCs. Error bars denote SD. (C) Frequency of genomic modifications in erythroblasts derived from adult HSPCs electroporated using Cas9 RNP complexes containing gRNA pairs. Error bars denote SD. (D) Frequency of CFC in RNP-electroporated adult HSPCs. Untreated (UT), mock-transfected (Mock), and Cas9-only-transfected HSPCs served as controls (n = 4–8 donors with at least two replicates/donor). **p < 0.01, ***p < 0.001 (two-way ANOVA plus Tukey’s multiple comparison test). Error bars denote SD. (E) Correlation between frequency of genomic rearrangements and fold change in the proportion of F cells in edited cells compared to control samples. R2, line-of-best-fit equation and p value are indicated. For the Corfu-edited samples, we removed an outlier from the trend line (indicated with a gray circle).
Evaluation of Off-Target Activity Using Different Delivery Methods
We then evaluated the occurrence of off-target events in mature erythroblasts (day 13) derived from adult HSPCs treated with CRISPR/Cas9 using plasmid-, LV-, RNA-, and RNP-based delivery methods. The top-2 or top-3 predicted off-targets of the gRNAs targeting the 13.6-kb and the Corfu regions were analyzed by targeted deep sequencing (Figure 6; Tables S2 and S3). LV-mediated CRISPR/Cas9 expression was associated with the highest incidence of editing events at most of the predicted off-target loci. Importantly, despite the higher frequency of on-target genome editing, plasmid and RNP methods were associated with a lower frequency of off-target events compared to LV delivery (Figure 6; Table S3).
Figure 6.
Evaluation of gRNA Off-Target Activity Using Different Delivery Methods
Top-2 or top-3 predicted off-target sites were analyzed by targeted deep sequencing in mature erythroblasts (day 13). For plasmid delivery, we used samples derived from GFP+-sorted HSPCs; for LV delivery, samples derived from HSPCs treated with blasticidin; for RNA delivery, samples derived from HSPCs treated with the transient cold shock. The background level of indels was measured in non-edited cells (Ctr).
Discussion
Several therapeutic approaches to hemoglobinopathies have been developed by harnessing the potential of both HDR-mediated gene correction and NHEJ-based genome-editing approaches.9 Strategies aimed at correcting disease-causing mutations or achieving therapeutic levels of fetal γ-globin expression have the advantage of exploiting the use of endogenous β-like gene promoters to recreate physiological expression patterns, as compared to current vector-mediated gene-addition approaches.35 Gene-correction approaches may revert single-point mutations, e.g., the E6V mutation causing SCD, even though the rates of correction in long-term HSCs are often below the levels required to achieve therapeutic benefit.24, 36 The introduction of selectable markers in the HDR donor template allows for enrichment of edited HSCs,3 although cell selection in vitro is hardly a clinically translatable procedure. Given the apparent dominance of the NHEJ DNA repair pathway in HSCs, NHEJ-based editing strategies, such as downregulation of the BCL11A HbF repressor21, 37 or recreation of small or large deletions associated with naturally occurring HPFH,8, 10, 23 are more likely to achieve clinically relevant efficiencies in HSCs.
We and others have previously demonstrated that CRISPR/Cas9 is a suitable tool for reproducing HPFH deletions in the β-globin locus.8, 10 In particular, we identified a large, 13.6-kb HPFH-like genomic region encompassing the γ-β intergenic region and part of the β-globin gene, deletion or inversion of which causes robust HbF reactivation and amelioration of the SCD cell phenotype.10 Development of minimally toxic and efficient delivery methods of the CRISPR/Cas9 machinery and optimization of HSPC manipulation protocols are nonetheless necessary to translate these results into clinical applications. In this study, we aimed at optimizing different CRISPR/Cas9 delivery techniques for achieving high frequency of genomic rearrangements in adult, mobilized CD34+ HSPCs with minimal toxicity. We selected two candidate therapeutic targets, i.e., the Corfu and the 13.6-kb genomic regions and assessed the efficiency and toxicity of plasmid-, LV-, RNA-, or RNP-mediated delivery methods by evaluating the frequency of deletion, inversion, and scarring events in the target regions and the effect of each procedure on the clonogenic potential of HSPCs.
Plasmid-mediated delivery of Cas9 and gRNA expression cassettes achieved remarkable editing efficiency (∼30%) in primary HSPCs, although only after cell selection and at the price of high cell toxicity and of a just as remarkable reduction of clonogenic potential, even under optimized conditions. Nevertheless, deletions or inversions of the 13.6-kb region led to robust reactivation of γ-globin gene expression and a concomitant decrease in β-globin expression that correlated with the overall frequency of genomic rearrangements, confirming results previously obtained in HSPCs from healthy and SCD donors.10 Plasmid delivery remains therefore an easy and relatively efficient method for obtaining CRISPR/Cas9-mediated genome rearrangements in primary hematopoietic cells for proof-of-principle studies but appears to be unsuited for clinical application. We therefore tested and optimized additional CRISPR/Cas9 delivery methods, in an attempt to reduce cytotoxicity and increase efficiency.
LVs are extensively used in clinical applications entailing genetic modification of HSPCs, with minimal transduction-related toxicity and minimal, if any, genotoxicity. We designed and tested a dual LV-based platform to independently express Cas9 and gRNAs upon co-transduction of target cells. For proof-of-principle purposes, we coupled Cas9 expression with a selectable marker and a dual gRNA expression cassette with a GFP reporter gene. The relative orientation of the gRNA cassettes in the LV affected transduction frequency in both K562 and primary HSPCs. We can envision two mechanisms that can possibly explain these findings: (1) generation of double-stranded RNA, possibly deriving from the hybridization of two U6 transcripts (inward configuration) or U6 transcripts and long terminal repeat (LTR)-driven LV genomic transcripts (inward and outward configurations), could activate innate immune pathway and affect vector production38, 39, 40; (2) stable secondary structure of gRNA sequences in the vector RNA (inward and outward configurations) could impede efficient LV genome retrotranscription and affect vector transduction.41, 42 Co-transduction was minimally toxic for HSPCs but resulted in poor deletion/inversion and scarring efficiency even upon selection of Cas9-expressing cells. The VCN obtained in HSPCs (∼3) might not be sufficient to achieve the Cas9 and/or gRNA expression levels required for efficient editing. Integrating LVs containing selectable markers have been used in other proof-of-concept studies to express the Cas9 and a single gRNA disrupting an erythroid-specific enhancer of the HbF repressor BCL11A gene or recreating a small HPFH deletion.21, 23 However, this LV-based approach would likely not be applicable in a clinical setting, since Cas9 transient expression is highly desirable for several reasons: (1) expression of Cas9 in HSPCs can potentially trigger an host immune response against edited cells; (2) prolonged expression of the CRISPR/Cas9 system can aggravate off-target effects.43, 44 Indeed, our results show that LV-mediated persistent CRISPR/Cas9 expression is associated with the highest incidence of off-target events. An integration-deficient LV (IDLV)-based approach or the use of self-inactivating Cas945, 46 could circumvent these issues. However, considering the poor efficiency of generation of large rearrangements obtained using integrating LVs, these latter approaches would unlikely lead to clinically relevant HbF levels.
DNA-free delivery approaches, such as RNA and RNP electroporation, achieved transient and efficient delivery of Cas9 in up to 60% and 80% of unselected HSPCs, respectively. However, RNA transfection showed limited genome-editing efficiency, particularly in generating the expected deletion/inversion events, and caused a reduction of the HSPC clonogenic potential that was positively correlated with the amount of transfected RNA. On the contrary, transfection of RNP complexes led to high genome-editing efficiency with minimal toxicity, as reported in other studies,25 allowing to achieve up to 24% and 20% of deletion and inversion of the 13.6-kb region in unselected HSPC-derived erythroid precursors. Importantly, despite of the persistence of Cas9 expression in a fraction of HSPC-derived erythroblasts, we observed a low frequency of off-targets events. Should the observed frequency of deletion/inversion be confirmed in long-term repopulating HSCs, RNP-mediated editing would approach the minimal efficiency required to achieve clinical benefit in SCD and β-thalassemia in an autologous HSC transplantation setting. The clinical history of allogeneic HSC transplantation in SCD or β-thalassemia patients would in fact suggest that 20%–30% of genetically corrected HSCs would be sufficient to achieve a therapeutic benefit given the in vivo selective survival of corrected RBCs or erythroid precursors.47, 48, 49
Compared to other NHEJ-based editing strategies, such as downregulation of the BCL11A HbF repressor21, 37 or recreation of a small deletion associated with naturally occurring HPFH,23 our approach requires the generation of two DSBs, which might decrease the overall efficiency of genome editing and potentially increase DSB toxicity and off-target frequency. However, our strategy targeting the 13.6-kb region might be more effective as a therapeutic approach for SCD by mimicking HPFH large rearrangements that permanently disrupt the sickle β-globin gene, thus potently ameliorating the SCD cell phenotype.10
Overall, our results support the use of RNP-mediated CRISPR/Cas9 delivery as a non-viral, DNA-free genome-editing platform for generation of large rearrangements in human HSCs. Further optimization, e.g., the use of alternative protospacers and optimized scaffolds in the gRNAs, may further increase gene-editing efficiency and reduce potential off-target effects. Clinical translation of this technology will ultimately require the development of large-scale transfection protocols based on clinical-grade reagents and demonstration of precise editing in a proportion of repopulating HSCs comparable with those currently achievable with classical, LV-mediated gene addition technology.
Materials and Methods
Plasmid Construction
Plasmids expressing a SpCas9 (Streptococcus pyogenes Cas9)-GFP fusion protein (Cas-GFP) (pMJ920) and gRNA (MLM3636) were purchased from Addgene (plasmids #42234 and #43860; Addgene, Cambridge, MA, USA). We used ZIFIT50 to select sequence-specific gRNAs in the genomic sequences flanking the 5′ and 3′ ends of the Corfu and 13.6-kb regions. All gRNA protospacers were screened in silico with COSMID51 to select the ones with minimal potential off-target sites. We used truncated gRNAs (18–19 bp long) to increase on target specificity.30 Oligonucleotide duplexes containing the gRNA protospacers were ligated into BsmBI (New England Biolabs, Ipswich, MA, USA)-digested MLM3636. A guanine was added to the 5′ end of all gRNA sequences to allow their expression from the U6 promoter (Table S1). The list of predicted off-targets is provided in Table S2.
An LV harboring the SpCas9 gene (Cas9), followed by the porcine 2A self-cleaving peptide (derived from porcine teschovirus-1) and the blasticidin resistance gene (Blast), was purchased from Addgene (lentiCas9-Blast, plasmid #52962). The Cas9-2A-Blast sequence was cloned downstream of the SFFV promoter to generate the LV.Cas9 construct.
LV constructs expressing paired-gRNAs were generated adapting the multiplex gRNA platform developed by Kabadi et al.13 Protospacer sequences were cloned in BbsI-digested expression vectors (BpiI, Thermo Scientific, Waltham, MA, USA) downstream of the human (phU6-gRNA, Addgene, #53188) or murine (pmU6-gRNA, Addgene, #53187) U6 promoters. Each gRNA expression cassette was excised using BsmBI (Esp3I, Thermo Scientific, Waltham, MA, USA) and inserted in different orientations with respect to each other (inward, tandem, and outward configuration) in a self-inactivating LV backbone expressing a reporter GFP under the control of EF1α promoter.
pMJ920 was a gift from Jennifer Doudna (Addgene plasmid #4223452). MLM3636 was a gift from Keith Joung (Addgene plasmid #43860; unpublished data). LentiCas9-Blast was a gift from Feng Zhang (Addgene plasmid #5296253). phU6-gRNA and pmU6-gRNA were a gift from Charles Gersbach (Addgene plasmid #53188 and #5318713).
LV Production and Titration
Third-generation LVs were produced by 293T calcium phosphate transient transfection and titrated in HCT116 cells, as previously described.54 LV particles were measured using the HIV-1 Gag p24 antigen immunocapture assay (Perkin-Elmer, Waltham, MA, USA) according to manufacturer’s instructions. LV infectivity was calculated as the ratio between infectious titer and ng HIV-1 Gag p24 per mL.
Cell Culture
Human erythroleukemia cell line K562 was maintained in RPMI 1640 medium (Gibco-Thermo Fisher Scientific, Waltham, MA, USA) containing 2 mM glutamine (Sigma, St. Louis, MO, USA) and supplemented with 10% fetal bovine serum (FBS) (Gibco-Thermo Fisher Scientific, Waltham, MA, USA), 10 mM HEPES (Sigma, St. Louis, MO, USA), 1 mM sodium pyruvate (Life Technologies, Carlsbad, CA, USA), and 100 U/mL each of penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA).
Human primary HSPCs were obtained in compliance with French national bioethics law. Human umbilical cord blood samples were provided by the Centre Hospitalier Sud-Francilien (CHSF, Evry, France) after uncomplicated births with informed consent of the mother, then processed and stored anonymously at Genethon (declaration DC-2012-1655 to the French Ministry of Higher Education and Research). Umbilical cord blood HSPCs were purified by immunomagnetic selection with AUTOMACS PRO (Miltenyi Biotec, Paris, France) after immunostaining with CD34 MicroBead Kit (Miltenyi Biotec, Paris, France). Peripheral blood G-CSF-mobilized HSPCs from healthy donors were obtained by Institut Gustave Roussy Villejuif (Paris, France, agreement #011117-2WPIT-00) and Centre Hospitalier Amiens (Amiens, France, agreement #013127-00) and were purified by immunomagnetic selection with CLINIMACS or CLINIMACS Prodigy (Miltenyi Biotec, Paris, France) after immunostaining with CD34 MicroBead Kit (Miltenyi Biotec, Paris, France). HSPCs were thawed and pre-activated (for 24 hr or 48 hr, depending on the protocol) in StemSpan serum-free expansion medium (SFEM) (StemCell Technologies, Vancouver, Canada), containing 2 mM glutamine, 100 U/mL each of penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA), Flt3-ligand (300 ng/mL, Cellgenix, Freiburg, Germany), stem cell factor (SCF) (300 ng/mL, Cellgenix, Freiburg, Germany), thrombopoietin (TPO) (100 ng/mL, Cellgenix, Freiburg, Germany), interleukin 3 (IL-3; 60 ng/mL, Cellgenix, Freiburg, Germany). HSPCs were differentiated toward the erythroid lineage in liquid culture for 14 days as previously described.55 The number of hematopoietic progenitors was evaluated in colony-forming cell (CFC) assays, where HSPCs were plated at a concentration that ranged between 0.5 and 2 × 103 cells/mL in methylcellulose medium (#H4435, Stem Cell Technologies, Vancouver, Canada) under conditions supporting both erythroid and granulo-monocytic differentiation. We scored colonies (BFU-Es, CFU-G/GMs, and CFU-GEMMs) after 14 days; BFU-Es and CFU-G/GMs were randomly picked, and pools of at least 100 colonies were analyzed for the efficiency of genome editing.
Electroporation
Plasmid Electroporation
K562 were transfected using Amaxa Nucleofector 4D (Lonza, Basel, Switzerland), following the manufacturer instructions and the SF-cell line 4D Nucleofector X Kit (#V4XC-2032). HSPCs were transfected using Amaxa Human CD34 Cell Nucleofector Kit (#VPA-1003) and Nucleofector I (U-08 program, Lonza), given the poor efficiency of transfection obtained using the Amaxa Nucleofector 4D (data not shown). We electroporated 4 μg of a Cas9-GFP plasmid (Addgene, pMJ920) and 0.8 to 3.2 μg of each gRNA-expressing plasmid (Addgene, MLM3636). HSPCs were transfected 48 hr after thawing and resuspended in the pre-activation medium supplemented with StemRegenin 1 (750 nM, StemCell Technologies, Vancouver, Canada) and Z-VAD-FMK (120 μM, InvivoGen, Toulouse, France). After 18 hr (day 3), GFP+ and GFP− HSPCs were sorted using SH800 Cell Sorter (Sony Biotechnology, Tokyo, Japan). In brief, 2 to 6 million cells were collected, washed once with PBS, resuspended in 1 mL of PBS containing 1% FBS, filtered by using a 35-μm nylon mesh cell strainer snap cap (Corning, New York, NY, USA), and kept in ice until the sorting procedure. After first gating on single-cell live populations according to size and granularity, GFP+ cells were identified as previously described.10 Cells transfected only with Tris-EDTA (TE) were used as negative control to define the gating strategy. Sorted cells were cultured for 24 hr in the same medium. 2.5–5 × 105 cells were maintained in culture as unsorted samples. On day 4, both unsorted and GFP-sorted HSPCs were differentiated toward the erythroid lineage in liquid culture and were plated in methylcellulose medium (#H4435, StemCell Technologies, Vancouver, Canada) for CFC assay.
RNA and RNP Electroporation
HSPCs were thawed and resuspended in the pre-activation medium supplemented with StemRegenin 1 (750 nM, StemCell Technologies, Vancouver, Canada) and were electroporated using the Amaxa Nucleofector 4D Kit (#V4XP-3032; Lonza, Basel, Switzerland) following manufacturer’s instructions. Synthetic GFP and SpCas9 mRNA with modified nucleotides (Cap0, 5-Methylcytosine and PseudoUridine) were purchased from Trilink (Trilink Biotechnologies, San Diego, CA, USA; #L6101 and #L-6125, respectively). Synthetic gRNAs with modified nucleotides (5′-2′O-methyl (N(ps)N(ps) N(ps)) N-(∼90ny)-N 2′O-methyl (U(ps)U(ps) U(ps))-U-3′), resistant to base hydrolysis20 were purchased from Trilink. Cas9 protein was purchased from Feldan (Feldan Therapeutics, Québec, Canada) and assembled in RNP complexes together with modified gRNA using a ratio 1:2.5 (Cas9:gRNA). Electroporation was performed using the CA137 nucleofector program (the best performing program according to Figure S3). For RNA transfection, we used 1 × 105 HSPCs and delivered 2.5 μg and 5 μg (1.2 mM and 2.5 mM, respectively) of Cas9 mRNA and 5 μg of each gRNA, and for RNP electroporation we used 2 × 105 HSPCs and 5 μg of Cas9 protein (1.3 μM, 31 pmol) pre-assembled with 2.5 μg of each gRNA (78 pmol).
LV Transduction
K562 were LV-transduced at MOI of 20 overnight in the presence of 4 μg/mL polybrene (Sigma, St. Louis, MO, USA). HSPCs were pre-activated for 24 hr and then transduced in retronectin-coated plates (5 μg/cm2; Takara, Japan) overnight in the presence of 4 μg/mL protamine sulfate (Sanofi Aventis, Paris, France). We performed a first transduction with LV.Cas9 at MOI 100 in the LV pre-activation medium containing rapamycin (10 μM, Sigma, St. Louis, MO, USA). Cells were transduced cells 24 hr later with different LV.gRNA at an MOI of 100 or 400. A fraction of HSPCs were exposed to blasticidin for 5 days. VCN was determined as previously described.26, 54, 56, 57, 58, 59, 60
Quantification of Genome-Editing Events
Genome-editing efficiency was measured in mature erythroblasts at day 13 of erythroid liquid culture and in pools of BFU-E and CFU-GM 14 days after the plating. Editing frequency was analyzed in erythroid liquid cultures also at earlier time points for RNP (day 3 and 7) and LV delivery (day 7). Genomic DNA was extracted using DNA extraction (QIAamp DNA Mini and Micro Kit, QIAGEN, Hilden, Germany), following manufacturer’s instructions. To evaluate NHEJ efficiency at each gRNA on-target site, we performed PCR followed by Sanger sequencing and TIDE analysis (tracking of indels by decomposition).61 To detect deletion and inversion events, we performed Droplet Digital PCR (QX200 Droplet Digital PCR System, Bio-Rad, Hercules, CA, USA), using QX200 EvaGreen ddPCR Supermix (Bio-Rad, Hercules, CA, USA), and control primers annealing to a genomic region on the same chromosome containing the β-globin locus (Chr11). To evaluate NHEJ efficiency at predicted off-target sites in mature erythroblasts (day 13), we performed PCR followed by deep sequencing. In brief, Illumina compatible barcoded DNA amplicon libraries were prepared using the TruSeq DNA PCR-Free kit (Illumina). Libraries were pooled and sequenced using Illumina HiSeq2500 (paired-end sequencing 130 × 130 bases). A total of 0.59 to 1.12 million passing filter reads per sample were produced. Targeted deep-sequencing data were analyzed using CRISPRESSO.62 Primer and probe sequences are listed in the Supplemental Materials and Methods.
qRT-PCR
Total RNA was extracted using RNeasy mini or micro kit (QIAGEN, Hilden, Germany) or MagNA Pure 96 Cellular RNA Large Volume kit (Roche, Basel, Switzerland) and DNase treated (DNA-free kit; Ambion-Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions. To quantify gRNA expression, qPCR was performed using qScript XLT One-Step RT-qPCR ToughMix (Quanta Bioscience, Beverly, MA, USA; #95132-100). gRNA primers and probe sequences were kindly provided by Rasmus Bak (Prof. Matthew Porteus’ lab). The amount of Cas9 transcript was quantified by one-step qPCR using the kit LightCycler Multiplex RNA Virus Master (Roche, Basel, Switzerland). GAPDH was used as reference gene. For globin mRNA quantification, total RNA was reverse ranscribed using Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland), and qPCR was performed using Syber Green/Rox (Life Scientific, Thermo Fisher Scientific, Waltham, MA, USA). Primer and probe sequences are listed in Supplemental Materials and Methods.
Flow Cytometry Analysis
We performed flow cytometry analyses using Cytomics FC-500 flow cytometer (Beckman Coulter). We used 7-aminoactinomycin D (7-AAD; Sigma, St. Luis, MO, US) to detect non-viable cells. HSPC-derived erythroblasts were stained with an antibody against the erythroid surface molecule Glycophorine A (CD235a-PECY7, BD Pharmingen, Franklin Lakes, NJ, USA), fixed and permeabilized using BD Cytofix/Cytoperm solution (BD Pharmingen, Franklin Lakes, NJ, USA), and then stained with an antibody recognizing HbF (HbF-APC, MHF05, Life Technologies, Carlsbad, CA, USA), following the manufacturer’s instructions. We measured the percentage of F cells in the Glycophorine Ahigh population containing highly differentiated erythroblasts.
Reversed-Phase HPLC Analysis of Globin Chains
HPLC analysis was performed using a NexeraX2 SIL-30AC chromatograph (Shimadzu) and the LC Solution software. HSPC-derived erythroblasts were lysed in water, and globin chains were separated using a 250 × 4.6-mm, 3.6-μm Aeris Widepore column (Phenomenex). Samples were eluted with a gradient mixture of solution A (water/acetonitrile/trifluoroacetic acid, 95:5:0.1) and solution B (water/acetonitrile/trifluoroacetic acid, 5:95:0.1). The absorbance was measured at 220 nm. β-like globin expression was normalized to α-globin. Relative abundance of β-like chains was calculated as percentage of total β-like (β + γ + δ) globins.
Immunofluorescence
Cells were seeded on glass slides (cytospin) and fixed with 4% paraformaldehyde (Sigma, St. Louis, MO, USA) Samples were permeabilized for 20 min with PBS containing 0.01% Triton and 10% goat serum. After three washes (5 min each) with PBS, samples were incubated (overnight at 4°C) with an antibody specifically recognizing Cas9 (Diagenode, Liege, Belgium; monoclonal anti-Cas9, #C15200203/C15200216). After three washes (5 min each) with PBS, samples were incubated with a fluorochrome-conjugated secondary antibody (anti-mouse IgG-Cy3, Roche, Basel, Switzerland) for 1 hr, washed with PBS, and DAPI counterstained (Dapi-Fluoromount-G, CliniSciences, Nanterre, France). Cas9+ cells over a total number of 100–300 nuclei were scored manually using a Leica fluorescence microscope coupled with a Retiga2000R camera (Qimaging, Surrey, Canada).
Statistics
Statistical analyses were performed when the total number of replicates for each group was ≥3. We used Shapiro-Wilk test to evaluate if data follow a Gaussian distribution followed by F test to compare variances. For comparison between two groups, if data followed a Gaussian distribution and variances were not statistically different, we used the parametric unpaired or paired Student’s t test, whereas if variances were different, we used the Welch’s t test. If data did not follow a Gaussian distribution, we used non-parametric tests (Kolmogorov-Smirnov test for comparison between two groups). For comparison between more than two groups, if data followed a Gaussian distribution, we used the parametric two-way ANOVA plus Tukey’s multiple comparison test. If data did not follow a Gaussian distribution, we used a non-parametric test (Kolmogorov-Smirnov test) to compare the untreated control and each treated sample. Data were analyzed with GraphPad Prism software (version 6.0; GraphPad Software, La Jolla, CA, USA) and RStudio63 and expressed as mean ± SD. Linear regression analysis was performed to assess the potential correlation between frequency of genome editing and percentage of F cells in edited HSPC-derived erythroblasts.
Author Contributions
A.L. designed and conducted experiments and wrote the paper; V.M., G.P., S.R., C.A., C.L., and O.A. designed and conducted experiments; F.A. and T.F. conducted experiments; C.M. analyzed data; M.H.P., G.B., and M.A. contributed to the design of the experimental strategy. F.M. conceived the study and wrote the paper. A.M. conceived the study, designed experiments, and wrote the paper.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was supported by grants from AFM-Telethon (17224), the European Research Council (ERC-2010-AdG, GT-SKIN), the Agence nationale de la recherché (ANR-16-CE18-0004 and ANR-10-IAHU-01 “Investissements d’avenir” program), EU Marie Curie-COFUND (PRESTIGE_2015_2_0015), Genopole (Chaire junior fondagen), the EU H2020 research and innovation program (SCIDNET consortium, GA no. 666908), the Cancer Prevention and Research Institute of Texas (RR14008 and RP170721), and a collaboration with CRISPR Therapeutics. We thank Samia Martin for the LV production, Christine Bole for the deep sequencing, and Patrick Nitschke for help with the bioinformatics analysis. We are grateful to the consenting mothers, to Dr. Rigonnot and staff of the Maternity at the Centre Hospitalier Sud-Francilien (CHSF, Evry, France) and to Dr. Anne Galy and Dr. Sabine Charrier (Genethon, Evry, France) for providing umbilical cord blood and peripheral blood G-CSF mobilized HSPCs.
Footnotes
Supplemental Information includes five figures, three tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.10.008.
Supplemental Information
References
- 1.Morgan R.A., Gray D., Lomova A., Kohn D.B. Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell. 2017;21:574–590. doi: 10.1016/j.stem.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sather B.D., Romano Ibarra G.S., Sommer K., Curinga G., Hale M., Khan I.F., Singh S., Song Y., Gwiazda K., Sahni J. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015;7:307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamatoyannopoulos G. The molecular basis of hemoglobin disease. Annu. Rev. Genet. 1972;6:47–70. doi: 10.1146/annurev.ge.06.120172.000403. [DOI] [PubMed] [Google Scholar]
- 5.Platt O.S., Rosenstock W., Espeland M.A. Influence of sickle hemoglobinopathies on growth and development. N. Engl. J. Med. 1984;311:7–12. doi: 10.1056/NEJM198407053110102. [DOI] [PubMed] [Google Scholar]
- 6.Madigan C., Malik P. Pathophysiology and therapy for haemoglobinopathies. Part I: sickle cell disease. Expert Rev. Mol. Med. 2006;8:1–23. doi: 10.1017/S1462399406010659. [DOI] [PubMed] [Google Scholar]
- 7.Carroll D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 8.Ye L., Wang J., Tan Y., Beyer A.I., Xie F., Muench M.O., Kan Y.W. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: An approach for treating sickle cell disease and β-thalassemia. Proc. Natl. Acad. Sci. USA. 2016;113:10661–10665. doi: 10.1073/pnas.1612075113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazzana M., Antoniani C., Miccio A. Gene Therapy for β-Hemoglobinopathies. Mol. Ther. 2017;25:1142–1154. doi: 10.1016/j.ymthe.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniani C., Meneghini V., Lattanzi A., Felix T., Romano O., Magrin E., Weber L., Pavani G., El Hoss S., Kurita R. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the β-globin locus. Blood. 2018;131:1960–1973. doi: 10.1182/blood-2017-10-811505. [DOI] [PubMed] [Google Scholar]
- 11.Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 12.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabadi A.M., Ousterout D.G., Hilton I.B., Gersbach C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J.G., Dang Y., Abraham S., Ma H., Zhang J., Guo H., Cai Y., Mikkelsen J.G., Wu H., Shankar P., Manjunath N. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23:627–633. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 15.Yu S., Yao Y., Xiao H., Li J., Liu Q., Yang Y., Adah D., Lu J., Zhao S., Qin L., Chen X. Simultaneous Knockout of CXCR4 and CCR5 Genes in CD4+ T Cells via CRISPR/Cas9 Confers Resistance to Both X4- and R5-Tropic Human Immunodeficiency Virus Type 1 Infection. Hum. Gene Ther. 2017;29:51–67. doi: 10.1089/hum.2017.032. [DOI] [PubMed] [Google Scholar]
- 16.Ortinski P.I., O’Donovan B., Dong X., Kantor B. Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing. Mol. Ther. Methods Clin. Dev. 2017;5:153–164. doi: 10.1016/j.omtm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu K.R., Natanson H., Dunbar C.E. Gene Editing of Human Hematopoietic Stem and Progenitor Cells: Promise and Potential Hurdles. Hum. Gene Ther. 2016;27:729–740. doi: 10.1089/hum.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porteus M.H. Towards a new era in medicine: therapeutic genome editing. Genome Biol. 2015;16:286. doi: 10.1186/s13059-015-0859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendel A., Fine E.J., Bao G., Porteus M.H. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015;33:132–140. doi: 10.1016/j.tibtech.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendel A., Bak R.O., Clark J.T., Kennedy A.B., Ryan D.E., Roy S., Steinfeld I., Lunstad B.D., Kaiser R.J., Wilkens A.B. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canver M.C., Smith E.C., Sher F., Pinello L., Sanjana N.E., Shalem O., Chen D.D., Schupp P.G., Vinjamur D.S., Garcia S.P. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoban M.D., Cost G.J., Mendel M.C., Romero Z., Kaufman M.L., Joglekar A.V., Ho M., Lumaquin D., Gray D., Lill G.R. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C., Weiss M.J. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeWitt M.A., Magis W., Bray N.L., Wang T., Berman J.R., Urbinati F., Heo S.J., Mitros T., Muñoz D.P., Boffelli D. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gundry M.C., Brunetti L., Lin A., Mayle A.E., Kitano A., Wagner D., Hsu J.I., Hoegenauer K.A., Rooney C.M., Goodell M.A., Nakada D. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep. 2016;17:1453–1461. doi: 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mettananda S., Fisher C.A., Hay D., Badat M., Quek L., Clark K., Hublitz P., Downes D., Kerry J., Gosden M. Editing an α-globin enhancer in primary human hematopoietic stem cells as a treatment for β-thalassemia. Nat. Commun. 2017;8:424. doi: 10.1038/s41467-017-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakalova L., Osborne C.S., Dai Y.F., Goyenechea B., Metaxotou-Mavromati A., Kattamis A., Kattamis C., Fraser P. The Corfu deltabeta thalassemia deletion disrupts gamma-globin gene silencing and reveals post-transcriptional regulation of HbF expression. Blood. 2005;105:2154–2160. doi: 10.1182/blood-2003-11-4069. [DOI] [PubMed] [Google Scholar]
- 29.Camaschella C., Serra A., Gottardi E., Alfarano A., Revello D., Mazza U., Saglio G. A new hereditary persistence of fetal hemoglobin deletion has the breakpoint within the 3′ beta-globin gene enhancer. Blood. 1990;75:1000–1005. [PubMed] [Google Scholar]
- 30.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Levetzow G., Spanholtz J., Beckmann J., Fischer J., Kögler G., Wernet P., Punzel M., Giebel B. Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev. 2006;15:278–285. doi: 10.1089/scd.2006.15.278. [DOI] [PubMed] [Google Scholar]
- 32.Uchida S., Kataoka K., Itaka K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics. 2015;7:137–151. doi: 10.3390/pharmaceutics7030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyon Y., Choi V.M., Xia D.F., Vo T.D., Gregory P.D., Holmes M.C. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 34.DiGiusto D.L., Cannon P.M., Holmes M.C., Li L., Rao A., Wang J., Lee G., Gregory P.D., Kim K.A., Hayward S.B. Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol. Ther. Methods Clin. Dev. 2016;3:16067. doi: 10.1038/mtm.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari G., Cavazzana M., Mavilio F. Gene Therapy Approaches to Hemoglobinopathies. Hematol. Oncol. Clin. North Am. 2017;31:835–852. doi: 10.1016/j.hoc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Hoban M.D., Lumaquin D., Kuo C.Y., Romero Z., Long J., Ho M., Young C.S., Mojadidi M., Fitz-Gibbon S., Cooper A.R. CRISPR/Cas9-Mediated Correction of the Sickle Mutation in Human CD34+ cells. Mol. Ther. 2016;24:1561–1569. doi: 10.1038/mt.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang K.H., Smith S.E., Sullivan T., Chen K., Zhou Q., West J.A., Liu M., Liu Y., Vieira B.F., Sun C. Long-Term Engraftment and Fetal Globin Induction upon BCL11A Gene Editing in Bone-Marrow-Derived CD34+ Hematopoietic Stem and Progenitor Cells. Mol. Ther. Methods Clin. Dev. 2017;4:137–148. doi: 10.1016/j.omtm.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gantier M.P., Williams B.R. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pernod G., Fish R., Liu J.W., Kruithof E.K. Increasing lentiviral vector titer using inhibitors of protein kinase R. Biotechniques. 2004;36:576–578, 580. doi: 10.2144/04364BM02. [DOI] [PubMed] [Google Scholar]
- 40.Maetzig T., Galla M., Brugman M.H., Loew R., Baum C., Schambach A. Mechanisms controlling titer and expression of bidirectional lentiviral and gammaretroviral vectors. Gene Ther. 2010;17:400–411. doi: 10.1038/gt.2009.129. [DOI] [PubMed] [Google Scholar]
- 41.Harrison G.P., Mayo M.S., Hunter E., Lever A.M. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 1998;26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klasens B.I., Huthoff H.T., Das A.T., Jeeninga R.E., Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 43.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaj T., Guo J., Kato Y., Sirk S.J., Barbas C.F., 3rd Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merienne N., Vachey G., de Longprez L., Meunier C., Zimmer V., Perriard G., Canales M., Mathias A., Herrgott L., Beltraminelli T. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 2017;20:2980–2991. doi: 10.1016/j.celrep.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 46.Petris G., Casini A., Montagna C., Lorenzin F., Prandi D., Romanel A., Zasso J., Conti L., Demichelis F., Cereseto A. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat. Commun. 2017;8:15334. doi: 10.1038/ncomms15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akinsheye I., Alsultan A., Solovieff N., Ngo D., Baldwin C.T., Sebastiani P., Chui D.H., Steinberg M.H. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powars D.R., Weiss J.N., Chan L.S., Schroeder W.A. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63:921–926. [PubMed] [Google Scholar]
- 49.Steinberg M.H., Chui D.H., Dover G.J., Sebastiani P., Alsultan A. Fetal hemoglobin in sickle cell anemia: a glass half full? Blood. 2014;123:481–485. doi: 10.1182/blood-2013-09-528067. [DOI] [PubMed] [Google Scholar]
- 50.Sander J.D., Maeder M.L., Reyon D., Voytas D.F., Joung J.K., Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lattanzi A., Duguez S., Moiani A., Izmiryan A., Barbon E., Martin S., Mamchaoui K., Mouly V., Bernardi F., Mavilio F., Bovolenta M. Correction of the Exon 2 Duplication in DMD Myoblasts by a Single CRISPR/Cas9 System. Mol. Ther. Nucleic Acids. 2017;7:11–19. doi: 10.1016/j.omtn.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankaran V.G., Menne T.F., Xu J., Akie T.E., Lettre G., Van Handel B., Mikkola H.K., Hirschhorn J.N., Cantor A.B., Orkin S.H. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 56.Mandal P.K., Ferreira L.M., Collins R., Meissner T.B., Boutwell C.L., Friesen M., Vrbanac V., Garrison B.S., Stortchevoi A., Bryder D. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Shou J., Guo Y., Tang Y., Wu Y., Jia Z., Zhai Y., Chen Z., Xu Q., Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J. Mol. Cell Biol. 2015;7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park C.Y., Kim D.H., Son J.S., Sung J.J., Lee J., Bae S., Kim J.H., Kim D.W., Kim J.S. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Park A.I., Mou H., Colpan C., Bizhanova A., Akama-Garren E., Joshi N., Hendrickson E.A., Feldser D., Yin H. A versatile reporter system for CRISPR-mediated chromosomal rearrangements. Genome Biol. 2015;16:111. doi: 10.1186/s13059-015-0680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Development Core Team . R Foundation for Statistical Computing; 2016. R: A language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.