Abstract
Endophytic fungi are plant tissue-associated fungi that represent a rich resource of unexplored biological and chemical diversity. As part of an ongoing effort to characterize Amazon rainforest-derived endophytes, numerous fungi were isolated and cultured from plants collected in the Yasuní National Park in Ecuador. Of these samples, phylogenetic and morphological data revealed a previously undescribed fungus in the order Pleosporales that was cultured from the tRopical tRee Duroia hirsuta. ExtRacts from this fungal isolate displayed activity against Staphylococcus aureus and were thus subjected to detailed chemical studies. Two compounds with modest antibacterial activity were isolated, and their stRuctures were elucidated using a combination of NMR spectRoscopic analysis, LC-MS studies, and chemical degradation. These efforts led to the identification of stelliosphaerols A (1) and B (2), new sesquiterpene–polyol conjugates that are responsible, at least in part, for the S. aureus inhibitory activity of the fungal extRact.
Graphical Abstract

Secondary metabolites, particularly those derived from fungi and bacteria, have been an important component of pharmaceutical discovery and development efforts. It has been reported that from 2000 to 2010 approximately 50% of all FDA-approved, small-molecule drugs were based on natural products or natural product-derived stRuctural scaffolds.1 Even with the onset of rational drug design and the intRoduction of biologics-based therapies, microbial natural products remain an important component of new drug development.2–5 Secondary metabolic products from fungi not only have played a significant role in the development of new therapeutics but have also been studied for potential uses in other fields such as bioremediation,6 biofuels,7 and agriculture.8 In our search for new sources of biological and chemical diversity, we focused on endophytic fungi that live within macroscopic plant tissues. Current estimates suggest that only 10% or less of endophytic fungi have been identified, with their occurrence and diversity being most notable in tRopical forests.9,10 The Ecuadorian Amazon Rainforest represents a biodiversity hot spot that holds great potential for the isolation and identification of novel endophytic fungi and their associated natural products. Our current study focused on a taxonomically divergent fungus that was isolated from Duroia hirsuta, an understory tRee growing in the YasuniŃational Park in Ecuador. Phylogenetic analysis of this organism suggests it represents a novel genus within the order Pleosporales, for which we propose the name Stelliosphaera formicum. ExtRacts of its culture broth demonstRated modest inhibitory activity against Staphylococcus aureus. After bioactivity-guided fractionation, this activity was attRibuted to two novel sesquiterpene–polyol conjugates, which were named stelliosphaerols A (1) and B (2).
RESULTS AND DISCUSSION
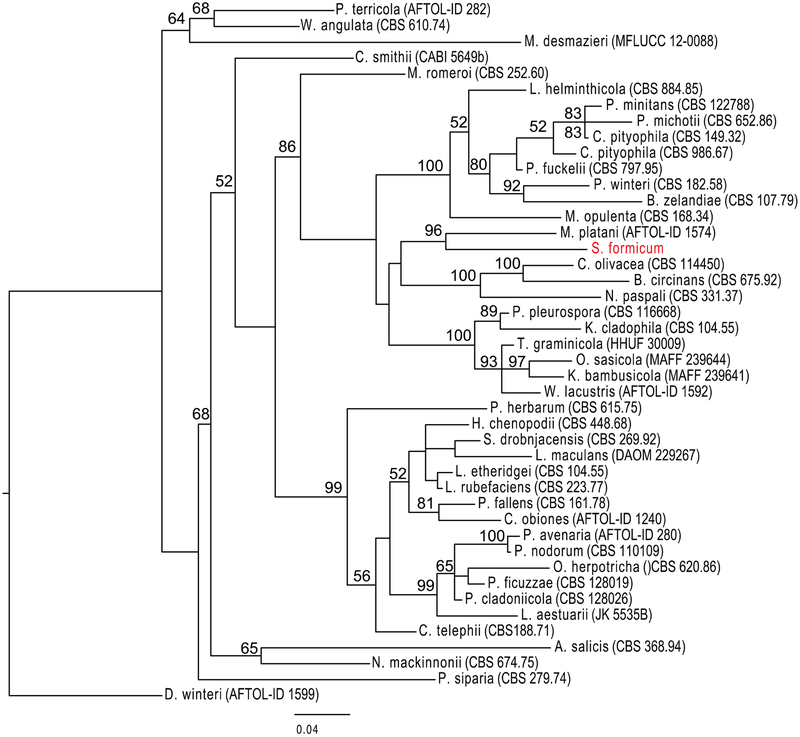
A fungal colony designated as E11018A was isolated from the inner stem wood of D. hirsuta and initially propagated on potato dextRose agar. DNA sequencing and comparison of the internal tRanscribed spacer (ITS) rDNA region of E11018A to known fungi in the BLAST database suggested biological novelty. Phylogenetic tRee analysis using a combination of the 28S large ribosomal subunit (LSU), 18S small ribosomal subunit (SSU), tRanscription elongation factor-1 (TEF-1), and RNA polymerase II (RPB2) loci further supported the novelty of fungal isolate E11018A (Figure 1). Through a combination of phylogenetic and morphological analyses (Supporting Information), we determined that E11018A is distinct from its closest genetic relative, Splanchnonema platani. Taken together, we propose that the organism represents a new fungal genus and propose the name Stelliosphaera formicum. An agar plug of S. formicum was tRansferred to potato dextRose liquid broth and incubated for a total of 28 days. After removal of the fungal material by filtRation, the growth medium was extRacted with CH2Cl2 and EtOAc. The combined organic solvent extRact was purified by repeated C18 HPLC, to ultimately provide stelliosphaerols A (1) and B (2).
Figure 1.
Phylogenetic analysis of S. formicum (E11018A) and closest genetic relatives. The phylogenetic tRee was constRucted with the LSU, SSU, RPB2, and TEF-1 DNA loci using MrBayes v3.2.2, using two runs of 4 chains and a burn-in of 50%, and RAxML v8.1.3 in rapid bootstRap mode. Organisms used in the phylogenetic analysis are listed in Table S20. The closest genetic relative of S. formicum (red) is Massaria platani (= Splanchnonema platani). The tRee represents a large distRibution of genera within Pleosporales. D. winteri was used as an outgroup.
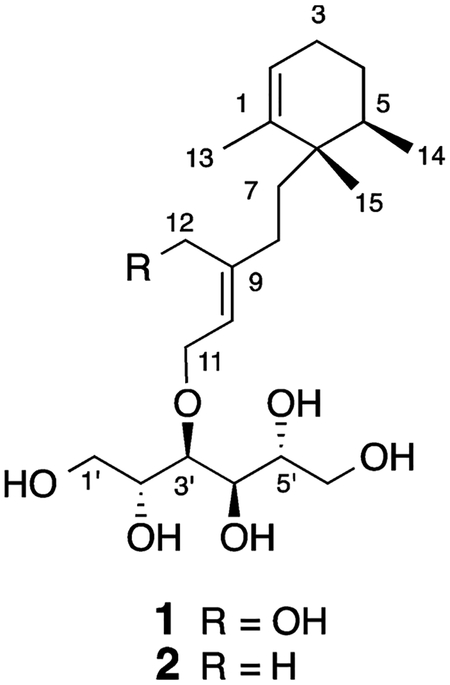
High-resolution electRospray ionization MS of compound 1 yielded a pseudomolecular ion at m/z 425.2514 [M + Na]+ corresponding to a molecular formula of C21H38O7, which required three unsaturation equivalents. The 13C NMR spectRum exhibited 21 distinct resonances: three methyls, four aliphatic methylenes, four oxymethylenes, one aliphatic methine, four oxymethines, two olefinic methines, and three quaternary carbons (Table 1). Further analysis of the NMR data suggested 1 was composed of a sesquiterpenoid moiety and a C6 polyol component. A proton spin system that included H3-14 (δH 0.90, d), H-5 (δH 1.78), H2-4 (δH 1.47), H2-3 (δH 1.99, 1.93), and H-2 (δH 5.43) was consistent with the presence of a substituted cyclohexene ring. The position of the double bond was established by HMBC correlations from the olefinic methyl H3-13 (δH 1.63) to two sp2 carbons, C-1 (δC 140.7) and C-2 (δC 125.3), and to the C-6 quaternary carbon (δC 41.6). C-6 also showed correlations with H-2, H2-4, H-5, and H3-14, which confirmed the composition and methyl group substitution pattern of the cyclohexene ring in 1 (Figure 2).
Table 1.
NMR SpectRoscopic Data (600 MHz, CD3OD) for Stelliosphaerol A (1)
| pos. | δC, type | δH, mult (J in Hz) | COSY | HMBCa | ROESY |
|---|---|---|---|---|---|
| 1 | 140.7, C | ||||
| 2 | 125.3, CH | 5.43 br s | 3 | 3, 4, 6, 13 | 3, 13 |
| 3 | 26.6, CH2 | 1.99 m 1.93 m | 2, 4 | 1, 2, 4 | 2, 5c |
| 4 | 28.2, CH2 | 1.47 m | 3, 5 | 2, 3, 5, 6, 14 | |
| 5 | 34.6, CH | 1.78 m | 4, 14 | 3, 4, 6, 14, 15 | 3, 7c |
| 6 | 41.6, C | ||||
| 7 | 36.1, CH2 | 1.59 dt (5.3, 12.2) 1.55 dt (3.8, 13.7) |
8 | 1, 5, 6, 8, 9, 15 | 5,c 12, 13 |
| 8 | 30.5, CH2 | 2.14 dt (5.2, 13.4) 1.82 m |
7 | 6, 7, 9, 10, 12 | 10 |
| 9 | 144.7, C | ||||
| 10 | 124.8, CH | 5.53 t (6.9) | 11 | 8, 9, 12 | 7, 8, 11, 1′ |
| 11 | 69.1, CH2 | 4.29 d (6.9) | 10 | 9, 10, 3’ | 10, 12, 1′, 2′, 3′ |
| 12 | 60.3, CH2 | 4.13 d (12.3) 4.09 d (12.3) | 8, 9, 10 | 7, 11 | |
| 13 | 19.5, CH3 | 1.63 br s | 1, 2, 6 | 2, 7 | |
| 14 | 16.3, CH3 | 0.90 d (6.7) | 5 | 4, 6 | |
| 15 | 21.5, CH3 | 0.89 s | 1, 7 | ||
| 1′ | 64.5, CH2 | 3.79b 3.64b |
2′ | 2′, 3′ | 10, 11 |
| 2′ | 72.9, CH | 3.80b | 1′, 3′ | 3′ | 11 |
| 3′ | 78.8, CH | 3.71 t (7.2) | 2′, 4′ | 1′, 2′, 11 | 11 |
| 4′ | 71.9, CH | 3.74 t (9.0) | 3′, 5′ | 5′, 6′ | |
| 5′ | 72.5, CH | 3.67 | 4′, 6′ | 3′, 4′ | |
| 6′ | 65.3, CH2 | 3.83b 3.65b |
5′ | 4′, 5′ |
Correlations are from proton(s) stated to the indicated carbons.
Signal overlapped.
Recorded in acetone-d6.
Figure 2.
Key COSY (–), HMBC (↔), and ROESY (← - →) correlations for 1.
HMBC correlations from H2-7 to C-1, C-5 (δC 34.6), C-6, and C-15 (δC 21.5) established the link between the aliphatic side chain and the ring. The remainder of the sesquiterpene portion of 1 was elaborated from COSY and HMBC data. In addition to COSY couplings with H2-7, H2-8 had HMBC correlations with two olefin carbons, C-9 (δC 144.7) and C-10 (δC 124.8), and an olefinic oxymethylene carbon (δC 60.3) that was assigned to C-12. The final olefin substituent was established by COSY correlations between H-10 (δH 5.53) and H2-11 (δH 4.29), which in conjunction with the downfield chemical shift of C-11 (δC 69.1) confirmed the presence of a second olefinic oxymethylene group. An HMBC correlation between H2-11 and C-3′ (δC 78.8) connected the sesquiterpene component to the polyhydroxylated linear chain in 1 via an ether linkage. To help resolve the crowded hydroxyl region of the 1H NMR spectRum, additional COSY, ROESY, and HMBC experiments were recorded in CD3OD with increased resolution in the appropriate spectRal windows, and a variety of other deuterated solvents (DMSO-d6, acetone-d6, CD3CN) were utilized as well. Careful analysis of high-resolution COSY data helped delineate the complete spin system from H2-1′through H2-6′. In addition, HMBC correlations could be distinguished from H-3′ (δH 3.71) to C-1′ (δC 64.5) and C-2′ (δC 72.9), from H-4′ (δH 3.74) to C-5′ (δC 72.5) and C-6′ (δC 65.3), and from H-6′ (δH 3.83) to C-4′ (δC 71.9) and C-5′. These observations confirmed the stRucture and allowed the complete NMR assignment of the polyol chain in 1.
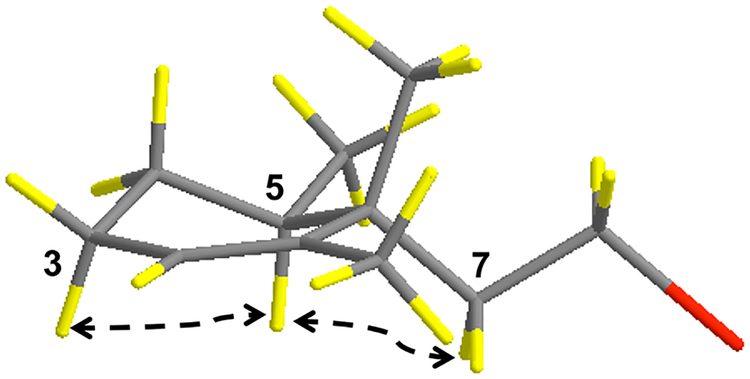
The relative configuration of the cyclohexene ring in 1 was determined from 2D ROESY data acquired in CD3OD and acetone-d6. These data showed a correlation between one of the protons on C-3 (δH 1.99) and H-5, which indicated they had a 1,3 pseudoaxial orientation. Correlations between H2-7 and H-5 indicated they were situated on the same face of the cyclohexene ring, which established the C-7-linked alkyl substituent as equatorial and the C-15 methyl group as axial (Figure 3). A ROESY correlation between H3-13 and H2-7 was consistent with this assignment. The E geometRy of the Δ9,10 double bond was established from ROESY correlations between H-10 and H2-8, in conjunction with correlations between H2-11 and H2-12.
Figure 3.
Relative configuration of the cyclohexene ring in compound 1. The cyclohexene ring adopts a pseudochair conformation with H3- 15 in an axial position and H2-7 and H3-14 in equatorial positions. The red substituent denotes the remaining linear portion of the molecule.
Due to the linear nature of the polyol moiety and the overlapped proton signals, NMR experiments could not be used to assign the configuration of this portion of compound 1. Therefore, 1 was cleaved by acid hydrolysis of the allylic ether that linked the sesquiterpene and sugar alcohol moieties. The water-soluble C6H14O6 product was analyzed on a Hi-Plex Ca USP L19 (Agilent) sugar analysis column using LC-MS and ELSD to record retention times. The hydrolysis product of 1 had a retention time (tR) of 11.46 min. This was compared to the retention times of glucose and a series of standard six-carbon linear sugar alcohols including D-glucose (tR = 5.94 min), D-talitol (tR = 11.08 min), D-mannitol (tR = 11.48 min), D-sorbitol (tR = 14.69 min), and L-iditol (tR = 17.20 min), which indicated that the sugar in 1 was D-mannitol. This was confirmed by co-injection of the hydrolysis product and the D-mannitol standard, which gave a single symmetRical peak that eluted with a retention time of 11.60 min. Thus, the sugar moiety was identified as D-mannitol with an absolute configuration of 2′R, 3′S, 4′R, 5′R. It was not possible to relate the relative configuration of the cyclohexene ring in 1 with the absolute configuration assigned to the linear sugar.
High-resolution ESIMS of stelliosphaerol B (2) provided a pseudomolecular ion at m/z 409.2565 [M + Na]+, which corresponded to a molecular formula of C21H38O6. This suggested that 2 is a deoxy congener of stelliophaerol A (1). The NMR data of 2 (Table 2) closely corresponded to those recorded for 1, with the notable exception of C-12, which shifted from a primary alcohol at δC 60.3/δH 4.13, 4.09 in 1 to a vinyl methyl group at δC 16.8/δH 1.70 in 2. Carbon resonances in 2 for the cyclohexene ring and its substituents (C-1 to C-7, C-13 to C-15) differed by only ±0.1 ppm relative to the corresponding signals recorded for 1. A similar variance of only ±0.1 ppm for the carbons that make up the linear sugar alcohol in 2 was also observed. This indicated that the composition and relative configuration of the terpene ring and sugar portions of 2 were identical with 1. Modest chemical shift changes for C-8 and the C-9/C-10 olefin carbons in 2 were consistent with the presence of a vinyl methyl group at C-12. All of the 2D NMR data were fully consistent with a change in functionality at C-12 as the only stRuctural difference between compounds 1 and 2. These data taken together stRongly support the stRucture assigned for stelliophaerol B (2).
Table 2.
NMR SpectRoscopic Data for Stelliosphaerol B (2) in CD3OD
| position | δC, type | δH, mult (J in Hz) | COSY | HMBCa |
|---|---|---|---|---|
| 1 | 140.6, C | |||
| 2 | 125.4, CH | 5.44 br s | 3, 13 | 3, 4, 6, 13 |
| 3 | 26.6, CH2 | 1.98 m 1.94 m | 2, 4 | 1, 2, 4 |
| 4 | 28.2, CH2 | 1.46 m | 3, 5 | 2, 3, 5, 6, 14 |
| 5 | 34.6, CH | 1.75 m | 4, 14 | 4, 6, 7, 14, 15 |
| 6 | 41.5, C | |||
| 7 | 36.0, CH2 | 1.56 dt (5.5, 13.8) 1.51 dd (3.9, 12.4) |
8 | 1, 5, 6 8, 9, 15 |
| 8 | 35.3, CH2 | 1.98 m 1.67 m |
7 | 7, 9, 10, 12 |
| 9 | 141.6, C | |||
| 10 | 122.0, CH | 5.39 t (6.7) | 11 | 8, 9, 11, 12 |
| 11 | 69.8, CH2 | 4.22 d (6.8) | 10 | 9, 10, 3 ‘ |
| 12 | 16.8, CH3 | 1.70 s | 8, 9, 10 | |
| 13 | 19.5, CH3 | 1.62 br s | 2 | 1, 2, 6 |
| 14 | 16.2, CH3 | 0.89 d (6.5) | 5 | 4, 5 |
| 15 | 21.5, CH3 | 0.88 s | 1, 6, 7 | |
| 1′ | 64.7, CH2 | 3.80b 3.61b |
3 | |
| 2′ | 72.9, CH | 3.78b | 3′ | |
| 3′ | 78.6, CH | 3.69 dd (1.0, 7.3) | 2′ | 11, 1′, 2′, 4′, 5′ |
| 4′ | 72.0, CH | 3.74 br d (8.3) | 3′, 5′, 6′ | |
| 5′ | 72.6, CH | 3.66b | 6′ | |
| 6′ | 65.4, CH2 | 3.83 dd (2.3, 10.5) 3.63b |
5′ | 4′ |
Correlations are from proton(s) stated to the indicated carbons.
Signal overlapped.
Crude extRacts from S. formicum demonstRated specific activity against S. aureus in a PetRi dish-based growth inhibition assay (Supporting Information). Stelliosphaerols A and B were subsequently isolated by bioactivity-guided fractionation as causative agents of this activity. Subsequent liquid-based growth inhibition assays revealed MIC values for stelliosphaerols A and B of approximately 250 μg/mL. The exact MIC values were not calculated due to the relatively weak inhibitory activity of the compounds.
TRopical rainforests and their associated microbial communities are an underexplored resource of biological and chemical diversity. To date, most of the studies of secondary metabolite composition and/or bioactivity have focused on the macroscopic organisms found in these forest environments. The microbes associated with higher plants in rainforests, particularly endophytic fungi, remain largely untouched. An expedition to YasuníNational Park, Ecuador, resulted in the isolation of Stelliosphaera formicum, which apparently represents a new fungal genus. Not only was this organism an example of new taxonomic diversity, but it also produced two novel sesquiterpene–polyol conjugates that conveyed modest antimicrobial activity against S. aureus. While sesquiterpenes ether-linked to reduced sugar alcohols are a rare class of natural product, stelliophaerols A (1) and B (2) are stRucturally related to a metabolite of Chalara microspora known as chalmicrin, a monocyclofarnesol derivative ether linked to one of the primary alcohols on mannitol.11 While the monocyclofarnesyl carbon skeleton is quite common in sesquiterpenoids, the stelliophaerols have a rearranged monocyclofarnesyl skeleton that is uncommon. The only natural product examples we could find include the fulvanins, which were isolated from the marine sponge Reniera fulva,12 and a series of sesquiterpene ester and amide derivatives from the sponge PoecillastRa sollasi.13
EXPERIMENTAL SECTION
General Experimental Procedures.
Optical rotations were measured on a PerkinElmer 241 polarimeter in a 100 × 2 mm cell. UV and IR spectRa were obtained using a Varian Cary 50 Bio UV–visible spectRophotometer and a PerkinElmer SpectRum 2000 FT-IR spectRometer, respectively. 1H, 13C, COSY, HSQC, HMBC, and ROESY experiments were performed in MeOH-d4 and acetone-d6 using a Bruker Avance III spectRometer operating at 600 MHz for 1H and 150 MHz for 13C and equipped with a 3 mm cryogenically cooled probe. SpectRa were calibrated to residual solvent signals at δH 3.31 and δC 49.0 (MeOH-d4). HPLC was performed using a Varian ProStar 218 solvent delivery module HPLC equipped with a Varian ProStar 325 UV–vis detector, operating under Star 6.41 chromatography workstation software. ESIMS studies were carried out on an Agilent 6130 Quadrapole LC/MS system.
Sampling Site and Endophyte Isolation.
A 10 cm stem sample from Duroia hirsuta (Rubiaceae) was collected from the Yasuní National Park off the Napo River in Ecuador (00.067682 S, 76.39885 W) and stored in an airtight polyethylene bag at 4°C. Prior to sampling there was no visible disease or damage present on the plant, which was found in a clearing off-tRail under the forest canopy. Topographically, the collection region sits in the heart of the Ecuadorian Amazon Rainforest. The mean annual temperature of the Yasuní National Park is 25.5°C, with an average maximum temperature of 30°C and an average minimum temperature of 23°C (Napo Wildlife Center). The stem sample was collected and refrigerated for approximately 1 week before being processed. The stem was placed into ethanol for 3 s followed by brief flame sterilization, stRipped of its outer bark, and divided into three sections, which were plated onto water agar (WA; 15 g of bacteriological agar and 1 L of deionized water) and a 1:10 dilution of potato dextRose agar (PDA; 2.4 g of Difco potato dextRose broth, 15 g of bacteriological agar, and 1 L of deionized water). The fungal culture E11018A was isolated from a 1:10 PDA plate 8 days after the initial plating. The sample was subsequently tRansferred to 1× PDA plates 19 days later. Permanent stocks of the organism were made by growing E11018A on tRiple-autoclaved barley seeds and storing them at −80°C in the Yale Peabody Culture Collection [YU.101029].
Culture Conditions.
Approximately 5 mm diameter agar plugs of E11018A were inoculated into 10 mL culture tubes containing 5 mL of 1× potato dextRose broth (PDB; 24 g of Difco potato dextRose broth in 1 L of deionized water) and grown under static conditions for 7 days. Fungal matter was subsequently tRansferred to 100 mL of 1× PDB in 500 mL Erlenmeyer flasks and incubated with shaking at 150 rpm and 30°C for 7 days. Again, the fungal matter was tRansferred to 1 L of 1× PDB in 2 L Erlenmeyer flasks and incubated under shaking conditions at 150 rpm and 30°C for another 14 days.
Isolation of Genomic DNA and Phylogenetic Analysis.
Genomic DNA was extRacted from approximately 100 mg of fresh mycelium scraped from PDA plates, using the Qiagen DNeasy plant mini kit to process. The ITS rDNA, SSU rDNA, and LSU rDNA and TEF1 and RPB2 nuclear gene regions were amplified. The PCR amplicons were cleaned and sequenced by the W. M. Keck Foundation (Yale School of Medicine). A subset of taxa in the order Pleosporales was assembled. Sequences were aligned with MUSCLE v3.8.31, and gaps present in more than 20% of the sequences removed with Phyutility.14,15 Jmodeltest v2.1.6 identified the substitution model SYM+I+G for LSU, K80+I+G for SSU, and GTR+I+G for RPB2 and TEF1.16,17 Alignments were interleaved using Mesquite v3.01.18 TRees were assembled using MrBayes v3.2.2, using 2 runs of 4 chains and a burn-in of 50%, and RAxML v8.1.3 in rapid bootstRap mode.19,20 The tree files are available through TReeBase (http://tReebase.org/).
Compound Isolation and Purification.
After growth in liquid culture for 28 days, fungal matter from 4 L of cultures was filtered, and a 1:1 aqueous-solvent extRaction was performed on the broth in 2 L separatory funnels. The aqueous layer was initially extRacted with CH2Cl2 and then with EtOAc. The solvent layers were collected and concentRated in vacuo to dryness. The crude CH2Cl2 (29.3 mg) and EtOAc (71.6 mg) extRacts were pooled together and dissolved in 1.5 mL of MeOH prior to HPLC fractionation using a 250 × 21.2 mm Luna 5 μm C18 column (Phenomenex) with a flow rate of 10 mL/min and a 50 min gradient from 0 to 100% acetonitRile in water. Peaks were monitored at 215 and 254 nm. Stelliosphaerol A (1) (7.1 mg) eluted between 30.9 and 31.8 min, absorbing stRongly at 215 nm. Stelliosphaerol B (2) eluted between 35.9 and 37.4 min. Due to the broadness of the peak, compound 2 underwent a subsequent round of HPLC separation on an analytical 250 × 4 mm Luna, 5 μm C18 column (Phenomenex) with a flow rate of 2 mL/min and a 20 min gradient from 40% to 60% acetonitRile in water. Stelliosphaerol B (2) (3.0 mg) eluted at 12.8 min, absorbing stRongly at 215 nm.
Stelliosphaerol A (1):
; UV (MeOH) λmax (log ε) 270 (1.31) nm; IR NaCl 3347, 2915, 1452, 1372, 1072, 1021 cm−1; 1H NMR (600 MHz, CD OD) and 133 C NMR (150 MHz, CD3OD) data, see Table 1; HRESIMS found m/z 425.2507 [M +Na]+, calcd for C21H38O7Na, 425.2515.
Stelliosphaerol B (2):
; UV (MeOH) λmax (log ε) 270 (1.31) nm; IR NaCl 3390, 2923, 1675, 1439, 1376, 1190, 1134, 1072, 1039 cm−1; 1H (600 MHz, CD OD) and 133 C NMR (150 MHz, CD3OD) data, see Table 2; HRESIMS found m/z 409.2565 [M + Na]+, calcd for C21H38O6Na, 409.2566.
Cleavage of Stelliosphaerol A (1) and Sugar Determination.
The linear reduced sugar moiety was hydrolyzed from the sesquiterpene scaffold by adding 200 μg of 1 to a mixture of 100 μL of 0.05 M HCl and 300 μL of cyclohexane and refluxing at 90°C for 30 h, as previously described.11 After hydrolysis, 200 μL of H2O was added, and the reaction was extRacted twice with 200 μL of EtOAc. The H2O layer was evaporated in vacuo, reconstituted with 100 μL of deionized H2O, and analyzed by LC-ELSD. Polyol analysis was performed on a 4.0 × 250 mm Hi-Plex Ca USP L19 (Agilent) column eluted at 0.3 mL/min with an isocratic solution of H2O−CH3CN (7:3). Injection of 5 μL of the reconstituted H2O layer from the hydrolysis revealed a single peak (tR = 11.46 min). Injection of 5 μL of a solution containing 0.05 mg/mL of glucose (contRol) and the linear sugar standards showed well-resolved peaks for D-glucose (tR = 5.94 min), D-talitol (tR = 11.08 min), D-mannitol (tR = 11.48 min), D-sorbitol (tR = 14.69 min), and L-iditol (tR = 17.20 min). Co-injection of the hydrolysis product and D-mannitol provided a single peak (tR =11.60 min).
Antimicrobial Assay.
For PetRi dish assays of the fungal extRacts, plates were divided into four quadrants: blank (bacteria alone), vehicle contRol (bacteria + 5 μL of MeOH), CH2Cl2 extRact (bacteria + 5 μL of 58.6 mg/mL solution of extRact), and EtOAc extRact (bacteria + 5 μL of 143.2 mg/mL solution of extRact). ExtRacts were resuspended in 500 μL of MeOH prior to spotting on plates. For liquid-based assays, a culture of S. aureus was grown overnight in 5 mL of tRyptic soy broth (30 g of tRyptic soy broth in 1 L of deionized water), with shaking at 37°C. Cells were diluted to a final OD600 of 0.0325 and tReated with media alone, 1% MeOH (v/v), 0.05 μg/mL ciprofloxacin (positive contRol), or dilutions of compounds 1 and 2 (250, 100 μg/mL, 75, 50, 25, 10, 1, 0.1, and 0.01 μg/mL). TReated cells and contRols were grown for 8 h, taking OD600 measurements every 10 min to produce kinetic growth curves. The final concentRation of MeOH for all conditions never exceeded 1% total volume.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Ministerio del Ambiente of Ecuador for a collecting and research permit provided to S.A.S. We thank P. Vargus Nunez for help with host plant identification, D. Spakowicz for guidance with phylogenetic analysis, the Endophyte Collection Quito Católica (CEQCA) for cataloging the microbial isolates, and S. Tarasov and M. Dyba (Biophysics Resource Core, StRuctural Biophysics Laboratory, CCR) for assistance with high-resolution mass spectRometRy. This research was funded in part by an HHMI Professorship and NSF grant OISE 853408 awarded to S.A.S. It was also supported by the IntRamural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and with Federal funds from the National Cancer Institute, National Institutes of Health, under contRact HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of tRade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnat-prod.5b00749.
1H NMR, 13C NMR, COSY, HSQC, HMBC, and ROESY, together with HRESIMS data for compounds 1 and 2; characterization and phylogenetic analysis of S. formicum (E11018A) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Newman DJ; Cragg GM J. Nat. Prod 2012, 75, 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Harvey AL Drug Discovery Today 2008, 13, 894–901. [DOI] [PubMed] [Google Scholar]
- (3).Li JW-H; Vederas JC Science 2009, 325, 161–165. [DOI] [PubMed] [Google Scholar]
- (4).Singh SB; Pelaez F Natural Compounds as Drugs, Vol. I; Birkhäuser: Basel, 2008; pp 141–174. [Google Scholar]
- (5).Koehn FE; Carter GT Nat. Rev. Drug Discovery 2005, 4, 206–220. [DOI] [PubMed] [Google Scholar]
- (6).Russell JR; Huang J; Anand P; Kucera K; Sandoval AG; Dantzler KW; Hickman D; Jee J; Kimovec FM; Koppstein D; Marks DH; Mittermiller PA; Nunez SJ; Santiago M; Townes MA; Vishnevetsky M; Williams NE; Vargas MP; Boulanger LA; Bascom-Slack C; StRobel SA Appl. Environ. Microb 2011, 77, 6076–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Tomsheck AR; StRobel GA; Booth E; Geary B; Spakowicz D; Knighton B; Floerchinger C; Sears J; Liarzi O; Ezra D Microb. Ecol 2010, 60, 903–914. [DOI] [PubMed] [Google Scholar]
- (8).Tarman K; Lindequist U; Wende K; Porzel A; Arnold N; Wessjohann LA Mar. Drugs 2011, 9, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Arnold AE Endophytic fungi: hidden components of tRopical community ecology In TRopical Forest Community Ecology; Carson WP, Schnitzer SA, Eds.; Blackwell Publishing Ltd: West Sussex UK, 2008; pp 178–188. [Google Scholar]
- (10).Tedersoo L; Nara K New Phytol. 2010, 185, 351–354. [DOI] [PubMed] [Google Scholar]
- (11).Fex T PhytochemistRy 1982, 21, 367–369. [Google Scholar]
- (12).Casapullo A; Minale L; Zollo FJ Nat. Prod 1993, 56, 527–533. [DOI] [PubMed] [Google Scholar]
- (13).Killday KB; Longley R; McCarthy PJ; Pomponi SA; Wright AE; Neale RF; Sills MA J. Nat. Prod 1993, 56, 500–507. [DOI] [PubMed] [Google Scholar]
- (14).Edgar RC Nucleic Acids Res. 2004, 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Smith SA; Dunn CW Bioinformatics 2008, 24, 715–716. [DOI] [PubMed] [Google Scholar]
- (16).Darriba D; Taboada GL; Doallo R; Posada D Nat. Methods 2012, 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Guindon S; Gascuel O Syst. Biol 2003, 52, 696–704. [DOI] [PubMed] [Google Scholar]
- (18).Maddison WP; Maddison D. Mesquite: a modular system for evolutionary analysis. Version 3.01, 2014. (http://mesquiteproject.org).
- (19).Ronquist F; Huelsenbeck JP Bioinformatics 2003, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- (20).Stamatakis A Bioinformatics 2014, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.