Abstract
Overexpression of Oct3/4, Klf4, Sox2, and c-Myc (OKSM) transcription factors can de-differentiate adult cells in vivo. While sustained OKSM expression triggers tumorigenesis through uncontrolled proliferation of toti- and pluripotent cells, transient reprogramming induces pluripotency-like features and proliferation only temporarily, without teratomas. We sought to transiently reprogram cells within mouse skeletal muscle with a localized injection of plasmid DNA encoding OKSM (pOKSM), and we hypothesized that the generation of proliferative intermediates would enhance tissue regeneration after injury. Intramuscular pOKSM administration rapidly upregulated pluripotency (Nanog, Ecat1, and Rex1) and early myogenesis genes (Pax3) in the healthy gastrocnemius of various strains. Mononucleated cells expressing such markers appeared in clusters among myofibers, proliferated only transiently, and did not lead to dysplasia or tumorigenesis for at least 120 days. Nanog was also upregulated in the gastrocnemius when pOKSM was administered 7 days after surgically sectioning its medial head. Enhanced tissue regeneration after reprogramming was manifested by the accelerated appearance of centronucleated myofibers and reduced fibrosis. These results suggest that transient in vivo reprogramming could develop into a novel strategy toward the acceleration of tissue regeneration after injury, based on the induction of transiently proliferative, pluripotent-like cells in situ. Further research to achieve clinically meaningful functional regeneration is warranted.
Keywords: plasmid DNA, OKSM, in vivo reprogramming, pluripotency, muscle, regeneration
Graphical Abstract
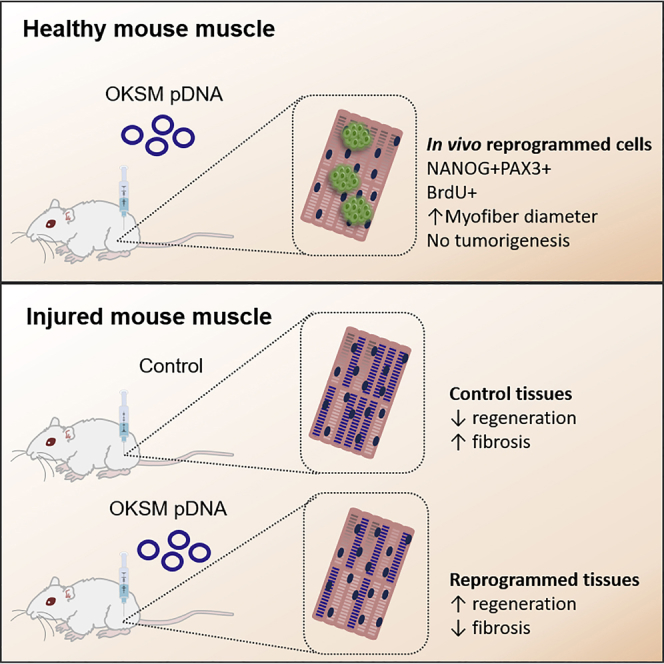
de Lázaro et al. force the expression of four exogenous reprogramming factors in mouse skeletal muscle, and they report transient proliferation of de-differentiated, progenitor-like cells in the absence of tumorigenesis. After physical injury to the muscle, this strategy leads to decreased fibrosis and accelerated regeneration at the histological level.
Introduction
Forced expression of Oct3/4, Klf4, Sox2, and cMyc transcription factors, dubbed Yamanaka or OKSM reprogramming factors, reverts a wide variety of differentiated cell types to pluripotency, albeit with differing efficiencies.1, 2, 3 Such conversion can be not only triggered in vitro but also within living organisms.4, 5, 6, 7, 8, 9, 10, 11, 12 In vivo cell reprogramming to pluripotency has been described in a variety of transgenic and wild-type (WT) animal models, in spite of pro-differentiation signals present in the tissue microenvironment, but with outcomes that vary depending on the pattern of OKSM overexpression.13 Ubiquitous and/or sustained expression of reprogramming factors leads to uncontrolled proliferation of toti- and pluripotent cells and widespread tumorigenesis.6, 7, 8, 9, 10, 11 On the contrary, transient OKSM expression generates pluripotent intermediates that proliferate only temporarily, preventing dysplasia and teratoma formation.4, 5, 7, 12 We proved this effect in WT mouse liver, using a non-viral approach based on hydrodynamic tail vein (HTV) injection of plasmid DNA (pDNA) encoding OKSM (pOKSM).5, 12 In vivo-reprogrammed cells, also known as in vivo-induced pluripotent stem cells (i2PSCs), demonstrated all the main hallmarks of functional pluripotency.14 Interestingly, other studies have shown that the frequency and duration of OKSM overexpression can be tuned to halt cell reprogramming at a proliferative but non-pluripotent stage that also avoids tumorigenesis.15, 16
In the adult mammalian organism, muscle repair relies mainly on the self-renewal and myogenic potential of resident stem cells, primarily satellite cells.17 The number and regenerative capacity of such cells varies across species and dramatically decreases with age;18, 19 hence, it can be exhausted if the injury is severe or repeated or occurs in elderly muscle. In such scenarios, various resident cell types differentiate into myofibroblasts that generate a fibrotic scar unable to meet the contractile requirements of the tissue, thus impeding the complete functional rehabilitation of the injured muscle.20, 21
Different strategies are currently being explored to enhance muscle regeneration after severe injuries, including surgical suturing22 or the administration of anti-fibrotic drugs,20, 21, 23, 24, 25 growth factors,26 replacement cells,27, 28, 29 combinations of these,30, 31, 32 and microRNAs (miRNAs) involved in muscle development.33 However, none of these approaches has yet reached routine clinical practice, and the treatment and management of major muscle injuries continues to be primarily conservative.34, 35 At the same time, the skeletal muscle offers an excellent platform for the expression of foreign genes in vivo, given the capacity of myofibers and interstitial cells to uptake naked pDNA after a simple intramuscular (i.m.) administration.36, 37
We have previously hypothesized that the generation of pluripotent or pluripotent-like cells in vivo, which are able to transiently proliferate and eventually re-differentiate to the appropriate phenotype within a damaged tissue, may assist its cellular repopulation, boosting its capacity to regenerate after a traumatic insult or injury.38 In the present work, we aimed first to explore whether transient, teratoma-free reprogramming could be achieved in mouse skeletal muscle via i.m. administration of pOKSM. We then interrogated whether cell reprogramming would enhance the regenerative capacity of the muscle after a surgically induced injury.
Results
Gene Expression in Mouse Skeletal Muscle after i.m. Administration of Reprogramming Factors
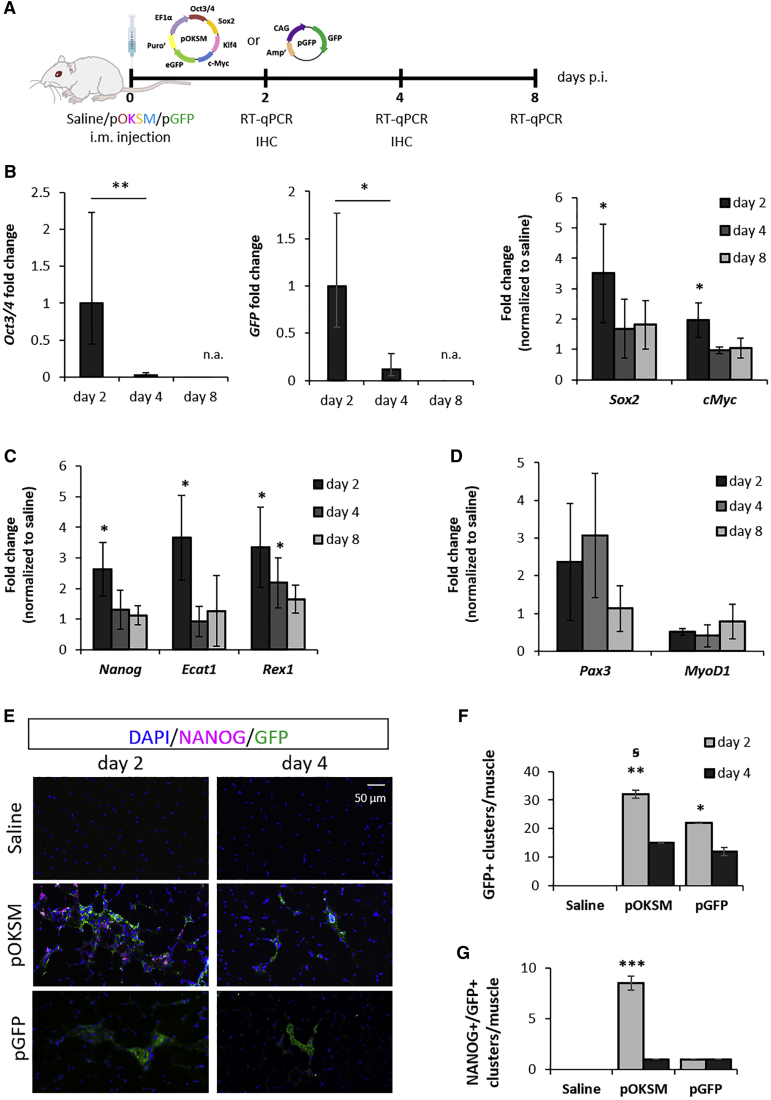
To test whether the expression of pluripotency-related genes could be induced by OKSM in mouse skeletal muscle, we first administered BALB/c mice with 50 μg of a single pDNA cassette, pOKSM, that encodes OKSM factors and a GFP reporter, in the gastrocnemius (GA). The contralateral hind limb was injected with the same volume of saline solution and used as a control (Figure 1A). Changes in gene expression in the GA were investigated at different time points after injection by real-time qRT-PCR (Figures 1B–1D). At 2 days after injection, Sox2 and c-Myc mRNAs were significantly upregulated in muscles administered with pOKSM, compared to saline-injected controls (p = 0.043 for Sox2 and p = 0.035 for cMyc). However, by day 4 we did not observe significant differences between groups (Figure 1B). Oct3/4 and GFP expression was not detected in saline-injected muscles; therefore, the relative expression was normalized to the values of pOKSM-injected muscles dissected on day 2. We observed a significant decrease in the levels of both mRNAs from day 2 to day 4 after injection (p = 0.003 for Oct3/4 and p = 0.042 for GFP), and none of such targets was amplified at a later time point (day 8 after injection; Figure 1B).
Figure 1.
Changes in Gene and Protein Expression after i.m. Administration of Reprogramming pDNA in Mouse GA
(A) BALB/c mice were i.m. injected into the GA with 50 μg pOKSM or pGFP in 50 μL 0.9% saline or with 50 μL saline alone. (B–D) GAs were dissected 2, 4, and 8 days after injection with pOKSM or saline, and real-time qRT-PCR was performed to determine changes in gene expression of (B) reprogramming factors and GFP transgene encoded in pOKSM, (C) endogenous pluripotency markers, and (D) genes involved in myogenesis. Gene expression levels were normalized to day 2 for Oct3/4 and GFP mRNA. **p < 0.01 and *p < 0.05 indicate statistically significant differences between day 2 and day 4 post-injection (p.i.), assessed by one-way ANOVA; n.a. indicates no amplification of the target. Expression levels of other transcripts were normalized to saline-injected controls and *p < 0.05 indicates statistically significant differences in gene expression between pOKSM- and saline-injected groups, assessed by one-way ANOVA or Welch ANOVA. Data are presented as 2ˆ-ΔΔCt ± propagated error, n = 3. (E) 10-μm-thick GA sections, obtained 2 or 4 days after saline, pOKSM, or pGFP injection, were stained with anti-NANOG and anti-GFP antibodies. Images were taken with a slide scanner at 20× magnification; scale bars represent 50 μm. (F) Number of GFP+ cell clusters per GA. *p < 0.05 and **p < 0.01 indicate statistically significant differences in the number of GFP+ clusters compared to saline-injected controls, and ꞩ for p < 0.05 indicates statistically significant differences between 2 and 4 days after pOKSM injection, assessed by one-way ANOVA and Tukey's post-hoc test (n = 2 GAs, 3 whole sections/muscle). (G) Number of NANOG+GFP+ cell clusters per GA. ***p < 0.001 indicates statistically significant differences in the number of NANOG+GFP+ clusters between pOKSM-injected muscles (2 days p.i.) and the rest of the groups, assessed by one-way ANOVA and Tukey’s test (n = 2 GAs, 3 whole sections/muscle).
Nanog, Ecat1, and Rex1 are genes expressed in the pluripotent state but repressed in adult tissues. Significant upregulation of these pluripotency markers was confirmed as early as 2 days after pOKSM administration (p = 0.021 for Nanog, p = 0.034 for Ecat1, and p = 0.04 for Rex1). Similar to the trend observed in the expression of reprogramming factors, their mRNA levels decreased afterward (Figure 1C). In addition, we investigated the expression of genes characteristically expressed at different stages during myogenesis. Higher mRNA levels of Pax3—expressed in myogenic progenitors, but not in differentiated myofibers39—were detected in the pOKSM-injected group. On the contrary, MyoD1, which is expressed in more committed cells during skeletal muscle development40 and is directly suppressed by Oct4 during myoblast-to-induced pluripotent stem cell (iPSC) reprogramming,41 was expressed at lower levels compared to saline-injected controls (Figure 1D). Again, these changes persisted only transiently.
To confirm that the above changes in pluripotency and myogenesis markers were indeed triggered by OKSM and not by the injection of pDNA itself, we also administered BALB/c mice with 50 μg pCAG-GFP (pGFP). This cassette encoded a GFP reporter but no OKSM factors (Figure S1A). As expected, Oct3/4 mRNA was not amplified by real-time qRT-PCR, and Sox2 and cMyc were expressed at the same levels as saline-injected controls (Figure S1B). In addition, the expression of pluripotency (Figure S1C) and myogenesis-related genes remained unaltered (Figure S1D). GFP mRNA levels remained stable for the duration of the study (8 days; Figure S1E).
Taken together, the changes in gene expression observed in pOKSM-injected tissues were compatible with a transient reprogramming event, whereby forced OKSM expression could activate an embryonic-like gene expression program, inducing the expression of pluripotency but also early myogenesis markers, in a subset of cells within the tissue.
Identification of Reprogrammed Cells within Muscle Tissue
The analysis of mRNA from bulk tissue did not allow us to determine whether the changes in the transcripts described above occurred in the same cells or to identify the specific cell subsets within the tissue that undertook reprogramming. Besides, the rapid decline in transgene mRNA levels upon pOKSM injection contrasted with the stable and long-term foreign gene expression that is normally observed after the uptake of pDNA in post-mitotic myofibers.42
To clarify this and to overcome the limitations of bulk tissue RNA analysis, we obtained muscle tissue sections 2 and 4 days after pDNA administration and identified transfected cells based on the expression of the GFP reporter (Figure 1E). Nanog is considered the archetypic marker of pluripotency,43 and it was significantly upregulated in pOKSM-injected muscles (Figure 1C); thus, we utilized an anti-NANOG antibody to distinguish reprogrammed cells within the skeletal muscle tissue. Interestingly, green fluorescence was found in both myofibers (Figure S2B) and interstitial mononucleated cells located among muscle fibers (Figure 1E), but we observed significant differences between pOKSM and pGFP-injected tissues. In pOKSM-injected muscles, 2 days after injection, some of such GFP+ interstitial cells were located in large cell clusters that stained positively for NANOG (Figure 1E). However, the overall number of GFP+ cell clusters (Figure 1F), and especially that of NANOG+GFP+ double-positive cell clusters, decreased significantly by day 4 in this group (p = 0.03 and p = 0.0002, respectively; Figures 1E–1G). The number of GFP+ cell clusters in pGFP-injected tissues did not change in such time frame (p = 0.62). This observation agrees with our mRNA data on transgene and pluripotency marker expression (Figures 1B and 1C; Figure S2E), and it could be explained if transient reprogramming induced cell division with the consequent loss of the transfected plasmid. Of note, this vector is known to remain mainly as an episome.44
NANOG was only found in pOKSM-injected tissues, always within clusters of GFP+ mononucleated cells among myofibers (Figure 1E). This observation further confirmed that the upregulation of the pluripotency marker was triggered by OKSM overexpression. It is worth noting that not all GFP+ cells within NANOG+GFP+ clusters expressed NANOG and that some GFP+ cell clusters in pOKSM-injected tissues contained no NANOG+ cells at all (Figures 1E–1G). Considering the well-known low efficiency of cell reprogramming, even under more controlled in vitro conditions,44 this result was expected. Figure S3 shows further details of representative NANOG+GFP+ cell clusters found on pOKSM-injected tissues, including orthogonal views that confirmed NANOG+GFP+ co-localization. The expression of NANOG was cytoplasmic, instead of its most common localization in the cell nucleus. However, such an event has been previously described by others.45, 46 NANOG was not detected from day 4 after i.m. injection (Figures 1E and 1F), which supported our observations at the mRNA level and confirmed that de-differentiation to a pluripotent-like state occurred only transiently.
The expression of GFP protein can trigger a mild immune cell reaction, albeit significant differences have been found among mouse strains and depending on the route of administration.47 We aimed to rule out the possibility that the large NANOG+GFP+ cell clusters observed in pOKSM-injected tissues corresponded in fact to the recruitment of immune cells around the needle tract left by the i.m. injection. First, we confirmed that no NANOG expression was detected around the needle tract in saline-injected and pGFP-injected muscles (Figure S2C). In addition, we found an average of 8.5 ± 0.7 NANOG+GFP+ cell clusters per pOKSM-injected GA, and those were distributed in proximal and distal areas from the injection site (Figure 1G). We also ruled out the possibility that pOKSM resulted in being more immunogenic than pGFP or saline injection by comparing mRNA levels of Cd11b, a leukocyte marker, among saline-, pGFP-, and pOKSM-injected muscles. We did not find significant differences among such groups (Figure S2D). Staining pOKSM-injected tissues with an anti-CD11b antibody confirmed that a number of CD11b+ cells were present within NANOG+GFP+ cell clusters; however, CD11b and NANOG expression did not co-localize in the same cells (Figure S2E). This may indicate that immune cells are recruited to the sites of NANOG+GFP+ cell clusters, but it confirms that their identity is different from that of reprogrammed cells.
In Vivo Reprogramming in Nanog-GFP and Pax3-GFP Transgenic Mouse Skeletal Muscle
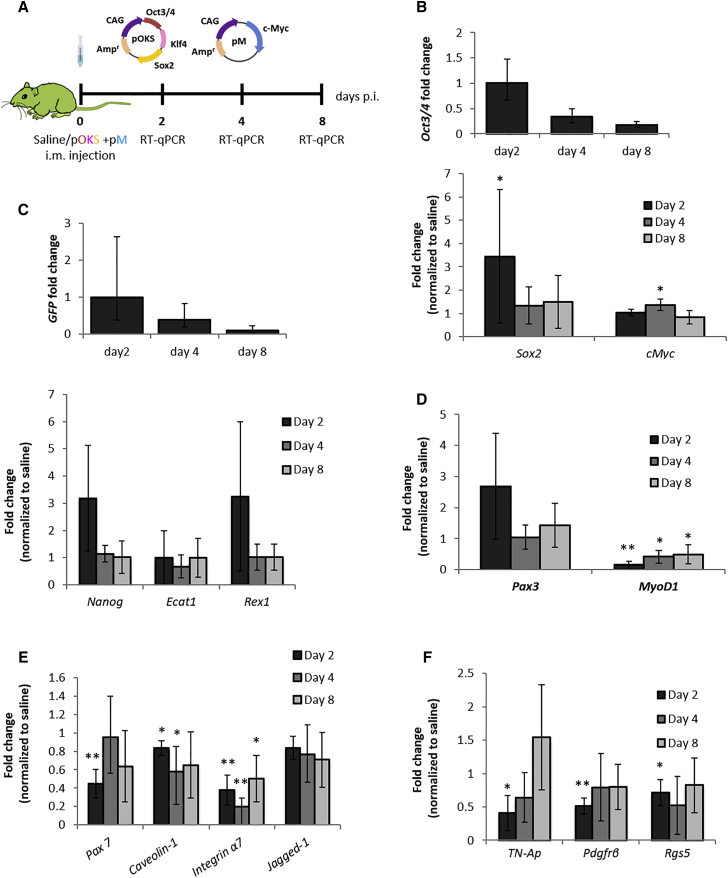
Sv129-Tg(Nanog-GFP) transgenic mice, referred to as Nanog-GFP for simplicity, have the reporter GFP sequence inserted in the Nanog locus.43 They were used to identify cells exhibiting pluripotency-like features in the tissue, thanks to the emission of green fluorescence. pOKS and pM plasmids, containing the OKSM reprogramming factors in two separate cassettes, were used instead of pOKSM to avoid overlap with the GFP reporter encoded in the latter (Figure 2A). Prior to any histological analysis, we confirmed that the expression of reprogramming factors, endogenous pluripotency genes, and myogenesis-related markers on days 2, 4, and 8 after i.m. injection was comparable to that observed in WT mice (Figures 2B–2D). As in the BALB/c strain, gene expression changes in Nanog-GFP mice GA suggested the transient reprogramming of a subset of cells within the muscle toward an embryonic-like state (Figures 2C and 2D).
Figure 2.
Changes in Gene Expression after i.m. Administration of Reprogramming pDNA in Nanog-GFP Mice
(A) Nanog-GFP transgenics were i.m. administered, 50 μg pOKS and 50 μg pM in 50 μL 0.9% saline or 50 μL saline alone, into the GA. (B–F) At 2, 4, and 8 days p.i. GA tissues were dissected, and real-time qRT-PCR was performed to determine the relative gene expression of (B) reprogramming factors, (C) endogenous pluripotency markers, (D) genes involved in myogenesis, (E) satellite cell markers, and (F) pericyte markers. Relative expression was normalized to day 2 values (Oct3/4 and GFP mRNAs) or to saline-injected controls (other genes). *p < 0.05 and **p < 0.01 indicate statistically significant differences in gene expression between pDNA and saline-injected groups, assessed by one-way ANOVA or Welch ANOVA. Data are presented as 2ˆ-ΔΔCt ± propagated error, n = 4.
In addition, we aimed to clarify the input of pericytes and satellite cells, since they are present in skeletal muscle and have myogenic potential.17, 48 Especially, satellite cells express Pax3 in certain muscles.39 Therefore, it was of particular importance to rule out the contribution of a hypothetical satellite cell proliferative response, which could be triggered by the injection trauma, toward the elevated mRNA levels of such transcription factor. With that aim, we investigated the expression of other satellite cell-specific markers (Pax7, Caveolin1, Integrin-α7, and Jagged1) and pericyte markers (TN-AP, Pdgfrβ, and Rgs5). A significant reduction of their mRNA levels was found in pOKSM-injected specimens compared to controls (Figures 2E and 2F), which ruled out an injection-induced proliferation of such cell types and rather pointed to them as being potential targets of reprogramming.
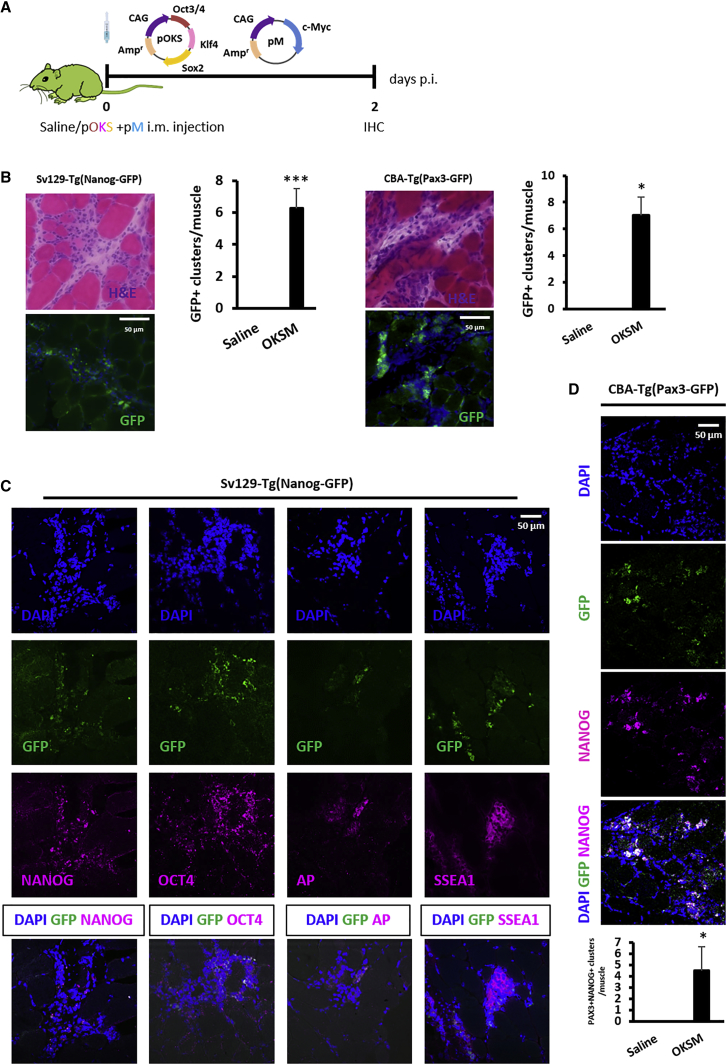
At the histological level (Figure 3), H&E staining revealed the presence of dense clusters of mononucleated cells among the myofibers in Nanog-GFP mice administered OKSM. The bright green fluorescence observed 2 days after injection (Figure 3B), which confirmed the expression of Nanog and hence the pluripotent-like identity of such cells, was not detected at later time points (data not shown), further confirming the transiency of reprogramming. An average of 6.3 ± 1.3 GFP+ clusters was identified per GA, in agreement with the number of NANOG+ clusters quantified in the BALB/c study (Figure 1G). In addition, GFP co-localized with the expression of several other pluripotency and embryonic stem cell-specific markers identified by immunohistochemistry (IHC) (NANOG, OCT4, AP, and SSEA1) (Figure 3C). The number of cell clusters positive for such markers is shown in Figure S4 and was in a similar order. No GFP signal or immunoreactivity for any of the pluripotency markers tested was found in saline-injected controls, excluding alkaline phosphatase (AP), which is also expressed by pericytes (Figures S5B and S5D).
Figure 3.
Characterization of In Vivo-Reprogrammed Cell Clusters in the GAs of Nanog-GFP and Pax3-GFP Mice
(A) Nanog-GFP and Pax3-GFP transgenics were administered, 50 μg pOKS and 50 μg pM in 50 μL 0.9% saline, into the GA and dissected 2 days p.i. for histological analysis. (B) Clusters of reprogrammed cells were identified by H&E and the green fluorescence resulting from either Nanog or Pax3 upregulation (100×; scale bars represent 50 μm). Bright-field and fluorescence images show the same region within the tissue. *p < 0.05 and ***p < 0.001 indicate statistically significant differences in the number of GFP+ clusters found in reprogrammed tissues compared to saline-injected controls, assessed by one-way ANOVA, n = 4 (Nanog-GFP mice) and n = 2 (PAX3-GFP mice). (C) IHC for the expression of pluripotency markers in Nanog-GFP GA administered with reprogramming pDNA (100×; scale bar represents 50 μm). (D) IHC for NANOG expression in Pax3-GFP GA administered with reprogramming pDNA (100×; scale bar represents 50 μm). *p < 0.05 indicates statistically significant differences in the number of NANOG+PAX3+ clusters found in reprogrammed tissues compared to saline-injected controls, one-way ANOVA; n = 2, 5 whole sections/muscle.
We also found cells staining positively for satellite cell and pericyte markers (PAX7 and PDGFrβ, respectively) within GFP+ cell clusters. However, similar to what we observed when we investigated the presence of immune cells in reprogrammed tissues, such cells did not express the green fluorescence characteristic of reprogrammed cells (Figure S5C). This observation confirmed that the appearance of large clusters of mononucleated cells within reprogrammed tissues was not due to the proliferation of satellite cells or pericytes, at least in the absence of a prior reprogramming event.
We then used the CBA-Tg(Pax3-GFP) transgenic mouse strain,49 shortened as Pax3-GFP, to confirm the upregulation of the early myogenesis marker Pax3. At 2 days after the injection of pOKS and pM, we found an average of 7 ± 1.4 GFP+ cell clusters per GA, similar to the number of GFP+ cell clusters observed in Nanog-GFP mouse tissues from the same treatment group (Figure 3B). Green fluorescence did not appear in saline-injected controls (Figure S5B). In addition, the staining of an anti-NANOG antibody in OKSM-injected tissues co-localized with the green fluorescence signal triggered by PAX3 expression (Figure 3D). This finding confirmed that reprogrammed cells expressing NANOG also expressed PAX3.
Overall, these data suggested that cells reprogrammed within the skeletal muscle tissue grew in clusters among myofibers and expressed pluripotency and early myogenic progenitor markers. While not all cells within the clusters in Nanog-GFP and Pax3-GFP specimens expressed the green reporter, this heterogeneity may again be explained by the limited efficiency of the reprogramming process. Taking also into account the transiency of the reprogramming event, we cannot rule out that some of such cells were already re-differentiating at the time point investigated (day 2). The occurrence of partial reprogramming events that do not reach the de-differentiation status of NANOG+PAX3+ cells may also be considered.
Short- and Long-Term Effects of In Vivo Reprogramming in Healthy Skeletal Muscle
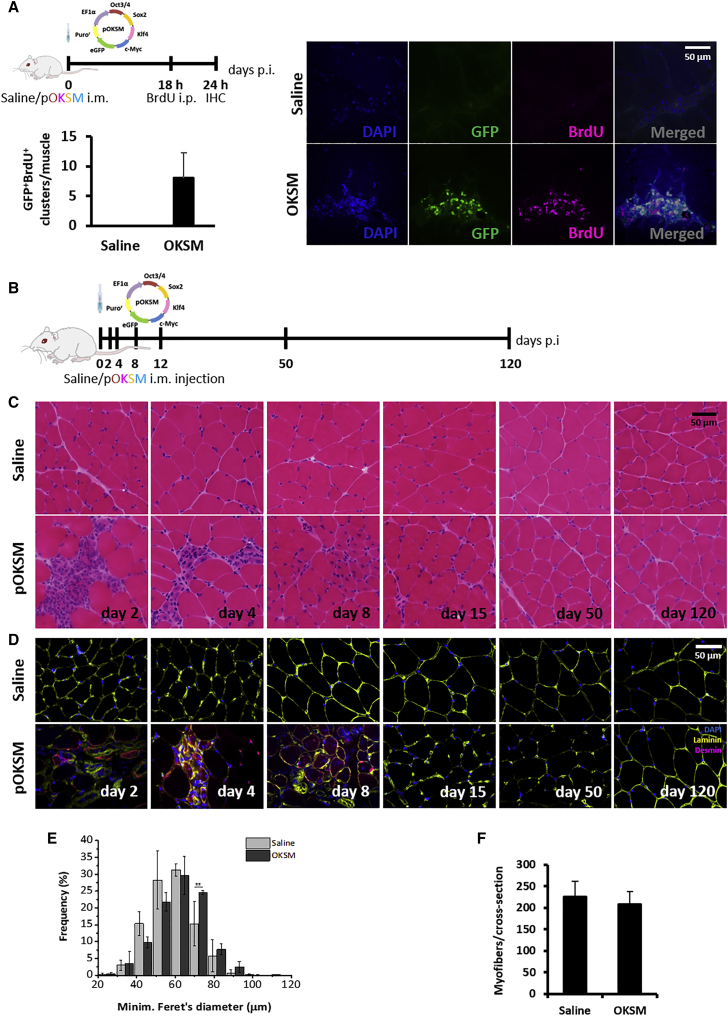
We next focused on the outcome of OKSM-mediated reprogramming in healthy BALB/c GA with an emphasis on signs of cell proliferation and of the potential appearance of dysplastic lesions or teratomas. Both short and long time points after reprogramming, from day 2 to 120 after pOKSM injection, were studied (Figures 4A and 4B). We first sought to obtain more stringent evidence of cell proliferation within the reprogrammed cell clusters, given that cell division is known as a distinct and indispensable step in the somatic-to-pluripotent conversion.50, 51 18 hr after i.m. administration of 50 μg pOKSM or saline control, BALB/c mice were intraperitoneally (i.p.) injected with 5-Bromo-2′deoxyuridine (BrdU), which can be incorporated into the DNA of proliferating cells.52 Tissues were collected 6 hr later for histological examination (Figure 4A). GA tissue sections were stained with an anti-BrdU antibody, while the GFP reporter in pOKSM was used to identify the cells transfected with reprogramming factors. We found co-localization of the two signals in cell clusters within the GA tissue in pOKSM-injected mice, which were morphologically identical to those observed in our previous studies. This finding confirmed that in vivo-reprogrammed cells not only acquired an embryonic-like gene expression profile but also proliferated actively.
Figure 4.
The Effect of In Vivo Reprogramming toward Pluripotency on Healthy Skeletal Muscle
(A) BALB/c mice were administered 50 μg pOKSM in 50 μL 0.9% saline or 50 μL saline alone in the GA. 18 hr later, BALB/c mice were i.p. administered 500 mg/kg BrdU. GAs were dissected 24 hr after pDNA injection. 10-μm-thick tissue sections were stained with anti-BrdU antibody. Green fluorescence corresponds to the GFP reporter encoded in the pDNA. Images were captured with a confocal microscope (100×). Scale bar represents 50 μm. (B) BALB/c mice were administered 50 μg pOKSM in 50 μL 0.9% saline or 50 μL saline alone in the GA. (C and D) Muscles were dissected 2, 4, 8, 12, 50, and 120 days p.i., sectioned, and stained with (C) H&E (100×; scale bar represents 50 μm) and (D) anti-desmin and anti-laminin antibodies (40×; scale bar represents 50 μm). (E) Fiber size distribution. *p < 0.05 indicates statistically significant differences in the frequency of 70-μm fibers between pOKSM and saline-injected groups, analyzed by one-way ANOVA; n = 3; data are presented as mean ± SD. (F) Number of myofibers per cross section. No statistically significant differences were found between pOKSM- and saline-injected animals, analyzed by one-way ANOVA; n = 3; data are presented as mean ± SD.
We then followed the evolution of such clusters in the reprogrammed tissues (Figure 4B). While they were very densely populated by small mononucleated cells on day 2 after injection, from day 4, small caliber, desmin-positive, centronucleated myofibers (characteristic features of regenerating, immature myofibers53, 54) started to predominate instead. By day 8, only a few desmin-positive, centronucleated myofibers remained, and none was noted at later time points (Figures 4C and 4D). Lower-magnification images of these observations are shown in Figure S6B. Very importantly, no teratomas or any signs of tissue abnormality were observed for the duration of the study (120 days). This observation, together with the progressive disappearance of cell clusters, suggests that the proliferative state confirmed by BrdU incorporation (Figure 4A) was only transient.
TUNEL staining indicated the presence of limited numbers of apoptotic nuclei in the GA tissue. However, those were only found at the earliest time points after injection (day 2) and in both conditions (saline and pOKSM injection); hence, their occurrence was attributed to the mild tissue damage caused by the injection along the needle tract (Figure S6C). The absence of significant cell death constitutes a first indication that in vivo-reprogrammed cells could successfully re-integrate into the muscle tissue. In the absence of an appropriate lineage-tracing system, we next performed morphometric analyses to indirectly follow the fate of reprogrammed cells. The distribution of myofiber diameter shifted to larger calibers compared to the saline-injected controls (Figure 4E), whereas the average number of myofibers per cross-sectional area remained unaltered (Figure 4F). These data could indicate that reprogrammed cells proliferate in clusters to then fuse with existing myofibers, enlarging their diameter, but do not form de novo fibers. If confirmed, this course of events would recapitulate those that occur during post-natal myogenesis.55 Nonetheless, ad hoc lineage-tracing studies must in the future be employed to confirm these findings.
Enhancement of Regeneration in a Surgically Induced Model of Skeletal Muscle Injury
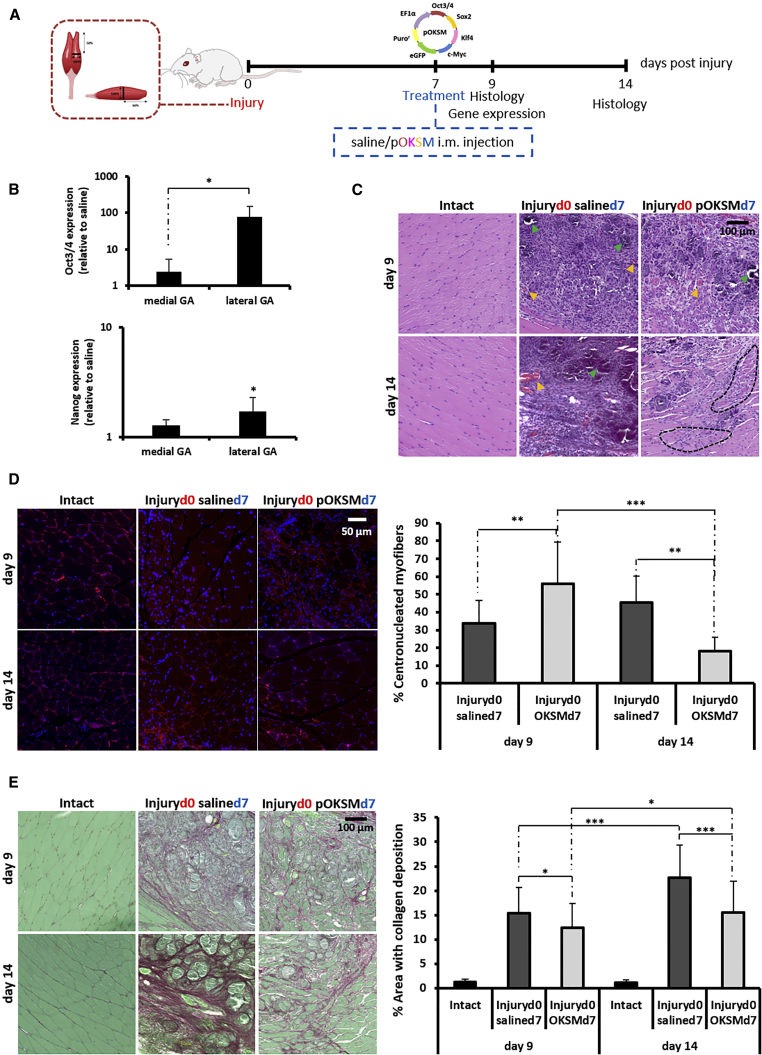
The capacity of in vivo reprogramming to enhance regeneration was interrogated in a clinically relevant model of skeletal muscle injury. The medial head of the left GA of BALB/c mice was surgically sectioned in the transverse plane (as represented in Figure S7A), and pOKSM was i.m. administered in the injured hind limb at the time of injury or 5 or 7 days later. We chose such timings so that significant endogenous regeneration would have not yet taken place, thus to explore if in vivo reprogramming would accelerate the process and to look for differences in the induction of reprogramming at different stages of muscle degeneration. Control mice bearing the same injury were injected with 0.9% saline solution alone and the contralateral (right) leg of each mouse was left intact (uninjured and uninjected) as an internal control. The analysis of Nanog mRNA levels in the tissue 2 days after the administration of pOKSM revealed that the maximum upregulation of the pluripotency marker was achieved when the Yamanaka factors were administered 7 days after injury (Figure S7B). We thus selected such a dose regimen for further studies, whereby muscle regeneration was investigated 9 and 14 days after injury (2 and 7 days after the administration of reprogramming factors or saline control) (Figure 5A). The uptake and expression of the reprogramming pDNA, evidenced by Oct3/4 expression, was confirmed on day 9. mRNA levels of the transgene were significantly higher in the lateral head of the GA compared to the directly injured medial head (p = 0.027), possibly due to the abundance of necrotic myofibers at the injury site. Similarly, the higher mRNA levels of Nanog were detected in the lateral head of the GA (p = 0.010 compared to saline control; Figure 5B).
Figure 5.
The Effect of In Vivo Reprogramming toward Pluripotency after Laceration of the Medial Head of the GA
(A) The medial head of the GA of BALB/c mice was surgically transected, and 100 μg pOKSM in 40 μL 0.9% saline or 40 μL saline alone were i.m. administered 7 days after surgery. GA muscles were dissected 9 and 14 days after injury. (B) Oct3/4 and Nanog gene expression was normalized to saline-injected controls. *p < 0.05 indicates statistically significant differences between Oct3/4 expression in the medial and lateral heads of the GA of pDNA-injected mice and in the Nanog expression between the lateral head of the GA of pDNA- and saline-injected animals, assessed by one-way ANOVA and Tukey’s test (n = 4). (C) H&E staining (40×; scale bar represents 100 μm). Orange arrowheads point to sites of likely neovascularization. Green arrowheads point to areas of likely mineralization. Black dashed lines delineate areas of randomly organized regenerating myofibers. (D) Laminin/DAPI staining (63×; scale bar represents 50 μm) and quantification of the percentage of centronucleated myofibers. **p < 0.01 and ***p < 0.001 indicate statistically significant differences between pDNA- and saline-injected groups and different time points, assessed by Welch ANOVA and Games Howell’s test (n = 4 GAs per group, 2 sections per muscle, 3 random fields per section). All data are presented as mean ± SD and numerical values are provided in Table S2. (E) Picrosirius red-fast green staining (40×; scale bar represents 100 μm) and measurement of areas with collagen deposition. *p < 0.05 and ***p < 0.001 indicate statistically significant differences between pDNA- and saline-injected groups and different time points, assessed by one-way ANOVA and Tukey’s test (n = 4 GAs per group, 2 sections per muscle, 5 random fields per section). All data are presented as mean ± SD and numerical values are provided in Table S3.
H&E staining confirmed the loss of normal muscle tissue architecture and its replacement by granulation tissue with a high degree of cellularity—most likely composed of interspersed inflammatory cells, among others—in the vicinity of the injury. Signs of mineralization (deeply basophilic deposits) and neovascularization were also observed. All such histological aberrations were evident in both groups on day 9 after injury, but they diminished considerably by day 14 in reprogrammed tissues (Figure 5C). Laminin/DAPI staining allowed the quantification of centronucleated myofibers (Figure 5D). On day 9, a higher percentage of myofibers surrounding the injured site were regenerating in the reprogrammed group (56.0% ± 23.4%), as evidenced by the centralized position of the nucleus, compared to saline controls (33.9% ± 12.9%) (p = 0.005). In the saline-injected group, the percentage of centronucleated myofibers continued to increase (45.6% ± 14.6% on day 14), an indication that the regeneration process was still ongoing. On the contrary, the number of regenerating fibers decreased significantly by day 14 in in vivo-reprogrammed tissues (18.2% ± 7.7% on day 14, p = 0.0001 compared to day 9).
The formation of a fibrotic scar is one of the hallmarks of muscle remodelling after injury that prevents functional rehabilitation of the tissue.20, 21 We investigated whether the administration of reprogramming factors would have any effect on the deposition of collagen after injury, and we found that collagen-positive areas (detected by picrosirius red-fast green staining) on day 9 were more extensive in the tissues from saline control animals (p = 0.029), such difference being more significant on day 14 (p = 0.0001). Between these two time points, the fibrotic area (calculated as a percentage of the total area in the cross-section) in the vicinity of the injury increased only slightly (12.3% ± 5.1% to 15.5% ± 6.5%, p = 0.012) in the OKSM-injected group. In contrast, the area of fibrotic tissue increased significantly, from 15.4% ± 5.2% on day 9 to 22.6% ± 6.7% on day 14, in mice that received saline only (p = 0.000; Figure 5E).
Finally, we tested whether the improvements observed at the histological level would translate into a significant improvement in muscle function in the sectioned GA. The recovery of muscle force was measured by in vivo myography 9 and 14 days after injury (Figure S8A). In brief, the GA and sciatic nerve were exposed in terminally anesthetized mice and connected to a myograph as shown in Figure S8B, so that muscle force under twitch and tetanus contractions was measured upon direct stimulation of the nerve. Fast twitch was produced by a single stimulation at optimal voltage and length, and isometric tensile strength was recorded (Figure S8C). Tetanus contraction was produced by a series of stimulations repeated with a frequency of 150 Hz (Figure S8D). At 7 days after i.m. administration of reprogramming pOKSM (14 days after injury), treated animals recovered a higher percentage of the force of the contralateral (intact) leg as compared to saline-injected controls, when both fast twitch (50.4% ± 12.3% for saline and 80.2% ± 18.2% for OKSM-injected animals) and tetanus (49.9% ± 19.2% for saline and 56.6% ± 20.9% for OKSM-injected animals) contractions were investigated. However, none of these differences was statistically significant (Figures S8E and S8F).
Discussion
This study demonstrates that the induction of transient cell reprogramming within the skeletal muscle tissue, accompanied by likewise transient proliferation, accelerates regeneration at the histological level in a model of severe muscle injury. Our laboratory was first to report in vivo cell reprogramming to pluripotency in an adult mammalian tissue (liver) via forced expression of OKSM factors.5, 12, 14 Here, transient expression of the same factors in healthy adult skeletal muscle also rapidly triggered the upregulation of pluripotency markers that are otherwise repressed in adult tissues (Figures 1C and 2C). With the data obtained here, we cannot confirm whether reprogrammed cells within the skeletal muscle reached a state of functional pluripotency, as we found in our previous work;14 hence, we have referred to them as reprogrammed or pluripotent-like cells. In fact, co-expression of PAX3, a marker of myogenic precursors, in the same cells that expressed the pluripotency marker NANOG, suggests that OKSM overexpression could have induced a partial reprogramming event in this tissue (Figure 3D). A recent work by Ocampo et al.15 also described the induction of partial reprogramming when OKSM expression was not sustained for long periods of time. In that study, reprogrammed cells did not activate the expression of pluripotency-related genes, such as Nanog, but they did proliferate. As in our study, proliferation was only transient and no teratomas were found within reprogrammed tissues. We have previously pointed to the duration of OKSM expression as a critical factor to avoid tumorigenesis upon in vivo reprogramming and, therefore, to preserve the potential of this technology to develop into a therapeutic strategy.13 The observations that we have made in the present study support our previous claims5, 12 and Ocampo et al.’s15 that teratoma-free in vivo cell reprogramming can be achieved when OKSM expression is limited in time.
i.m. injection of pDNA has been previously reported to result in long-term transgene expression that can last for 2 months and reach peak levels 14 days after injection, based on the post-mitotic status of muscle fibers.42 This contrasted with the rapid decrease in the expression of the transfected reprogramming factors observed in our study (Figures 1B and 2B), which we explain by the proliferative status of pOKSM-transfected cells evidenced by BrdU labeling (Figure 4A) and by the predominantly episomal state of the pDNA vectors utilized in this study, which can thus be lost with cell division.44 BrdU incorporation was found within GFP+ cell clusters in reprogrammed tissues, but not in saline-injected controls, and, therefore, proliferation was attributed to the reprogramming event. Indeed, active cell division is an early and mandatory step in the differentiated-to-pluripotent conversion.51 This finding also suggested that in vivo reprogramming could enhance the cellular repopulation of a tissue after injury, precisely by increased cell proliferation. In addition, our histological observations of the evolution of the reprogrammed cell clusters over time suggested that, after the proliferative phase, reprogrammed cells re-differentiated and successfully re-integrated into the muscle tissue (Figures 4B–4F; Figure S4). This was supported by the gradual disappearance of mononucleated cell clusters, the rapid decrease in the expression of pluripotency markers together with the emergence of desmin-positive centronucleated myofibers from days 4 to 8 after injection, the enlargement of myofiber diameter, and the absence of pronounced apoptosis and of dysplastic lesions or teratomas. Nonetheless, despite the consistency among these observations, specific lineage-tracing studies that allow permanent labeling of in vivo-reprogrammed cells must be pursued in the future to confirm this hypothesis and confidently determine the fate of reprogrammed cells within the tissue. We also noted that centralized nuclei persisted only for up to 8 days after injection, which differs from classic regeneration where such feature is observed for longer periods of time.17
One of the caveats in our study remains the elucidation of the specific cell types that are reprogrammed within the skeletal muscle tissue, a limitation that affects most of the in vivo-reprogramming studies published to date, regardless of the target tissue.5, 6, 7, 11, 15 The multinucleated and post-mitotic status of muscle fibers would require cell fission and cell cycle re-entry to achieve successful reprogramming toward a pluripotent-like state. While myofibers of urodele amphibians de-differentiate and cleave into mononucleated progenitors during regeneration,56 there is currently no evidence that mammalian myofibers can undergo such processes. Recent studies attempted to achieve mammalian myofiber de-differentiation and fission via ectopic expression of transcription factors that mediate such processes in amphibians57, 58, 59 or via muscle injury.60, 61 However, they also lacked robust lineage-tracing tools that could confirm the origin of the resulting mononucleated cells. Here, we confirmed the expression of the GFP reporter encoded in pOKSM in both myofibers and mononucleated interstitial cells, but NANOG was only co-expressed in the latter (Figure 1E; Figure S2B). This observation suggests that OKSM expression may not be able to induce cell reprogramming in differentiated myofibers. Myoblasts have, however, been successfully de-differentiated into iPSCs in vitro via Oct4-mediated MyoD1 downregulation.41, 62 Interestingly, we consistently observed a decrease in MyoD1 mRNA levels in the reprogrammed groups throughout our study (Figures 1D and 2D). We also confirmed downregulation of markers specific to satellite cells. In vitro, these cells have been shown to be more efficiently reprogrammed than other more committed cell types.62 In addition, a report from Chiche et al.,10 which also explored the in vivo induction of pluripotency in the mouse skeletal muscle, identified PAX7+ satellite cells as the major cell of origin of reprogrammed cells within such tissue. However, the contribution of other cell types was not investigated.10 In our study, given the probably random incorporation of pDNA in a variety of cells and the absence of cell type-specific promoters in the cassettes, it is conceivable that different interstitial cells were reprogrammed rather than a specific cell type, all downregulating their differentiated markers.
We have previously hypothesized that the generation of transiently proliferative, pluripotent-like cells in the context of an injured tissue might help its cellular repopulation and overall regeneration.38 Indeed, there are notorious limitations in the management of major skeletal muscle injuries, for which the clinically established treatment continues to be conservative and the often lack of complete recovery leads to fibrosis and loss of muscle contractility.63 In our study, the administration of pDNA encoding Yamanaka factors 7 days after severe muscle injury not only accelerated the recovery toward normal muscle architecture (Figure 5C) and the appearance of centronucleated myofibers (Figure 5D) but also induced a moderate decrease of collagen deposition compared to the control group (Figure 5E). During the peer review process of this paper, Doeser and colleagues16 found similar observations upon the induction of transient reprogramming with OKSM factors in cutaneous wounds. In vivo reprogramming reduced the levels of profibrotic markers (Tgfβ-1, Collagen I, and Vegf), and it resulted in reduced scar formation at the wound site. α-Sma expression, associated with myofibroblast proliferation, was also significantly diminished in comparison to control (non-reprogrammed) wounds.16 It is thus conceivable that the reduced fibrosis observed in Doeser et al.’s and our own study is, at least partly, due to an OKSM-induced decrease in post-injury fibroblast-to-myofibroblast transdifferentiation.
In our hands, the histological improvements above did not translate into significant differences in the recovery of muscle force (Figure S8). In fact, only the administration of anti-fibrotic drugs has demonstrated significant functional rehabilitation in mouse injury models of comparable severity.20, 23 Other strategies, including the administration of adipose-derived stem cells and surgical suturing, reported improvements at the histological level only,32 similar to us, or utilized milder injury models.22, 31 Thus, future work is warranted to identify optimal gene transfer vectors and dosage regimens that grant clinically relevant, functional rehabilitation upon in vivo reprogramming. Indeed, transfection efficiency following i.m. administration of pDNA is relatively low and highly variable,64 and, in vitro, the use of naked pDNA to force OKSM overexpression has shown lower reprogramming efficiency compared to other alternatives, especially viral vectors.65 This agrees with our observation that not all pOKSM-transfected cells within reprogrammed tissues expressed NANOG (Figure 1E; Figure S3). Other strategies in the current literature may have achieved higher in vivo-reprogramming efficiencies, but they lack clinical relevance based on the use of transgenic mouse models6, 15 or retroviral vectors that cause genomic integration of OKSM transgenes, followed by sustained reprogramming and tumorigenesis.8 We thus foresee that the search for appropriate gene transfer vectors that boost the efficiency of in vivo reprogramming without compromising the safety of the approach (i.e., preserving the transiency of reprogramming) will be a main priority for the field.
The teratoma-free approach that we have postulated in this work may also offer advantages to other reprogramming-based strategies. First, it bypasses the need for ex vivo manipulation and transplantation, and their associated risks, encountered by the in vitro generation of iPSCs. Such limitations have been exhaustively reviewed by others.66, 67 The presence of a transient proliferative stage (Figure 4A) offers a possibility for cellular expansion and repopulation that in vivo transdifferentiation strategies described to date (i.e., direct reprogramming between two differentiated cell types, avoiding pluripotent intermediates, and active cell division) lack.68, 69, 70, 71, 72, 73, 74, 75, 76 In addition, transdifferentiation strategies require the identification of specific reprogramming factors to mediate each particular cell type conversion, while pluripotency can be induced in a variety of starting cell types with the defined OKSM cocktail.1, 2, 77
Abundant necrosis and neutrophil infiltration occur rapidly (1–2 days) after muscle injury, and they are followed by prominent macrophage recruitment and the secretion of cytokines and damage signals, among them interleukin (IL)-6, for up to approximately the first week after injury.78 An interesting finding in our work is that Nanog upregulation was significantly higher when pOKSM was administered 7 days after injury compared to earlier interventions (Figure S7B). This observation agrees with recent findings by Mosteiro et al.11 and Chiche et al.,10 which demonstrated that signals of tissue damage and senescence, among which IL-6 plays a key role, promote reprogramming by boosting cell plasticity. Ocampo et al.15 also found that the efficiency or reprogramming increased in senescent tissues. Thus, it is conceivable that, in our study, reprogramming was compromised at the very early time points after injury, given the abundance of cells becoming necrotic, but on the contrary was favored by the later peak of inflammation and release of IL-6 by day 7 after injury. Regardless of the mechanism, this observation adds a further advantage to the potential use of in vivo reprogramming to enhance tissue regeneration, as reprogramming appears to be favored within injured and/or aged tissues. Besides, the synergy between damage and reprogramming in this time frame circumvents the need for a rapid post-injury intervention. This is indeed a caveat of many muscle regeneration experimental strategies, especially of cell-based therapies that are administered at the time of injury, while the time required to achieve sufficient cell numbers for implantation can reach several weeks.27, 28, 31, 32
We have proposed here a transient reprogramming strategy to generate pluripotent-like intermediates in situ that can assist with cellular repopulation of an injured tissue while circumventing the challenges faced by cell transplantation, including cell isolation; extensive culturing; and the inherent risks of genomic aberrations, delivery, and engraftment. We hypothesize that this strategy could benefit from the presence of differentiation cues in the tissue microenvironment able to re-differentiate the in vivo-reprogrammed cells to the appropriate phenotypes. The transient character of this approach has proven to be critical to avoid tumorigenesis, a burden that keeps other in vivo reprogramming to pluripotency strategies far from the road into the clinic.
Materials and Methods
pDNA
pCX-OKS-2A (pOKS) encoding Oct3/4, Klf4, and Sox2; pCX-cMyc (pM) encoding cMyc; and pGFP encoding GFP, all under the control of the constitutive CAG promoter, were purchased as bacterial stabs from Addgene (USA). pLenti-lll-EF1a-mYamanaka (pOKSM), encoding Oct3/4, Klf4, Sox2, cMyc, and GFP under the control of the constitutive EF1α promoter, was purchased from Applied Biological Materials (USA). Research-grade plasmid production was performed in Plasmid Factory (Germany).
Animals
All experiments were performed with prior approval from the UK Home Office under a project license (PPL 70/7763) and in strict compliance with the Guidance on the Operation of the Animals (Scientific Procedures) Act 1986. BALB/c mice were purchased from Harlan (UK). Sv129-Tg(Nanog-GFP) mice, which carry the GFP reporter inserted into the Nanog locus,43 were a kind gift from the Wellcome Trust Centre for Stem Cell Research, University of Cambridge (UK), and they were bred in heterozygosity and genotyped at the University of Manchester. CBA-Tg(Pax3-GFP), in which GFP replaces the Pax3-coding sequence of exon 1,49 were bred and genotyped at the University of Manchester. All mice used in this work were female of 7 weeks of age. Mice were allowed 1 week to acclimatize to the animal facilities prior to any procedure.
i.m. Administration of pDNA
Mice were anesthetized with isofluorane and the left GA was injected with 50 μg pOKSM, 50 μg pGFP, or 50 μg pOKS and 50 μg pM in 50 μL 0.9% saline solution. The contralateral (right) leg was injected with 50 μL 0.9% saline solution alone as an internal control. Mice were culled at different time points, including 2, 4, 8, 12, 24, 50, and 120 days after i.m. injection, as specified in each particular experiment.
RNA Isolation and Real-Time qRT-PCR Analysis
Aurum Fatty and Fibrous Kit (Bio-Rad, UK) was used to isolate total RNA from muscle tissue. cDNA synthesis from 1 μg RNA sample was performed with iScript cDNA synthesis kit (Bio-Rad, UK), according to the manufacturer’s instructions. 2 μL of each cDNA sample was used to perform real-time qRT-PCR reactions with iQ SYBR Green Supermix (Bio-Rad, UK). Primer sequences are shown in Table S1. Experimental duplicates were run on CFX-96 Real Time System (Bio-Rad, UK) with the following protocol: 95°C for 3 min, 1 cycle; and 95°C for 10 s and 60°C for 30 s, repeated for 40 cycles. β-actin was used as a housekeeping gene, and gene expression levels were normalized to saline-injected controls, unless otherwise specified, following the Livak (2ˆ−ΔΔCt) method. The qRT-PCR data are presented as median 2ˆ−ΔΔCT ± propagation of the error from Ct values. ΔCt values were used for statistical analysis.
IHC of BALB/c Muscle Tissue Sections
GAs were dissected 2 and 4 days after i.m. injection with pOKSM, pGFP, or saline control and immediately frozen in isopentane, pre-cooled in liquid nitrogen. 10-μm-thick sections (in the transverse plane) were prepared on a cryostat (Leica, CM3050S) and air dried for 1 hr at room temperature (RT) prior to storage at −80°C. Before staining, muscle sections were post-fixed with methanol, pre-cooled at −20°C, for 10 min, air dried for 15 min, and finally washed twice with PBS for 5 min. Sections were then incubated for 1 hr in blocking buffer (5% goat serum-0.1% Triton in PBS [pH 7.3]) at RT, followed by two washing steps with PBS (1% BSA- 0.1% Triton [pH 7.3]) and overnight incubation at +4°C with the following primary antibodies: rabbit polyclonal antibody (pAb) anti-NANOG (ab80892, 1/200, Abcam, UK) and chicken pAb anti-GFP (ab13970, 1/1,000, Abcam, UK). The next day, sections were washed twice (5 min each) with PBS and incubated (1.5 hr, RT) with the following secondary antibodies: goat polyclonal anti-rabbit immunoglobulin G (IgG) labeled with Cy3 (1/250, Jackson ImmunoResearch Laboratories) and goat polyclonal anti-chicken IgY labeled with Alexa-647 (1/500, ab150171, Abcam, UK). After two washes in PBS, slides were mounted with ProLong Gold anti-fade DAPI-containing medium (Life Technologies, UK). Whole-muscle sections were imaged with a 3D Histech Pannoramic 250 Flash slide scanner. The number of GFP+ and NANOG+GFP+ cell clusters and the number of GFP+ muscle fibers were counted in 2 GAs, 3 sections per muscle. Representative images were taken at 20× magnification with Pannoramic Viewer software. The z stacks and orthogonal views (Figures S2E and S3) were obtained at 100× magnification with a Leica TCS SP5 AOBS inverted confocal microscope.
Characterization of GFP+ Cell Clusters in Nanog-GFP and Pax3-GFP Transgenic Mice
To characterize the clusters of reprogrammed cells in Nanog-GFP and Pax3-GFP mice, GAs were administered with pOKS and pM vectors, to avoid overlapping of the GFP reporter in pOKSM, and sacrificed 2 and 4 days after injection. To observe the GFP signal, frozen tissue sections obtained as described before were simply mounted with ProLong Gold anti-fade DAPI-containing mountant (Life Technologies, UK) after fixation and imaged with 3D Histech Pannoramic 250 Flash slide scanner. The number of GFP+ clusters was counted from 4 GAs. Representative images (100×) were taken with Pannoramic Viewer. To investigate co-localization of the GFP signal with the expression of different markers, tissue sections were processed for IHC as described above. Rabbit pAb anti-OCT4 (ab19857,3 μg/mL, Abcam, UK), rabbit pAb anti-NANOG (ab80892, 1 μg/mL, Abcam, UK), rabbit pAb anti-AP (ab95462, 1:200, Abcam, UK), mouse monoclonal antibody (mAb) anti-SSEA1 (ab16285, 20 μg/mL, Abcam, UK), rabbit pAb anti-Pax7 (pab0435, 1:200, Covalab, France), and rabbit mAb anti-PDGFrβ (3169, 1:100, Cell Signaling Technology) were used as primary antibodies. Anti-PDGFrβ was a kind gift from G.C.’s lab (University of Manchester). Goat pAb anti-rabbit IgG labeled with Cy3 and goat pAb anti-mouse IgG labeled with Cy5 (1/250, Jackson ImmunoResearch Laboratories) were used as secondary antibodies. Images were taken at 100× magnification with a Leica TCS SP5 AOBS inverted confocal microscope.
Histological Evaluation of Muscle Tissue
BALB/c GAs were dissected on days 2, 4, 8, 15, 50, and 120 after i.m. injection with pOKSM or saline control, frozen, and sectioned as previously described. H&E staining was performed following a standard protocol. Tissue sections were imaged with a 3D Histech Pannoramic 250 Flash slide scanner, and representative images at 40× and 100× magnification were taken with Pannoramic Viewer software. The minimum myofiber diameter and number of myofibers per cross-sectional area were analyzed from 5 sections per muscle (n = 3 GAs per condition and time point), with ImageJ 1.48 software.
Desmin/Laminin/DAPI Staining
The 10-μm-thick cryosections were stained following the IHC protocol previously described, using rabbit pAb anti-laminin (ab11575, 1:200, Abcam, UK) and mouse mAb anti-desmin (ab6322, 1:200, Abcam, UK) as primary antibodies. Goat pAb anti-rabbit IgG labeled with Cy3 (1/250, Jackson ImmunoResearch Laboratories) and goat pAb anti-mouse IgG labeled with Cy5 (1/250, Jackson ImmunoResearch Laboratories) were used as secondary antibodies. 40× images were obtained with a Zeiss Axio Observer epi-Fluorescence microscope.
BrdU Labeling and the Detection of Proliferating Cells
BrdU assay was used to label proliferating cells in vivo, as previously described.79 BALB/c mice were i.p. administered with 500 mg/kg BrdU (B5002, Sigma, UK) in 0.9% saline 18 hr after i.m. injection with pOKSM or saline control. Then 6 hr later, GAs were dissected and processed for IHC as described above. Treatment with 2 N HCl (10 min at 37°C) after fixation was performed to denature the DNA. Mouse mAb anti-BrdU (B8434, 1:100, Sigma, UK) and goat pAb anti-mouse IgG labeled with Cy5 (1/250, Jackson ImmunoResearch Laboratories) were used. The 100× images were captured with a Leica TCS SP5 AOBS inverted confocal microscope.
Surgically Induced Skeletal Muscle Injury Model and In Vivo Reprogramming to Pluripotency
BALB/c mice were anesthetized with isoflurane and the left hind limb was shaved and prepared for surgery; 0.05 mg/kg buprenorphine was subcutaneously (s.c.) administered at the start of the intervention. A vertical skin incision (6 mm long) was made overlying the posterior compartment of the calf with a scalpel number 11, and the fascia was exposed and incised with fine scissors at the level of mid-GA to release the muscle belly. The medial head of the GA was blunt dissected (i.e., carefully separated from the lateral head and remaining fascia without cutting or damaging the tissue) and sectioned at its mid-point with a single incision, performed with a scalpel number 11, in the transverse plane. The medial head was sectioned in its full width and depth, as described in Figure S7A. The sural nerve and lateral head of the GA were preserved. The skin was sutured with 5 interrupted stitches (Vicryl 6-0 absorbable suture, Ethicon, UK), and mice were allowed to recover in a warm chamber. The contralateral (right) leg was left intact for internal control. At 7 days after surgery, 100 μg pOKSM in 40 μL 0.9% saline or 40 μL 0.9% saline alone were i.m. administered in the injured (left) leg. The contralateral (right) leg was left uninjected. Mice were sacrificed at days 9 and 14 after injury (2 and 7 days after pOKSM or saline administration) for histological and electromechanical investigations (n = 4). A group of animals was also culled at day 9 (day 2 after i.m. injection) for gene expression analysis.
Laminin/DAPI Staining and Analysis of the Percentage of Centronucleated Myofibers
The 10-μm-thick cryosections were obtained 9 and 14 days after surgical transection, and they were IHC stained following the protocol previously described, using rabbit pAb anti-laminin (ab11575, 1:200, Abcam, UK) and goat pAb anti-rabbit IgG labeled with Cy3 (1/250, Jackson ImmunoResearch Laboratories) antibodies. Whole-muscle sections were imaged with 3D Histech Pannoramic 250 Flash slide scanner, and 63× captures of random fields were collected with Pannoramic Viewer Software. The number of centronucleated myofibers and total number of fibers per cross-sectional area were quantified using ImageJ 1.48 software from 3 randomly selected fields per section. The data for each mouse were calculated from 2 sections (one from each half of the transected GA, close to the site of injury), and we observed 4 mice in each group. Measurements and calculations were conducted in a blinded manner.
Picrosirius Red-Fast Green Staining and Analysis of Fibrotic Area
GAs were dissected 9 and 14 days after injury and fixed in 10% buffered formalin solution (Sigma, UK). Tissues were embedded in paraffin wax, and 5-μm-thick sagittal and transverse sections obtained with a microtome (Leica RM2255) were left to dry overnight at 37°C prior to the staining procedure. Tissue sections were de-paraffinized following a standard protocol and stained for 1 hr in picrosirius red-fast green staining solution (0.1% Sirius red and 0.1% fast green in a saturated aqueous solution of picric acid). Tissue sections were then quickly immersed in 0.5% acetic acid for 6 s, dehydrated in 100% ethanol and xylene, and finally mounted with DPX (Sigma, UK). Sections were imaged on a 3D Histech Pannoramic 250 Flash slide scanner, and 40× images were obtained with Pannoramic Viewer Software. The percentage of collagen-positive areas was quantified with ImageJ 1.48 software in 5 randomly selected fields per section. The data for each mouse were calculated from 2 sections (one from each half of the transected GA, close to the site of injury), and 4 mice were included per group. Measurements and calculations were conducted in a blinded manner.
Sample Size Calculations
Sample size for gene expression studies was determined based on differences in Nanog expression in prelimary studies, given its hallmark role in cell reprogramming. We calculated sample size taking into account the means and SDs of Nanog ΔCt for control and OKSM-treated mice, power = 80% and Type I error = 0.05, and considering a one-way ANOVA comparison between the means. We also utilized the results observed in a previous work from our group,5 which explored the in vivo induction of pluripotency in mouse liver via the same episomal plasmid system, to inform the design and sample size determination of the studies in this work.
Statistical Analysis
Sample (n) numbers were specified for each particular experiment. Statistical analysis was performed first by Levene’s test to assess homogeneity of variance. When no significant differences were found in the variances of the different groups, statistical analysis was followed by one-way ANOVA and Tukey’s post hoc test. When variances were unequal, the analysis was followed with Welch ANOVA and Games-Howell’s post hoc test. Probability values < 0.05 were regarded as significant. SPSS software version 20.0 was used to perform this analysis.
Author Contributions
I.d.L., A.Y., and K.K. conceived the project and designed experiments. I.d.L. and A.Y. performed the majority of experiments. S.Q. contributed to some qPCR data. Y.N. and F.M.A.R. performed some IHC experiments. H.D. helped to set up in vivo myography. G.C. advised in experimental design and interpretation of data. I.d.L., A.Y., G.C., and K.K. wrote the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
I.d.L. and K.K. are the recipients of an award by the Royal College of Surgeons of Edinburgh (grant KAE WONJ4). I.d.L. would like to thank Obra Social LaCaixa and UCL School of Pharmacy for a jointly funded PhD Studentship. The authors would also like to acknowledge the Histology and Bioimaging Facilities at the University of Manchester for assistance, as well as Dr. Adam Reid (University of Manchester) for his help in the establishment of the injury model utilized in this study.
Footnotes
Supplemental Information includes eight figures, three tables, and Supplemental Materials and Methods and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.10.014.
Supplemental Information
References
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Aoi T., Yae K., Nakagawa M., Ichisaka T., Okita K., Takahashi K., Chiba T., Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 3.Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 4.Vivien C., Scerbo P., Girardot F., Le Blay K., Demeneix B.A., Coen L. Non-viral expression of mouse Oct4, Sox2, and Klf4 transcription factors efficiently reprograms tadpole muscle fibers in vivo. J. Biol. Chem. 2012;287:7427–7435. doi: 10.1074/jbc.M111.324368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmazer A., de Lázaro I., Bussy C., Kostarelos K. In vivo cell reprogramming towards pluripotency by virus-free overexpression of defined factors. PLoS ONE. 2013;8:e54754. doi: 10.1371/journal.pone.0054754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abad M., Mosteiro L., Pantoja C., Cañamero M., Rayon T., Ors I., Graña O., Megías D., Domínguez O., Martínez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- 7.Ohnishi K., Semi K., Yamamoto T., Shimizu M., Tanaka A., Mitsunaga K., Okita K., Osafune K., Arioka Y., Maeda T. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Gao X., Wang X., Xiong W., Chen J. In vivo reprogramming reactive glia into iPSCs to produce new neurons in the cortex following traumatic brain injury. Sci. Rep. 2016;6:22490. doi: 10.1038/srep22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi H.W., Kim J.S., Hong Y.J., Song H., Seo H.G., Do J.T. In vivo reprogrammed pluripotent stem cells from teratomas share analogous properties with their in vitro counterparts. Sci. Rep. 2015;5:13559. doi: 10.1038/srep13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiche A., Le Roux I., von Joest M., Sakai H., Aguín S.B., Cazin C., Salam R., Fiette L., Alegria O., Flamant P. Injury-Induced Senescence Enables In Vivo Reprogramming in Skeletal Muscle. Cell Stem Cell. 2017;20:407–414.e4. doi: 10.1016/j.stem.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Mosteiro L., Pantoja C., Alcazar N., Marión R.M., Chondronasiou D., Rovira M., Fernandez-Marcos P.J., Muñoz-Martin M., Blanco-Aparicio C., Pastor J. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354:aaf4445. doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- 12.Yilmazer A., de Lázaro I., Bussy C., Kostarelos K. In vivo reprogramming of adult somatic cells to pluripotency by overexpression of Yamanaka factors. J. Vis. Exp. 2013;17:e50837. doi: 10.3791/50837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lázaro I., Cossu G., Kostarelos K. Transient transcription factor (OSKM) expression is key towards clinical translation of in vivo cell reprogramming. EMBO Mol. Med. 2017;9:733–736. doi: 10.15252/emmm.201707650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lázaro I., Bussy C., Yilmazer A., Jackson M.S., Humphreys N.E., Kostarelos K. Generation of induced pluripotent stem cells from virus-free in vivo reprogramming of BALB/c mouse liver cells. Biomaterials. 2014;35:8312–8320. doi: 10.1016/j.biomaterials.2014.05.086. [DOI] [PubMed] [Google Scholar]
- 15.Ocampo A., Reddy P., Martinez-Redondo P., Platero-Luengo A., Hatanaka F., Hishida T., Li M., Lam D., Kurita M., Beyret E. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell. 2016;167 doi: 10.1016/j.cell.2016.11.052. P1719–1733.E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doeser M.C., Schöler H.R., Wu G. Reduction of Fibrosis and Scar Formation by Partial Reprogramming In Vivo. Stem Cells. 2018;36:1216–1225. doi: 10.1002/stem.2842. [DOI] [PubMed] [Google Scholar]
- 17.Chargé S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 18.Allbrook D.B., Han M.F., Hellmuth A.E. Population of muscle satellite cells in relation to age and mitotic activity. Pathology. 1971;3:223–243. doi: 10.3109/00313027109073739. [DOI] [PubMed] [Google Scholar]
- 19.Blau H.M., Cosgrove B.D., Ho A.T. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015;21:854–862. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukushima K., Badlani N., Usas A., Riano F., Fu F., Huard J. The use of an antifibrosis agent to improve muscle recovery after laceration. Am. J. Sports Med. 2001;29:394–402. doi: 10.1177/03635465010290040201. [DOI] [PubMed] [Google Scholar]
- 21.Sato K., Li Y., Foster W., Fukushima K., Badlani N., Adachi N., Usas A., Fu F.H., Huard J. Improvement of muscle healing through enhancement of muscle regeneration and prevention of fibrosis. Muscle Nerve. 2003;28:365–372. doi: 10.1002/mus.10436. [DOI] [PubMed] [Google Scholar]
- 22.Menetrey J., Kasemkijwattana C., Fu F.H., Moreland M.S., Huard J. Suturing versus immobilization of a muscle laceration. A morphological and functional study in a mouse model. Am. J. Sports Med. 1999;27:222–229. doi: 10.1177/03635465990270021801. [DOI] [PubMed] [Google Scholar]
- 23.Chan Y.S., Li Y., Foster W., Horaguchi T., Somogyi G., Fu F.H., Huard J. Antifibrotic effects of suramin in injured skeletal muscle after laceration. J. Appl. Physiol. (1985) 2003;95:771–780. doi: 10.1152/japplphysiol.00915.2002. [DOI] [PubMed] [Google Scholar]
- 24.Foster W., Li Y., Usas A., Somogyi G., Huard J. Gamma interferon as an antifibrosis agent in skeletal muscle. J. Orthop. Res. 2003;21:798–804. doi: 10.1016/S0736-0266(03)00059-7. [DOI] [PubMed] [Google Scholar]
- 25.Negishi S., Li Y., Usas A., Fu F.H., Huard J. The effect of relaxin treatment on skeletal muscle injuries. Am. J. Sports Med. 2005;33:1816–1824. doi: 10.1177/0363546505278701. [DOI] [PubMed] [Google Scholar]
- 26.Menetrey J., Kasemkijwattana C., Day C.S., Bosch P., Vogt M., Fu F.H., Moreland M.S., Huard J. Growth factors improve muscle healing in vivo. J. Bone Joint Surg. Br. 2000;82:131–137. doi: 10.1302/0301-620x.82b1.8954. [DOI] [PubMed] [Google Scholar]
- 27.Natsu K., Ochi M., Mochizuki Y., Hachisuka H., Yanada S., Yasunaga Y. Allogeneic bone marrow-derived mesenchymal stromal cells promote the regeneration of injured skeletal muscle without differentiation into myofibers. Tissue Eng. 2004;10:1093–1112. doi: 10.1089/ten.2004.10.1093. [DOI] [PubMed] [Google Scholar]
- 28.Shi M., Ishikawa M., Kamei N., Nakasa T., Adachi N., Deie M., Asahara T., Ochi M. Acceleration of skeletal muscle regeneration in a rat skeletal muscle injury model by local injection of human peripheral blood-derived CD133-positive cells. Stem Cells. 2009;27:949–960. doi: 10.1002/stem.4. [DOI] [PubMed] [Google Scholar]
- 29.Mori R., Kamei N., Okawa S., Nakabayashi A., Yokota K., Higashi Y., Ochi M. Promotion of skeletal muscle repair in a rat skeletal muscle injury model by local injection of human adipose tissue-derived regenerative cells. J. Tissue Eng. Regen. Med. 2015;9:1150–1160. doi: 10.1002/term.1659. [DOI] [PubMed] [Google Scholar]
- 30.Lee C.W., Fukushima K., Usas A., Xin L., Pelinkovic D., Martinek V., Somogyi G., Robbins P.D., Fu F.H., Huard J. Biological intervention based on cell and gene therapy to improve muscle healing after laceration. J. Musculoskelet. Res. 2000;04:265–277. [Google Scholar]
- 31.Park J.K., Ki M.R., Lee E.M., Kim A.Y., You S.Y., Han S.Y., Lee E.J., Hong I.H., Kwon S.H., Kim S.J. Losartan improves adipose tissue-derived stem cell niche by inhibiting transforming growth factor-β and fibrosis in skeletal muscle injury. Cell Transplant. 2012;21:2407–2424. doi: 10.3727/096368912X637055. [DOI] [PubMed] [Google Scholar]
- 32.Hwang J.H., Kim I.G., Piao S., Jung A.R., Lee J.Y., Park K.D., Lee J.Y. Combination therapy of human adipose-derived stem cells and basic fibroblast growth factor hydrogel in muscle regeneration. Biomaterials. 2013;34:6037–6045. doi: 10.1016/j.biomaterials.2013.04.049. [DOI] [PubMed] [Google Scholar]
- 33.Nakasa T., Ishikawa M., Shi M., Shibuya H., Adachi N., Ochi M. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J. Cell. Mol. Med. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gharaibeh B., Chun-Lansinger Y., Hagen T., Ingham S.J., Wright V., Fu F., Huard J. Biological approaches to improve skeletal muscle healing after injury and disease. Birth Defects Res. C Embryo Today. 2012;96:82–94. doi: 10.1002/bdrc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maffulli N., Oliva F., Frizziero A., Nanni G., Barazzuol M., Via A.G., Ramponi C., Brancaccio P., Lisitano G., Rizzo D. ISMuLT Guidelines for muscle injuries. Muscles Ligaments Tendons J. 2014;3:241–249. [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 37.Dupuis M., Denis-Mize K., Woo C., Goldbeck C., Selby M.J., Chen M., Otten G.R., Ulmer J.B., Donnelly J.J., Ott G., McDonald D.M. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J. Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 38.de Lázaro I., Kostarelos K. In vivo cell reprogramming to pluripotency: exploring a novel tool for cell replenishment and tissue regeneration. Biochem. Soc. Trans. 2014;42:711–716. [Google Scholar]
- 39.Collins C.A., Gnocchi V.F., White R.B., Boldrin L., Perez-Ruiz A., Relaix F., Morgan J.E., Zammit P.S. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS ONE. 2009;4:e4475. doi: 10.1371/journal.pone.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr. Opin. Genet. Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe S., Hirai H., Asakura Y., Tastad C., Verma M., Keller C., Dutton J.R., Asakura A. MyoD gene suppression by Oct4 is required for reprogramming in myoblasts to produce induced pluripotent stem cells. Stem Cells. 2011;29:505–516. doi: 10.1002/stem.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff J.A., Ludtke J.J., Acsadi G., Williams P., Jani A. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum. Mol. Genet. 1992;1:363–369. doi: 10.1093/hmg/1.6.363. [DOI] [PubMed] [Google Scholar]
- 43.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 44.Okita K., Hong H., Takahashi K., Yamanaka S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat. Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- 45.Ye F., Zhou C., Cheng Q., Shen J., Chen H. Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elatmani H., Dormoy-Raclet V., Dubus P., Dautry F., Chazaud C., Jacquemin-Sablon H. The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage. Stem Cells. 2011;29:1504–1516. doi: 10.1002/stem.712. [DOI] [PubMed] [Google Scholar]
- 47.Skelton D., Satake N., Kohn D.B. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther. 2001;8:1813–1814. doi: 10.1038/sj.gt.3301586. [DOI] [PubMed] [Google Scholar]
- 48.Sirabella D., De Angelis L., Berghella L. Sources for skeletal muscle repair: from satellite cells to reprogramming. J. Cachexia Sarcopenia Muscle. 2013;4:125–136. doi: 10.1007/s13539-012-0098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Relaix F., Rocancourt D., Mansouri A., Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 50.Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buganim Y., Faddah D.A., Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veronese S., Gambacorta M., Falini B. In situ demonstration of tissue proliferative activity using anti-bromo-deoxyuridine monoclonal antibody. J. Clin. Pathol. 1989;42:820–826. doi: 10.1136/jcp.42.8.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch U., Lehal R., Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 55.White R.B., Biérinx A.S., Gnocchi V.F., Zammit P.S. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 2010;10:21. doi: 10.1186/1471-213X-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison J.I., Lööf S., He P., Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J. Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Odelberg S.J., Kollhoff A., Keating M.T. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 58.McGann C.J., Odelberg S.J., Keating M.T. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc. Natl. Acad. Sci. USA. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Z., Liu Q., Mannix R.J., Xu X., Li H., Ma Z., Ingber D.E., Allen P.D., Wang Y. Mononuclear cells from dedifferentiation of mouse myotubes display remarkable regenerative capability. Stem Cells. 2014;32:2492–2501. doi: 10.1002/stem.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyoshi T., Nakano S., Nakamura K., Yamanouchi K., Nishihara M. In vivo electroporation induces cell cycle reentry of myonuclei in rat skeletal muscle. J. Vet. Med. Sci. 2012;74:1291–1297. doi: 10.1292/jvms.12-0195. [DOI] [PubMed] [Google Scholar]
- 61.Mu X., Peng H., Pan H., Huard J., Li Y. Study of muscle cell dedifferentiation after skeletal muscle injury of mice with a Cre-Lox system. PLoS ONE. 2011;6:e16699. doi: 10.1371/journal.pone.0016699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan K.Y., Eminli S., Hettmer S., Hochedlinger K., Wagers A.J. Efficient generation of iPS cells from skeletal muscle stem cells. PLoS ONE. 2011;6:e26406. doi: 10.1371/journal.pone.0026406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baoge L., Van Den Steen E., Rimbaut S., Philips N., Witvrouw E., Almqvist K.F., Vanderstraeten G., Vanden Bossche L.C. Treatment of skeletal muscle injury: a review. ISRN Orthop. 2012;2012:689012. doi: 10.5402/2012/689012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.André F.M., Cournil-Henrionnet C., Vernerey D., Opolon P., Mir L.M. Variability of naked DNA expression after direct local injection: the influence of the injection speed. Gene Ther. 2006;13:1619–1627. doi: 10.1038/sj.gt.3302827. [DOI] [PubMed] [Google Scholar]
- 65.Mochiduki Y., Okita K. Methods for iPS cell generation for basic research and clinical applications. Biotechnol. J. 2012;7:789–797. doi: 10.1002/biot.201100356. [DOI] [PubMed] [Google Scholar]
- 66.Forsberg M., Hovatta O. Challenges for the Therapeutic use of Pluripotent Stem Derived Cells. Front. Physiol. 2012;3:19. doi: 10.3389/fphys.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowry W.E., Quan W.L. Roadblocks en route to the clinical application of induced pluripotent stem cells. J. Cell Sci. 2010;123:643–651. doi: 10.1242/jcs.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banga A., Akinci E., Greder L.V., Dutton J.R., Slack J.M. In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc. Natl. Acad. Sci. USA. 2012;109:15336–15341. doi: 10.1073/pnas.1201701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inagawa K., Miyamoto K., Yamakawa H., Muraoka N., Sadahiro T., Umei T., Wada R., Katsumata Y., Kaneda R., Nakade K. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ. Res. 2012;111:1147–1156. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- 73.Guo Z., Zhang L., Wu Z., Chen Y., Wang F., Chen G. In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell. 2014;14:188–202. doi: 10.1016/j.stem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinrich C., Bergami M., Gascón S., Lepier A., Viganò F., Dimou L., Sutor B., Berninger B., Götz M. Sox2-mediated conversion of NG2 glia into induced neurons in the injured adult cerebral cortex. Stem Cell Reports. 2014;3:1000–1014. doi: 10.1016/j.stemcr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Su Z., Niu W., Liu M.L., Zou Y., Zhang C.L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X.B. Cellular reprogramming of human peripheral blood cells. Genomics Proteomics Bioinformatics. 2013;11:264–274. doi: 10.1016/j.gpb.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huard J., Li Y., Fu F.H. Muscle injuries and repair: current trends in research. J. Bone Joint Surg. Am. 2002;84-A:822–832. [PubMed] [Google Scholar]
- 79.Curran J. In vivo assay of cellular proliferation. In: Wilson J.B., May G.H.W., editors. Volume 174. Humana Press; 2001. pp. 379–389. (Epstein-Barr Virus Protocols). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.