Abstract
The efficacy of 4 commercial porcine reproductive and respiratory syndrome virus (PRRSV) modified-live vaccines (MLV), against PRRSV-1 and PRRSV-2 was evaluated and compared in growing pigs. Two of the vaccines were based on PRRSV-1 and two on PRRSV-2. There were no significant differences between each of the two PRRSV-1 MLV vaccines and the two PRRSV-2 MLV vaccines respectively based on virology, immunological, and pathological evaluations. Vaccination with either of the PRRSV-1 MLV vaccines resulted in reduced PRRSV-1 but not PRRSV-2 viremia. Additionally, vaccination with either of the PRRSV-1 MLV vaccines resulted in reduction of lung lesions and PRRSV-1 positive cells in PRRSV-1 challenged pigs but had no significant effect in PRRSV-2 challenged pigs. In contrast, vaccination with either of the two PRRSV-2 MLV vaccines resulted in the reduction of both PRRSV-1 and PRRSV-2 viremia. The PRRSV-2 MLV vaccines were also able to effectively reduce lung lesions and PRRSV positive cells after challenge with either PRRSV-1 or PRRSV-2. Our data suggest that while vaccination with PRRSV-1 MLV vaccines can be effective against PRRSV-1, only PRRSV-2 MLV vaccines can protect against both Korean PRRSV-1 and PRRSV-2 challenges in this study.
Résumé
L’efficacité de quatre vaccins vivants modifiés (VVM) commerciaux contre le virus du syndrome reproducteur et respiratoire porcin (VSRRP), a été évaluée et comparée chez des porcs en croissance infectés expérimentalement avec VSRRP-1 et VSRRP-2. Deux des vaccins étaient à base de VSRRP-1 et deux à base de VSRRP-2. Il n’y avait pas de différence significative entre chacun des deux VVM VSRRP-1 et les deux VVM VSRRP-2 respectivement sur la base des évaluations virologiques, immunologiques et pathologiques. La vaccination avec l’un ou l’autre des VVM VSRRP-1 a entrainé une réduction de la virémie causée par VSRRP-1 mais pas par VSRRP-2. De plus, la vaccination avec l’un ou l’autre des VVM VSRRP-1 a permis une réduction des lésions pulmonaires et des cellules positives pour VSRRP-1 chez les porcs infectés avec VSRRP-1 mais n’a pas eu d’effet significatif chez les porcs infectés avec VSRRP-2. La vaccination avec l’un ou l’autre des VVM SRRP-2 a résulté en une réduction de la virémie autant pour VSRRP-1 que VSRRP-2. Les VVM VSRRP-2 étaient également en mesure de réduire efficacement les lésions pulmonaires et les cellules positives pour VSRRP après l’infection soit par le VSRRP-1 ou VSRRP-2. Nos résultats suggèrent que bien que la vaccination avec les VVM VSRRP-1 peuvent être efficaces envers VSRRP-1, seulement les VVM VSRRP-2 peuvent protéger contre les infections par les VSRRP-1 et VSRRP-2 coréens utilisés dans la présente étude.
(Traduit par Docteur Serge Messier)
Introduction
Porcine reproductive and respiratory syndrome (PRRS) was first described as a “mystery swine disease” in 1987 in USA (1) and as “blue ear disease” in 1990 in Europe (2). The first cases of PRRS in Korea were detected in 1994. Since then, PRRS has rapidly become one of the most impactful global swine diseases causing devastating economical losses to the swine industry worldwide. Porcine reproductive and respiratory syndrome is characterized by reproductive failure in breeding females and respiratory disease in pigs of all ages (3). The causative agent for PRRS is the PRRS virus (PRRSV). Porcine reproductive and respiratory syndrome virus is a single-stranded positive sense RNA virus that belongs to the family of Arteriviridae in the order Nidovirales with 2 distinct species based on antigenic and pathogenic differences, PRRSV-1 of European origin and PRRSV-2 of North American origin (4,5).
In most European countries PRRSV-1 is the prevalent species (6), while the majority of PRRS cases in North American countries are caused by PRRSV-2 only (7). The PRRSV-1 is rarely considered an important economic pathogen in the US (Dr. Aaron J. Lower, Carthage Veterinary Service, personal communication, 2017). In contrast, the Korean farmland appears to be a region in which both PRRSV-1 and PRRSV-2 are equally prevalent with both species causing serious clinical problems (8). Currently on Korean farms, the most common method of controlling PRRSV infection is through vaccination. Although modified-live virus (MLV) vaccines so far have provided limited protection against heterologous field strains (9,10), they are widely used and considered to be the most effective tool in controlling PRRSV infection.
Currently, 4 MLV, 2 based on PRRSV-1 and 2 based on PRRSV-2 are commercially available in the Korean market. However no studies have been conducted to assess the efficacy of these 4 PRRSV MLV vaccines against heterologous PRRSV-1 and PRRSV-2 field viruses. The objective of this study was to evaluate and compare the efficacy of these 4 PRRSV MLV vaccines against single heterologous PRRSV-1 and PRRSV-2 challenge based on clinical, virological, immunological, and pathological criteria under the same experimental conditions.
Materials and methods
Virus
The PRRSV-1 (SNUVR090485, pan-European subtype 1) and PRRSV-2 (SNUVR090851, lineage 1) were used as challenge inocula (11,12). Nucleotide homology of the open reading frame 5 genome from PRRSV challenge viruses was compared with the vaccine viruses from 4 PRRSV MLV vaccines (Table I).
Table I.
Percentage nucleotide homology of open reading frame 5 genome from PRRSV challenge viruses used in this study compared with the vaccine viruses from 4 PRRSV modified-live virus vaccines.
| PRRSV-1 (JN315686) | PRRSV-2 (JN315685) | Porcilis PRRS (AY743931) | UNISTRAIN PRRS (GU067771) | Ingelvac PRRS MLV (AF066183) | Fostera PRRS (AF494042) | |
|---|---|---|---|---|---|---|
| PRRSV-1 | 100 | 59 | 87.9 | 88.1 | 61.1 | 61.1 |
| PRRSV-2 | 59 | 100 | 59.5 | 59.3 | 85.9 | 87.2 |
| Porcilis PRRS | 87.9 | 59.5 | 100 | 93.3 | 61.4 | 61.6 |
| UNISTRAIN PRRS | 88.1 | 59.3 | 93.3 | 100 | 61.1 | 60.6 |
| Ingelvac PRRS MLV | 61.1 | 85.9 | 61.4 | 61.1 | 100 | 91.3 |
Experimental design
A total of 132 colostrum-fed, cross-bred, conventional piglets were purchased at 14 d of age from a commercial PRRSV-free farm. All piglets were negative for PRRSV according to routine serological testing and real-time reverse transcriptase polymerase chain reaction (PCR) as previously described (13).
Pigs were divided into 11 groups (12 pigs per group) and assigned into 11 rooms using the random number generation function (Excel, Microsoft Corporation, Redmond, Washington, USA). The pigs within each group were housed in same room (Table II). Five groups (in 10 separate rooms) were challenged with PRRSV-1, 5 groups (in 10 separate rooms) were challenged with PRRSV-2, and 1 group (in 2 separate rooms) was used as control using the random number generation function (Excel; Microsoft Corporation). At −35 d post challenge (dpc, 28 d of age), pigs were injected intramuscularly on the right side of the neck with 2 mL of Porcilis PRRS (Vac1A/Ch1 and Vac1A/Ch2 groups; Lot No. D353A07, MSD Animal Health, Boxmeer, the Netherlands), UNISTRAIN PRRS (Vac1B/Ch1 and Vac1B/Ch2 groups; Lot No. 61WK-B, Hipra, Amer, Spain), Ingelvac PRRS MLV (Vac2A/Ch1 and Vac2A/Ch2 groups; Lot No. 245-659A, Boehringer Ingelheim Vetmedica, St. Joseph, Missouri, USA), and Fostera PRRS (Vac2B/Ch1 and Vac2B/Ch2 groups; Lot No. A405013B, Zoetis, Parsippany, New Jersey, USA) according to the manufacturer’s instructions. Pigs in the UnVac/Ch1, UnVac/Ch2, and UnVac/UnCh groups were administered an equal volume of phosphate-buffered saline (PBS, 0.01M, pH 7.4).
Table II.
Experimental design and results of lesion score and porcine reproductive and respiratory syndrome virus (PRRSV) RNA within lung lesion at 7 and 14 d post challenge (dpc).
| Groups | PRRSV | dpc | Lung lesion score | PRRSV-positive cells within lung lesion | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Vaccination (28 d) | Challenge (63 d) | Macroscopic | Microscopic | PRRSV-1 | PRRSV-2 | ||
| Vac1A/Ch1 | Porcilis PRRS | PRRSV-1 | 7 | 14.2 ± 4.9b | 0.7 ± 0.8a,b | 3.7 ± 1.0b | 0 ± 0 |
| 14 | 7.5 ± 2.7b | 0.5 ± 0.8a,b | 0.7 ± 0.8b | 0 ± 0 | |||
| Vac1B/Ch1 | UNISTRAIN PRRS | PRRSV-1 | 7 | 16.7 ± 7.5b | 0.8 ± 0.8a,b | 3.7 ± 0.8b | 0 ± 0 |
| 14 | 8.3 ± 4.1b | 0.7 ± 0.5a,b | 0.5 ± 0.5b | 0 ± 0 | |||
| Vac2A/Ch1 | Ingelvac PRRS MLV | PRRSV-1 | 7 | 18.3 ± 4.1b | 0.8 ± 0.6b | 0 ± 0c | 0 ± 0 |
| 14 | 4.2 ± 4.9b | 0.3 ± 0.6b | 0.5 ± 0.5b | 0 ± 0 | |||
| Vac2B/Ch1 | Fostera PRRS | PRRSV-1 | 7 | 16.7 ± 5.2b | 0.7 ± 0.5b | 0 ± 0c | 0 ± 0 |
| 14 | 3.3 ± 5.2b | 0.2 ± 0.4b | 0.3 ± 0.8b | 0 ± 0 | |||
| UnVac/Ch1 | None | PRRSV-1 | 7 | 30.8 ± 8.0a | 1.6 ± 0.5a | 20.3 ± 3.7a | 0 ± 0 |
| 14 | 24.2 ± 6.6a | 1.4 ± 0.5a | 9.7 ± 3.5a | 0 ± 0 | |||
| UnVac/UnCh | None | None | 7 | 0.8 ± 2.0c | 0.3 ± 0.5b | 0 ± 0c | 0 ± 0 |
| 14 | 1.7 ± 4.1b | 0.2 ± 0.4b | 0 ± 0b | 0 ± 0 | |||
| Vac1A/Ch2 | Porcilis PRRS | PRRSV-2 | 7 | 58.3 ± 14.7a | 3.7 ± 0.4a | 0 ± 0 | 45.5 ± 12.2a |
| 14 | 45.8 ± 6.6a | 3.3 ± 0.8a | 0 ± 0 | 34.8 ± 6.2a,b | |||
| Vac1B/Ch2 | UNISTRAIN PRRS | PRRSV-2 | 7 | 55.8 ± 10.2a | 3.8 ± 0.4a | 0 ± 0 | 43.8 ± 7.0a |
| 14 | 47.5 ± 7.6a | 3.3 ± 0.4a | 0 ± 0 | 35.3 ± 6.3a,b | |||
| Vac2A/Ch2 | Ingelvac PRRS MLV | PRRSV-2 | 7 | 32.5 ± 5.2b | 2.0 ± 0.6b | 0 ± 0 | 30.8 ± 9.2a |
| 14 | 21.7 ± 6.8b | 1.8 ± 0.8b | 0 ± 0 | 23.8 ± 5.0a,b | |||
| Vac2B/Ch2 | Fostera PRRS | PRRSV-2 | 7 | 31.7 ± 7.5b | 2.1 ± 0.2b | 0 ± 0 | 29.2 ± 7.0a |
| 14 | 18.3 ± 7.5b | 1.6 ± 0.5b | 0 ± 0 | 22.2 ± 6.7b | |||
| UnVac/Ch2 | None | PRRSV-2 | 7 | 63.3 ± 8.2a | 3.8 ± 0.4a | 0 ± 0 | 45.2 ± 13.7a |
| 14 | 46.7 ± 8.2a | 3.3 ± 0.5a | 0 ± 0 | 36.0 ± 14.0a | |||
| UnVac/UnCh | None | None | 7 | 0.8 ± 2.0c | 0.3 ± 0.5c | 0 ± 0 | 0 ± 0b |
| 14 | 1.7 ± 4.1c | 0.2 ± 0.4c | 0 ± 0 | 0 ± 0c | |||
Indicate significant (P < 0.05) difference among groups.
At 0 dpc (63 d of age), pigs in the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1, and UnVac/Ch1 groups were inoculated intranasally with 3 mL of PRRSV-1 inoculum (105 TCID50/mL of SNUVR090485, second passage in alveolar macrophages). Pigs in the Vac1A/Ch2, Vac1B/Ch2, Vac2A/Ch2, Vac2B/Ch2, and UnVac/Ch2 groups were inoculated intranasally with 3 mL of PRRSV-2 inoculum (105 TCID50/mL of SNUVR090851, second passage in alveolar macrophages). Pigs in the UnVac/UnCh group served as negative controls and were not exposed to either the vaccine or virus. Upon challenge, pigs in the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1, and UnVac/Ch1 groups were randomly assigned into 10 out of 22 rooms. Pigs in the Vac1A/Ch2, Vac1B/Ch2, Vac2A/Ch2, Vac2B/Ch2, and UnVac/Ch2 groups were randomly assigned into 10 out of 22 rooms. Each room contained 6 pens and each pig was housed individually in a pen. In each of the 10 rooms, allocation of pens to treatment was in accordance with a randomized complete block design with a 1-way treatment structure. Blocking was based on pen location. A block comprised of 4 pens located near each other. The experimental unit for treatment was the individual animal. Within each block, 1 pen was randomly assigned to each treatment group. Pigs in the UnVac/UnCh group were randomly placed into 12 pens in the 2 remaining rooms.
Following PRRSV challenge, the physical condition and the rectal temperature of each pig was monitored daily. Blood samples were collected at −35, −21, 0, 7, 10, and 14 dpc. Pigs were sedated by an intravenous injection of sodium pentobarbital and then euthanized by electrocution at 7 and 14 dpc as previously described (14). All methods were previously approved by the Seoul National University Institutional Animal Care and Use, and Ethics Committee.
Clinical observation
Clinical observation of respiratory symptoms was recorded daily using scores ranging from 0 (normal) to 6 (severe dyspnea and abdominal breathing) (15). Observers were blinded to vaccination and challenge status. Rectal temperatures were also recorded daily at the same time by the same personnel.
Serology
Serum samples that were collected were tested using a commercially available PRRSV enzyme-linked immunosorbent assay (ELISA) (HerdCheck PRRS X3 Ab test; IDEXX Laboratories, Westbrook, Maine, USA). Serum samples were considered positive for PRRSV antibody if the S/P ratio was ≥ 0.4, according to the manufacturer’s instructions.
Quantification of PRRSV RNA
RNA was extracted from serum samples that were collected to quantify PRRSV genomic cDNA copy numbers, as previously described (13). The PRRSV-1 forward and reverse primers were 5′-TGGCCAGTCAGTCAATCAAC-3′ and 5′-AATCGATTGCAAGCAGAGGGAA-3′, respectively. The PRRSV-2 forward and reverse primers were 5′-TGGCCAGTCAGTCAATCAAC-3′ and 5′-AATCGATTGCAAGCAGAGGGAA-3′, respectively (13).
For the Porcilis PRRS vaccine virus, the forward and reverse primers were 5′-TGTAGACAACCGGGGGAGAG-3′ and 5′-CTAGGCCTCCCATTGCTCAG-3′, respectively. For the UNISTRAIN vaccine virus, the forward and reverse primers were 5′-GTTGCCCAGCCATTTTGAC-3′ and 5′-CACGCTGCTGAGTACATACC-3′, respectively (16). For the Ingelvac PRRS MLV vaccine virus, the forward and reverse primers were 5′-CTAACAAATTTGATTGGGCAG-3′ and 5′-AGGACATGCAATTCTTTGCAA-3′, respectively (16). For the Fostera PRRS vaccine virus, the forward and reverse primers were 5′-CTTGACACAGTTGGTCTGGTTACT-3′ and 5′-GTTCTTCGCAAGCCTAATAACG-3′, respectively (17). Real-time PCR was done to quantify PRRSV genomic cDNA copies (13,16–18).
Enzyme-linked immunospot (ELISPOT) assay
The numbers of PRRSV-specific interferon-γ secreting cells (IFN-γ-SC) were determined in peripheral blood mononuclear cells (PBMC) using challenging PRRSV-1 and PRRSV-2 as previously described (17,19,20).
Pathology and in situ hybridization
The total amount of microscopic lesions in the lung sections was scored blindly ranging from 0 (normal) to 4 (severe) by 2 pathologists (15). In situ hybridization (ISH) for the detection was completed and analyzed morphometrically using computer software (NIH Image J 1.51r Program; http://imagej.nih.gov/ij/download.html), as previously described (21).
Statistical analysis
Continuous data included rectal temperature, PRRSV RNA (log10 of the number of PRRSV genomic copies per mL quantified by real-time PCR), PRRSV antibody titer, and number of IFN-γ-SC (measured by ELISPOT assay). Continuous data were analyzed using Tukey’s multiple comparisons test for comparison between groups in order to estimate the difference at each time point. Discrete data (clinical signs, lung lesion scores, and ISH scores) were analyzed using the Kruskal-Wallis test. When the Kruskal-Wallis test was significant, the Mann-Whitney test was done to determine the significant differences between the groups. A value of P < 0.05 was considered significant.
Results
Clinical observation
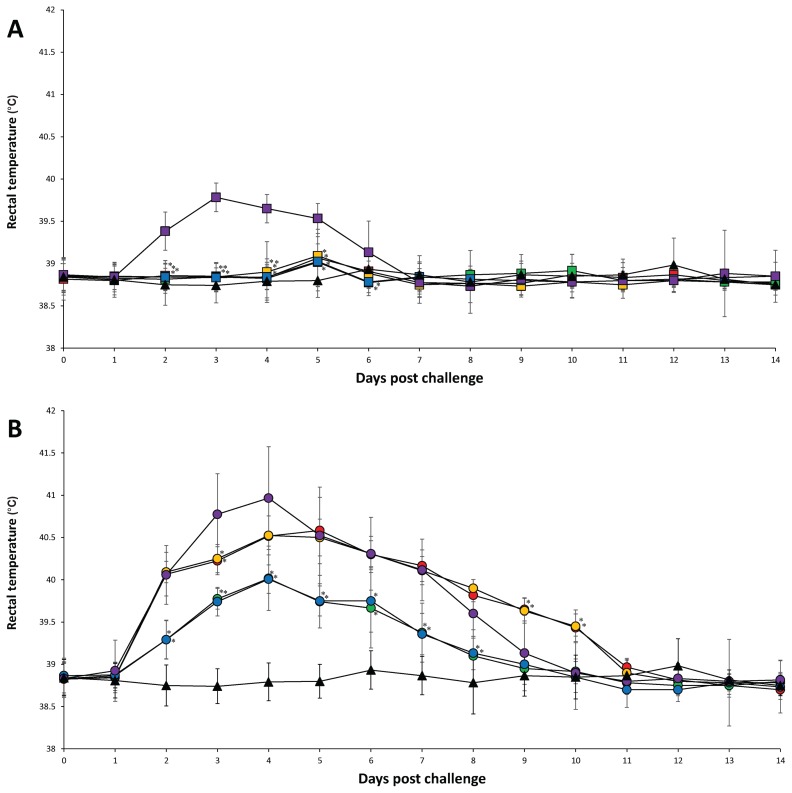
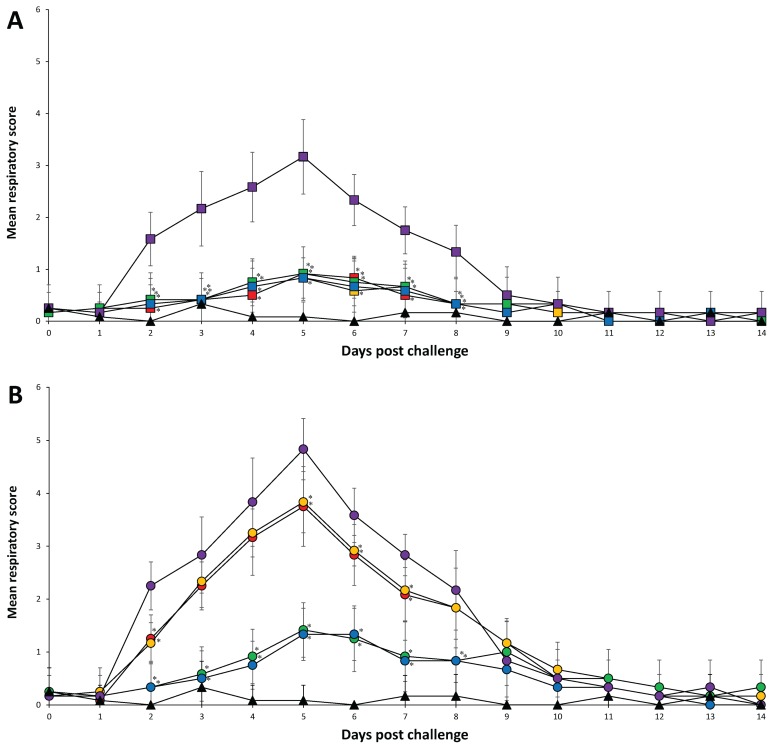
There were no observable clinical signs after vaccination and before challenge in any of the pigs from all 6 groups. In PRRSV-1 challenged groups, the mean rectal temperature was significantly (P < 0.05) lower in pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1, and UnVac/UnCh groups at 2 to 5 dpc compared to pigs from the UnVac/Ch1 group. The mean rectal temperature was significantly lower (P < 0.05) in pigs from the Vac2A/Ch1 and Vac2B/Ch1 groups at 6 dpc compared to pigs from the UnVac/Ch1 group (Figure 1A). The mean respiratory scores were significantly (P < 0.05) lower in pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1, and UnVac/UnCh groups at 2 to 8 dpc compared to pigs from the UnVac/Ch1 group. The mean respiratory scores were significantly lower (P < 0.05) in pigs from the UnVac/UnCh group compared to pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups at 5 and 6 dpc (Figure 2A).
Figure 1.
Mean rectal temperature. A — PRRSV-1 challenged groups from the Vac1A/Ch1 (
 ), Vac1B/Ch1 (
), Vac1B/Ch1 (
 ), Vac2A/Ch1 (
), Vac2A/Ch1 (
 ), Vac2B/Ch1 (
), Vac2B/Ch1 (
 ), UnVac/Ch1 (
), UnVac/Ch1 (
 ), and UnVac/UnCh (▲). B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
), and UnVac/UnCh (▲). B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
 ), Vac1B/Ch2 (
), Vac1B/Ch2 (
 ), Vac2A/Ch2 (
), Vac2A/Ch2 (
 ), Vac2B/Ch2 (
), Vac2B/Ch2 (
 ), UnVac/Ch2 (
), UnVac/Ch2 (
 ), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
Figure 2.
Mean respiratory score. A — PRRSV-1 challenged groups from the Vac1A/Ch1 (
 ), Vac1B/Ch1 (
), Vac1B/Ch1 (
 ), Vac2A/Ch1 (
), Vac2A/Ch1 (
 ), Vac2B/Ch1 (
), Vac2B/Ch1 (
 ), UnVac/Ch1 (
), UnVac/Ch1 (
 ), and UnVac/UnCh (▲). B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
), and UnVac/UnCh (▲). B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
 ), Vac1B/Ch2 (
), Vac1B/Ch2 (
 ), Vac2A/Ch2 (
), Vac2A/Ch2 (
 ), Vac2B/Ch2 (
), Vac2B/Ch2 (
 ), UnVac/Ch2 (
), UnVac/Ch2 (
 ), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
In PRRSV-2 challenge groups, the mean rectal temperature was significantly lower (P < 0.05) in pigs from the Vac2A/Ch2, Vac2B/Ch2, and UnVac/UnCh groups at 2 to 10 dpc compared to pigs from the Vac1A/Ch2 and Vac1B/Ch2 groups. The mean rectal temperature was significantly lower (P < 0.05) in pigs from the Vac2A/Ch2, Vac2B/Ch2, and UnVac/UnCh groups at 2 to 8 dpc compared to pigs from the UnVac/Ch2 group. The mean rectal temperature was significantly lower (P < 0.05) in pigs from the Vac1A/Ch2 and Vac1B/Ch2 groups at 3 dpc compared to pigs from the UnVac/Ch2 group. The mean rectal temperature was significantly lower (P < 0.05) in pigs from the UnVac/UnCh group at 2 to 7 dpc compared to pigs from the Vac2A/Ch2 and Vac2B/Ch2 groups (Figure 1B). The mean respiratory scores were significantly lower (P < 0.05) in pigs from the Vac2A/Ch2, Vac2B/Ch2, and UnVac/UnCh groups at 2 to 8 dpc compared to pigs from the Vac1A/Ch2, Vac1B/Ch2, and UnVac/Ch2 groups. The mean respiratory scores were significantly lower (P < 0.05) in pigs from the Vac1A/Ch2 and Vac1B/Ch2 groups at 2, 5, 6, and 7 dpc compared to pigs from the UnVac/Ch2 group. The mean respiratory scores were significantly lower (P < 0.05) in pigs from the UnVac/UnCh group at 2, 4, 5, 6, and 7 dpc compared to pigs from the Vac2A/Ch2 and Vac2B/Ch2 groups. Pigs in the UnVac/UnCh group maintained normal rectal temperatures and respiratory signs throughout the study (Figure 2B).
Quantification of PRRSV RNA
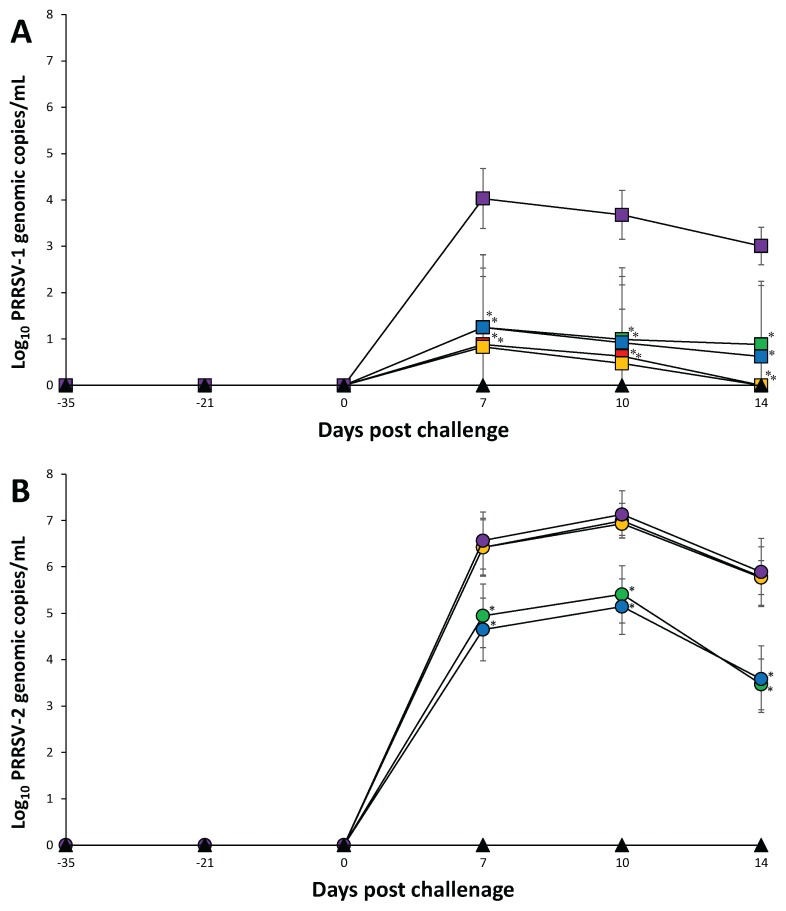
Genomic copies of the vaccine virus were detected in the sera of vaccinated pigs −21 dpc (14 d after vaccination) but, thereafter, no genomes of the vaccine strain were detected throughout the rest of the experiment. In the PRRSV-1 challenged groups, pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups had significantly lower (P < 0.05) genomic copies of PRRSV-1 in their sera compared to pigs from the UnVac/Ch1 group at 7 to 14 dpc. There was no significant difference in genomic copies of PRRSV-1 in the sera of pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups (Figure 3A).
Figure 3.
Mean values of the genomic copies number of PRRSV RNA. A — PRRSV-1 challenged groups in serum from the Vac1A/Ch1 (
 ), Vac1B/Ch1 (
), Vac1B/Ch1 (
 ), Vac2A/Ch1 (
), Vac2A/Ch1 (
 ), Vac2B/Ch1 (
), Vac2B/Ch1 (
 ), UnVac/Ch1 (
), UnVac/Ch1 (
 ), and UnVac/UnCh (▲). B — PRRSV-2 RNA in serum from the Vac1A/Ch2 (
), and UnVac/UnCh (▲). B — PRRSV-2 RNA in serum from the Vac1A/Ch2 (
 ), Vac1B/Ch2 (
), Vac1B/Ch2 (
 ), Vac2A/Ch2 (
), Vac2A/Ch2 (
 ), Vac2B/Ch2 (
), Vac2B/Ch2 (
 ), UnVac/Ch2 (
), UnVac/Ch2 (
 ), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
In the PRRSV-2 challenged groups, pigs from the Vac2A/Ch2 and Vac2B/Ch2 groups had significantly lower (P < 0.05) genomic copies of PRRSV-2 in their sera compared to pigs from the Vac1A/Ch2, Vac1B/Ch2, and UnVac/Ch2 groups at 7 to 14 dpc. There was no significant difference in genomic copies of PRRSV-2 in the sera of pigs from the Vac1A/Ch2, Vac1B/Ch2, and UnVac/Ch2 groups. The PRRSV-1 was not detected in PRRSV-2 challenged pigs and vice versa. No PRRSV of any genotype was detected in the sera of pigs from the UnVac/UnCh group throughout the experiment (Figure 3B).
Serology
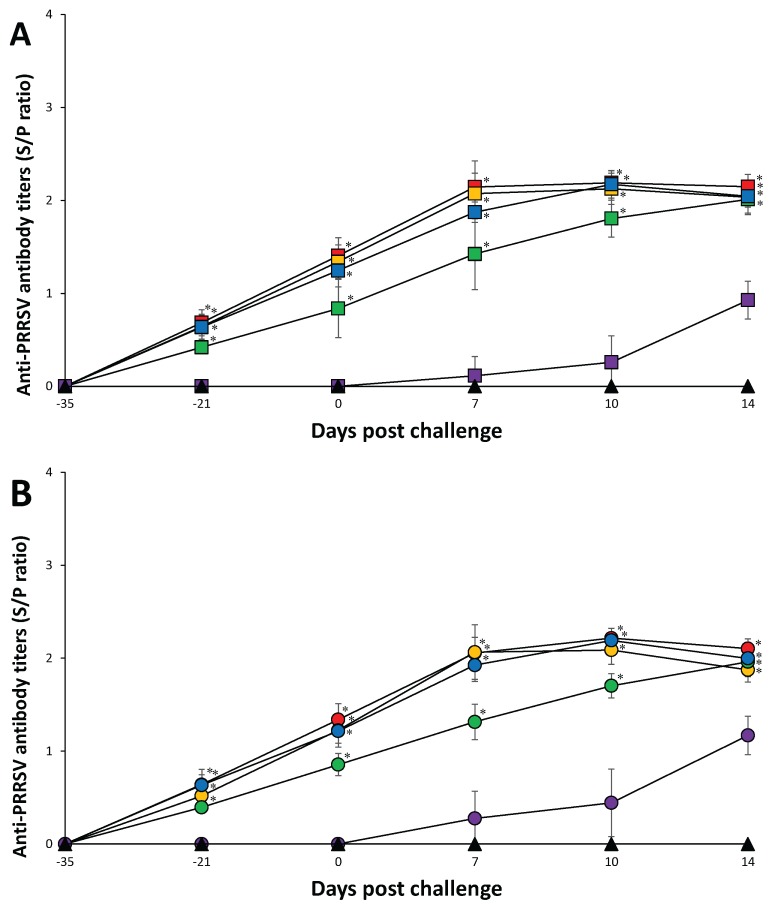
At the time of PRRSV vaccination (–35 dpc), pigs in all 11 groups were seronegative. Anti-PRRSV antibody titers were detected in vaccinated pigs only before challenge. In PRRSV-1 challenged groups, pigs from the Vac1A/Ch1, Vac1B/Ch1, and Vac2B/Ch1 groups had significantly higher (P < 0.05) anti-PRRSV antibody titers at 7 and 10 dpc compared to pigs from the Vac2A/Ch1 and UnVac/Ch1 groups. Pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups had significantly higher (P < 0.05) anti-PRRSV antibody titers at 14 dpc compared to pigs from the UnVac/Ch1 group (Figure 4A).
Figure 4.
Mean values of the anti-PRRSV antibodies. A — PRRSV-1 challenged groups from the Vac1A/Ch1 (
 ), Vac1B/Ch1 (
), Vac1B/Ch1 (
 ), Vac2A/Ch1 (
), Vac2A/Ch1 (
 ), Vac2B/Ch1 (
), Vac2B/Ch1 (
 ), UnVac/Ch1 (
), UnVac/Ch1 (
 ), and UnVac/UnCh (▲). B — RRSV-2 challenged groups from the Vac1A/Ch2 (
), and UnVac/UnCh (▲). B — RRSV-2 challenged groups from the Vac1A/Ch2 (
 ), Vac1B/Ch2 (
), Vac1B/Ch2 (
 ), Vac2A/Ch2 (
), Vac2A/Ch2 (
 ), Vac2B/Ch2 (
), Vac2B/Ch2 (
 ), UnVac/Ch2 (
), UnVac/Ch2 (
 ), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
In PRRSV-2 challenged groups, pigs from the Vac1A/Ch2, Vac1B/Ch2, Vac2A/Ch2, and Vac2B/Ch2 groups had significantly higher (P < 0.05) anti-PRRSV antibody titers at 7 to 14 dpc compared to pigs from the UnVac/Ch2 group. Pigs from the Vac1A/Ch2, Vac1B/Ch2, and Vac2B/Ch2 groups had significantly higher (P < 0.05) anti-PRRSV antibody titers at 7 and 10 dpc compared to pigs from the Vac2A/Ch2 group. Anti-PRRSV antibody titers were not detected in any of the pigs from the UnVac/UnCh group throughout the study (Figure 4B).
Interferon-γ secreting cells
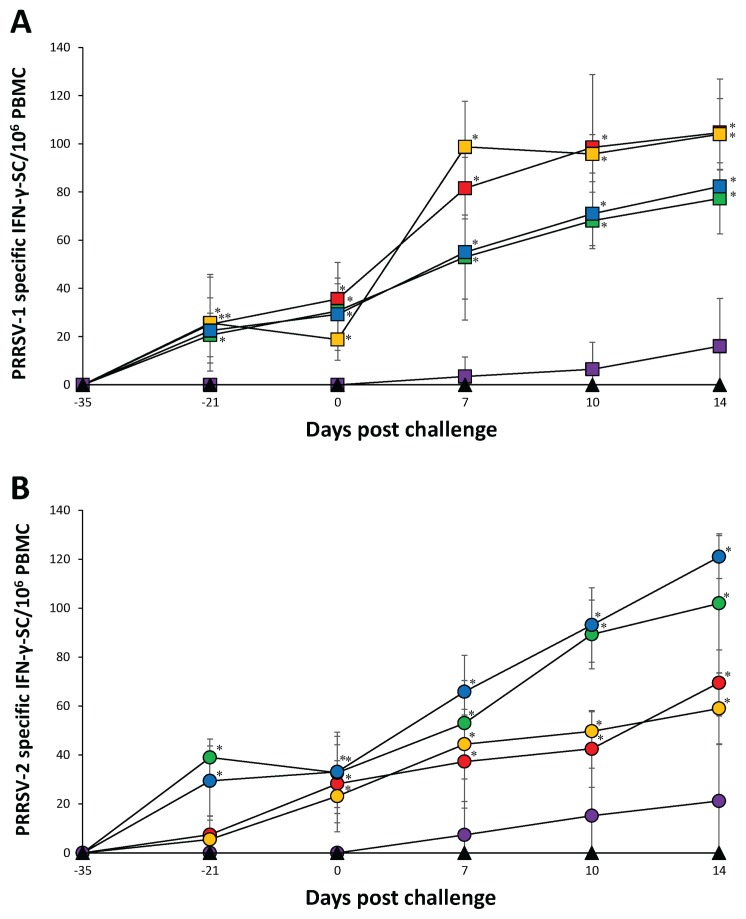
In PRRSV-1 challenged groups, pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups had a significantly higher (P < 0.05) number of PRRSV-1 specific IFN-γ-SC compared to pigs from the UnVac/Ch1 group at −21, 0, 7, 10, and 14 dpc. Pigs from the Vac1A/Ch1 and Vac1B/Ch1 groups had a significantly higher (P < 0.05) number of PRRSV-1 specific IFN-γ-SC compared to pigs from the Vac2A/Ch1 and Vac2B/Ch1 groups at 7 dpc. Pigs from the Vac1A/Ch1 and Vac1B/Ch1 groups had a significantly higher (P < 0.05) number of PRRSV-1 specific IFN-γ-SC compared to pigs from the Vac2A/Ch1 group at 10 and 14 dpc (Figure 5A).
Figure 5.
Frequency of PRRSV specific IFN-γ-SC/106 PBMC. A — PRRSV-1 challenged groups from the Vac1A/Ch1 (
 ), Vac1B/Ch1 (
), Vac1B/Ch1 (
 ), Vac2A/Ch1 (
), Vac2A/Ch1 (
 ), Vac2B/Ch1 (
), Vac2B/Ch1 (
 ), UnVac/Ch1 (
), UnVac/Ch1 (
 ), and UnVac/UnCh (▲) B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
), and UnVac/UnCh (▲) B — PRRSV-2 challenged groups from the Vac1A/Ch2 (
 ), Vac1B/Ch2 (
), Vac1B/Ch2 (
 ), Vac2A/Ch2 (
), Vac2A/Ch2 (
 ), Vac2B/Ch2 (
), Vac2B/Ch2 (
 ), UnVac/Ch2 (
), UnVac/Ch2 (
 ), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
), and UnVac/UnCh (▲). Variation is expressed as the standard deviation. Significant difference between vaccinated challenged and unvaccinated challenged groups within the same PRRSV type challenge is indicated as P < 0.05*.
In PRRSV-2 challenged groups, pigs from the Vac2A/Ch2 group had a significantly higher (P < 0.05) number of PRRSV-2 specific IFN-γ-SC compared to pigs from the Vac1A/Ch2, Vac1B/Ch2, Vac2B/Ch2, and UnVac/Ch2 groups at −21 dpc. Pigs from the Vac1A/Ch2, Vac1B/Ch2, Vac2A/Ch2, and Vac2B/Ch2 groups had a significantly higher (P < 0.05) number of PRRSV-2 specific IFN-γ-SC compared to pigs from the UnVac/Ch2 group at 0, 7, 10, and 14 dpc. Pigs from the Vac2B/Ch2 group had a significantly higher (P < 0.05) number of PRRSV-2 specific IFN-γ-SC compared to pigs from the Vac1A/Ch2 and Vac1B/Ch2 groups at 7 dpc. Pigs from the Vac2A/Ch2 and Vac2B/Ch2 groups had a significantly higher (P < 0.05) number of PRRSV-2 specific IFN-γ-SC compared to pigs from the Vac1A/Ch2 and Vac1B/Ch2 groups at 10 and 14 dpc. In pigs from the UnVac/UnCh group, the mean numbers of PRRSV-1 and PRRSV-2 specific IFN-γ-SC remained at basal levels (< 20 cells/106 PBMC) throughout the study (Figure 5B).
Pathology
In PRRSV-1 challenged groups, pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1, and UnVac/UnCh groups exhibited significantly (P < 0.05) lower mean macroscopic lung lesion scores at 7 and 14 dpc compared to pigs from the UnVac/Ch1 group. Pigs from the UnVac/UnCh group had significantly (P < 0.05) lower mean macroscopic lung lesion scores at 7 dpc compared to pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, and Vac2B/Ch1 groups. Pigs from the Vac2A/Ch1, Vac2B/Ch1, and UnVac/UnCh groups had significantly lower (P < 0.05) mean microscopic lung lesion scores at 7 and 14 dpc compared to pigs from the UnVac/Ch1 group. Pigs from the Vac1A/Ch1, Vac1B/Ch1, Vac2A/Ch1, Vac2B/Ch1 groups had a significantly lower (P < 0.05) number of PRRSV-1 positive cells per area unit of lung at 7 and 14 dpc compared to pigs from the UnVac/Ch1 group. Pigs from the Vac2A/Ch1 and Vac2B/Ch1 groups had a significantly lower (P < 0.05) number of PRRSV-1 positive cells per area unit of lung at 7 dpc compared to pigs from the Vac1A/Ch1 and Vac1B/Ch1 groups (Table II).
In PRRSV-2 challenged groups, pigs from the Vac2A/Ch2, Vac2B/Ch2, and UnVac/UnCh groups showed significantly lower (P < 0.05) mean macroscopic and microscopic lung lesion scores at 7 and 14 dpc compared to pigs from the Vac1A/Ch2, Vac1B/Ch2, and UnVac/Ch2 groups. Pigs from the Vac2B/Ch2 group also had a significantly lower (P < 0.05) number of PRRSV-2 positive cells per area unit of lung at 14 dpc compared to pigs from the UnVac/Ch2 group. No PRRSV of any genotype was detected in the lungs of pigs from the UnVac/UnCh group (Table II).
Discussion
In this study, we compared the efficacy of 2 PRRSV-1 MLV vaccines and 2 PRRSV-2 MLV vaccines against heterologous challenge with PRRSV-1 and PRRSV-2. There was no significant difference between the 2 PRRSV-1 MLV vaccines as they both can provide partial protection against a PRRSV-1 strain but only limited protection against a PRRSV-2 strain, during the acute phase. In contrast, 2 commercial PRRSV-2 MLV vaccines can provide partial protection against both PRRSV-1 and -2 strains. Our conclusions are based on clinical, virological, immunological, and pathological comparisons. These results are consistent with previous studies, in which PRRSV-1 MLV vaccines provide partial protection against respiratory disease caused by heterologous type 1 PRRSV challenge but confer no protection against heterologous type 2 PRRSV challenge in pigs (18,22,23). Similar to our results, previous studies have also shown that vaccination of pigs with a PRRSV-2 vaccine can protect pigs against both heterologous PRRSV-1 and PRRSV-2 challenge (17,24). However, our results should be interpreted cautiously because only 1 strain for each genotype was used as challenge. The type of strain used as challenge can have a significant impact on the efficacy of a vaccine. Our results do contrast with other studies in which vaccination of pigs with the same PRRSV-1 MLV vaccine provided partial protection against heterologous PRRSV-2 challenge (25,26). However, this study used a different PRRSV-2 strain suggesting that perhaps antigenicity plays a more important role on the efficacy of the PRRS MLV vaccine than genetic similarity between the vaccine and challenge strains. The PRRSV viremia plays a critical role in the development of respiratory disease. The levels of viremia are well-correlated with the severity of interstitial pneumonia (12). Therefore, the reduction of PRRSV viremia could be essential in preventing respiratory disease and an important indicator of the efficacy of a PRRSV vaccine (22,27). Vaccination of pigs with either of the PRRSV-2 MLV vaccines resulted in a significant reduction both of PRRSV-1 and PRRSV-2 viremia. Vaccination of pigs with the PRRSV-1 MLV vaccines could only significantly reduce PRRSV-1 viremia. In addition, duration of PRRSV-1 viremia in vaccinated and PRRSV-1-challenged (Vac1A/Ch1 and Vac2A/Ch1) groups in the present study is similar to that in a previous study (28). However, duration of PRRSV-2 viremia in vaccinated and PRRSV-2-challenged (Vac1A/Ch2 and Vac2A/Ch2) groups is longer in present study compared to a previous study (28). Altogether, these data suggest that PRRSV-2 (strain SNUVR090851) challenge virus used in this study is more virulent than PRRSV-2 (strain 19407b) challenge virus used in a previous study. The difference in protection between the PRRSV-1 and PRRSV-2 MLV vaccines may be due to the possibility that they elicit different cellular immune responses against the 2 PRRSV types. In our experimental conditions, vaccination of pigs with the PRRSV-2 MLV vaccines resulted in induction of equal levels of IFNγ-SC against PRRSV-1 and PRSV-2. Vaccination with the PRRSV-1 MLV vaccines induced higher levels of IFN-γ-SC against PRRSV-1 compared to PRRSV-2. T-cell cross reactivity has been previously shown with genetically distant PRRSVs (29,30). Evidence of correlation between the increase of PRRSV-1 specific IFN-γ-SC levels and reduction of PRRSV-1 viremia further supports the important role of T-cells in cross protection of PRRSV-2 vaccinated pigs after PRRSV-1 challenge. Therefore, T-cells activated by PRRSV-2 MLV vaccines respond against PRRSV-1 infection, resulting in partial cross protection. Even though the increase of IFN-γ-SC does not always correlate with protection (31,32), cell-mediated immunity seems to play an important role in cross protection against PRRSV infection. Since, in general, the PRRSV MLV vaccine provides good homologous protection but variable heterologous protection (9), the PRRSV challenge viruses used in this study should not originate from the vaccine virus. The PRRSV-1 (SNUVR090485) challenge virus was isolated from pigs in 2009 before two PRRSV-1 MLV vaccines were introduced in South Korea in 2014. The PRRSV-2 (SNUVR090851) challenge virus belongs to lineage 1 while two PRRSV-2 MLV vaccines belong to lineage 5 (Ingelvac PRRS MLV) and 8 (Fostera PRRS), respectively, based on the classification system (33). Therefore, the degree of heterologous protection by the PRRSV MLV vaccines is not influenced by the PRRSV challenge viruses, which are not derived from the vaccine virus. To the best of our knowledge, this is the first comparative study evaluating 4 commercial MLV vaccines, currently available on the Korean market, under the same experimental conditions. The results of this study are important because they provide swine producers and practitioners with valuable clinical information to better select future PRRSV vaccines.
Acknowledgments
This research was supported by contract research funds of the Research Institute for Veterinary Science from the College of Veterinary Medicine and by the BK 21 Plus Program (Grant no. 5360-20150100) for Creative Veterinary Science Research.
References
- 1.Keffaber KK. Reproductive failure of unknown etiology. Am Assoc Swine Pract Newsl. 1989;1:1–9. [Google Scholar]
- 2.Paton DJ, Brown IH, Edwards S, Wensvoort G. ‘Blue ear’ disease of pigs. Vet Rec. 1991;128:617. doi: 10.1136/vr.128.26.617. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman JJ, Benfield DA, Dee SA, et al. Porcine reproductive and respiratory syndrome virus (porcine arterivirus) In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of Swine. 10th ed. Ames, Iowa: Wiley-Blackwell; 2012. pp. 461–486. [Google Scholar]
- 4.Snijder EJ, Meulenberg JJM. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 5.Forsberg R, Storgaard T, Nielsen HS, et al. The genetic diversity of European type PRRSV is similar to that of the North American type but is geographically skewed within Europe. Virology. 2000;299:38–47. doi: 10.1006/viro.2002.1450. [DOI] [PubMed] [Google Scholar]
- 6.Stadejek T, Stankevicius A, Murtaugh MP, Oleksiewicz MB. Molecular evolution of PRRSV in Europe: Current state of play. Vet Microbiol. 2013;165:21–28. doi: 10.1016/j.vetmic.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Shi M, Lam TT-Y, Hon C-C, et al. Molecular epidemiology of PRRSV: A phylogenetic perspective. Virus Res. 2010;154:7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Choi K, Lee J, Park C, Jeong J, Chae C. Comparison of the pathogenesis of single or dual infections with type 1 and type 2 porcine reproductive and respiratory syndrome virus. J Comp Pathol. 2015;152:317–324. doi: 10.1016/j.jcpa.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Murtaugh MP, Genzow M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS) Vaccine. 2011;29:8192–8204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Galliher-Beckley A, Pappan L, et al. Comparison of host immune responses to homologous and heterologous type II porcine reproductive and respiratory syndrome virus (PRRSV) challenge in vaccinated and unvaccinated pigs. Biomed Res Int. 2014;2014 doi: 10.1155/2014/416727. 416727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han K, Seo HW, Oh Y, Kang I, Park C, Chae C. Pathogenesis of Korean type 1 (European genotype) porcine reproductive and respiratory syndrome virus in experimentally infected pigs. J Comp Pathol. 2012;147:275–284. doi: 10.1016/j.jcpa.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Han K, Seo HW, Park C, et al. Comparative pathogenicity of three Korean and one Lelystad type 1 porcine reproductive and respiratory syndrome virus (pan-European subtype 1) isolates in exeprimentally infected pigs. J Comp Pathol. 2013;149:331–340. doi: 10.1016/j.jcpa.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Wasilk A, Callahan JD, Christopher-Hennings J, et al. Detection of US, Lelystad, and European-like porcine reproductive and respiratory syndrome viruses and relative quantitation in boar semen and serum samples by realtime PCR. J Clin Microbiol. 2004;42:4453–446. doi: 10.1128/JCM.42.10.4453-4461.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaver BV, Reed W, Leary S, et al. 2002 Report of the AVMA panel on euthanasia. J Am Vet Med Assoc. 2001;218:669–696. doi: 10.2460/javma.2001.218.669. [DOI] [PubMed] [Google Scholar]
- 15.Halbur PG, Paul PS, Frey ML, et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32:648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 16.Han K, Seo HW, Park C, Chae C. Vaccination of sows against type 2 porcine reproductive and respiratory syndrome virus (PRRSV) before artificial insemination protects against type 2 PRRSV challenge but does not protect against type 1 PRRSV challenge in late gestation. Vet Res. 2014;45:12. doi: 10.1186/1297-9716-45-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park C, Seo HW, Han K, Kang I, Chae C. Evaluation of the efficacy of a new modified live porcine reproductive and respiratory syndrome virus (PRRSV) vaccine (Fostera PRRS) against heterologous PRRSV challenge. Vet Microbiol. 2014;172:432–442. doi: 10.1016/j.vetmic.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 18.Kim T, Park C, Choi K, et al. Comparison of two commercial type 1 porcine reproductive and respiratory syndrome virus (PRRSV) modified live vaccines against heterologous type 1 and type 2 PRRSV challenge in growing pigs. Clin Vaccine Immunol. 2015;22:631–640. doi: 10.1128/CVI.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier WA, Galeota J, Osorio FA, Husmann RJ, Schnitzlein WM, Zuckermann FA. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology. 2003;309:18–31. doi: 10.1016/s0042-6822(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 20.Diaz I, Mateu E. Use of ELISPOT and ELISA to evaluate IFN-gamma, IL-10 and IL-4 responses in conventional pigs. Vet Immunol Immunopathol. 2005;06:107–112. doi: 10.1016/j.vetimm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Han K, Seo HW, Oh Y, Kang I, Park C, Chae C. Comparison of the virulence of European and North American genotypes of porcine reproductive and respiratory syndrome virus in experimentally infected pigs. Vet J. 2013;195:313–318. doi: 10.1016/j.tvjl.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 22.van Woensel PA, Leifkens K, Demaret S. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Vet Rec. 1998;142:510–512. doi: 10.1136/vr.142.19.510. [DOI] [PubMed] [Google Scholar]
- 23.Labarque GG, Nauwynck HJ, van Woensel PAM, Visser N, Pensaert MB. Efficacy of an American and a European serotype PRRSV vaccine after challenge with American and European wild-type strains of the virus. Vet Res. 2000;31:97. [Google Scholar]
- 24.Park C, Choi K, Jeong J, Chae C. Cross-protection of a new type 2 porcine reproductive and respiratory syndrome virus (PRRSV) modified live vaccine (Fostera PRRS) against heterologous type 1 PRRSV challenge in growing pigs. Vet Microbiol. 2015;177:87–94. doi: 10.1016/j.vetmic.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 25.Roca M, Gimeno M, Bruguera S, et al. Effect of challenge with a virulent genotype II strain of porcine reproductive and respiratoiry syndrome virus on piglets vaccinated with an attenuated genotype I strain vaccine. Vet J. 2012;193:92–96. doi: 10.1016/j.tvjl.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Ko SS, Seo SW, Sunwoo SY, Yoo SJ, Lim MY, Lyoo YS. Efficacy of commercial genotype 1 porcine reproductive and respiratory virus (PRRSV) vaccine against field isolate of genotype 2 PRRSV. Vet Immunol Immunopathol. 2016;172:43–49. doi: 10.1016/j.vetimm.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Labarque G, Van Gucht S, Van Reeth K, Nauwynck H, Pensaert M. Respiratory tract protection upon challenge of pigs vaccinated with attenuated porcine reproductive and respiratory syndrome virus vaccines. Vet Microbiol. 2003;95:187–197. doi: 10.1016/s0378-1135(03)00157-3. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen CS, Kvisgaard LK, Pawlowski M, et al. Efficacy and safety of simultaneous vaccination with two modified live virus vaccines against porcine reproductive and respiratory syndrome virus types 1 and 2 in pigs. Vaccine. 2018;36:227–236. doi: 10.1016/j.vaccine.2017.11.059. [DOI] [PubMed] [Google Scholar]
- 29.Correas I, Osorio FA, Steffen D, Pattnaik AK, Vu HLX. Cross reactivity of immune responses to porcine reproductive and respiratory syndrome virus infection. Vaccine. 2017;35:782–788. doi: 10.1016/j.vaccine.2016.12.040. [DOI] [PubMed] [Google Scholar]
- 30.Vu HLX, Pattnaik AK, Osorio FA. Strategies to broaden the cross-protective efficacy of vaccine against porcine reproductive and respiratory syndrome virus. Vet Microbiol. 2017;206:29–34. doi: 10.1016/j.vetmic.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Charerntantanakul W, Platt R, Johnson W, Roof M, Vaughn E, Roth JA. Immune responses and protection by vaccine and various vaccine adjuvant candidates to virulent porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2006;109:99–115. doi: 10.1016/j.vetimm.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 32.Meier WA, Husmann RJ, Schnitzlein WM, Osorio FA, Lunney JK, Zuckermann FA. Cytokines and synthetic double-stranded RNA augment the T helper 1 immune response of swine to porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2004;102:299–314. doi: 10.1016/j.vetimm.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi M, Lam TT-Y, Hon C-C, et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010;84:8700–8711. doi: 10.1128/JVI.02551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]