Abstract
Background
Non-co-linear (NCL) transcripts consist of exonic sequences that are topologically inconsistent with the reference genome in an intragenic fashion (circular or intragenic trans-spliced RNAs) or in an intergenic fashion (fusion or intergenic trans-spliced RNAs). On the basis of RNA-seq data, numerous NCL event detectors have been developed and detected thousands of NCL events in diverse species. However, there are great discrepancies in the identification results among detectors, indicating a considerable proportion of false positives in the detected NCL events. Although several helpful guidelines for evaluating the performance of NCL event detectors have been provided, a systematic guideline for measurement of NCL events identified by existing tools has not been available.
Results
We develop a software, NCLcomparator, for systematically post-screening the intragenic or intergenic NCL events identified by various NCL detectors. NCLcomparator first examine whether the input NCL events are potentially false positives derived from ambiguous alignments (i.e., the NCL events have an alternative co-linear explanation or multiple matches against the reference genome). To evaluate the reliability of the identified NCL events, we define the NCL score (NCLscore) based on the variation in the number of supporting NCL junction reads identified by the tools examined. Of the input NCL events, we show that the ambiguous alignment-derived events have relatively lower NCLscore values than the other events, indicating that an NCL event with a higher NCLscore has a higher level of reliability. To help selecting highly expressed NCL events, NCLcomparator also provides a series of useful measurements such as the expression levels of the detected NCL events and their corresponding host genes and the junction usage of the co-linear splice junctions at both NCL donor and acceptor sites.
Conclusion
NCLcomparator provides useful guidelines, with the input of identified NCL events from various detectors and the corresponding paired-end RNA-seq data only, to help users selecting potentially high-confidence NCL events for further functional investigation. The software thus helps to facilitate future studies into NCL events, shedding light on the fundamental biology of this important but understudied class of transcripts. NCLcomparator is freely accessible at https://github.com/TreesLab/NCLcomparator.
Electronic supplementary material
The online version of this article (10.1186/s12859-018-2589-0) contains supplementary material, which is available to authorized users.
Keywords: RNA-seq, Non-co-linear RNA, Circular RNA, Trans-spliced RNA, Gene fusion, Alignment ambiguity
Background
Transcriptome-wide analyses of high-throughput RNA sequencing (RNA-seq) have discovered a large amount of ‘non-co-linear’ (NCL) transcripts, in which the exonic sequences are topologically inconsistent with the reference genome in an intragenic fashion (circular or intragenic trans-spliced RNAs) or in an intergenic fashion (fusion or intergenic trans-spliced RNAs) [1–4]. Although NCL transcripts were reported to be generally expressed at a rather low level compared with co-linear mRNAs, some NCL transcripts may be even more highly expressed than their corresponding co-linear isoforms [5] or evolutionarily conserved across species [6]. Accumulating evidence shows their biological importance in gene regulation and disease diagnosis [4, 7–9]. For fusion transcripts, some were demonstrated to correlate with malignant hematological disorders and sarcomas [10–13]. BCR-ABL1, a prominent example of fusion gene, was shown to be important in adult acute lymphoblastic leukemia cases and served as an effective biomarker for chronic myeloid leukemia [14–17]. For trans-spliced RNAs, some may play a role in anti-apoptotic function [3, 18, 19] and prostate cancer [3, 20]. A trans-spliced long non-coding RNA, tsRMST, can regulate pluripotency maintenance of human embryonic stem cells (hESCs) by repressing WNT5A [7, 21]. For circular RNAs (circRNAs), they are ubiquitous and have been observed in diverse species [5, 22–27]. The most famous function of circRNAs is their regulatory role in microRNA sponges [6, 28–32]. In addition, circRNAs can regulate their parent genes [4, 8, 33–35], or play a regulatory role in development [26, 36, 37], the aging nervous system [38], and cancer growth/metastasis [32, 39].
Nowadays, numerous RNA-seq-based NCL event detectors have been developed and employed to identify thousands of NCL transcript candidates in diverse species [40–50]. However, detection of NCL events is still hampered by the potentially false calls arising from sequencing errors, ambiguous alignment, and in vitro artifacts, which leads to great discrepancies in the detection results among tools [4, 51–53]. In addition, the biogenesis and functions of circRNAs and trans-spliced RNAs are mostly unclear. Even if the computationally identified NCL events are in vivo, it remains debatable whether most of them are merely side-products of imperfect pre-mRNA splicing [24, 54]. As accumulating NCL events are detected, the reliability and function of the identified NCL events become an unavoidable question for further investigation. Although several studies have provided helpful guidelines for evaluating the performance of various NCL event-detection tools [1, 4, 51, 55, 56], a systematic guideline for measurement of NCL events identified by different tools has not been available. To reduce the cost of further validation and functional analysis, it is essential to systematically evaluate the reliability of the detected NCL events.
To provide useful guidelines on screening the NCL events identified by various detectors for users, we develop an analysis package, NCLcomparator, for systematic comparisons of the outputs from different detectors. First, for each input NCL event, NCLcomparator concatenates the sequence flanking the NCL junction and then examines whether this NCL event is potentially false positives derived from ambiguous alignments by aligning the concatenated sequence against the reference genome. Next, on the basis of the number of the supporting NCL junction reads derived from the tools compared, NCLcomparator defines the NCL score, NCLscore, to evaluate the reliability of the input NCL events. To help selecting highly expressed NCL events, NCLcomparator provides expression levels of NCL events and their corresponding co-linear host genes and calculates the ratio of the number of reads spanning the NCL junction to that spanning the co-linear splice junctions at both NCL donor and acceptor sites. NCLcomparator further estimates the frequencies of occurrence of the co-linear junctions at the NCL donor and acceptor splicing sites in the host genes to examine the usage of the NCL junctions. NCLcomparator also provides the number of the mapped paired-end read with a read spanning outside the identified intragenic circle, which can be regarded as a good indicator for discrimination between circRNAs and intragenic trans-spliced RNAs [4]. Taken together, NCLcomparator is helpful not only for selecting highly confident and highly expressed NCL events but also for further investigating biogenesis and function of this important but understudied class of transcripts. Of note, NCLcomparator analyzes both intragenic and intergenic NCL events, allowing researchers for comparisons among circRNA detectors and among gene-fusion detectors.
Implementation
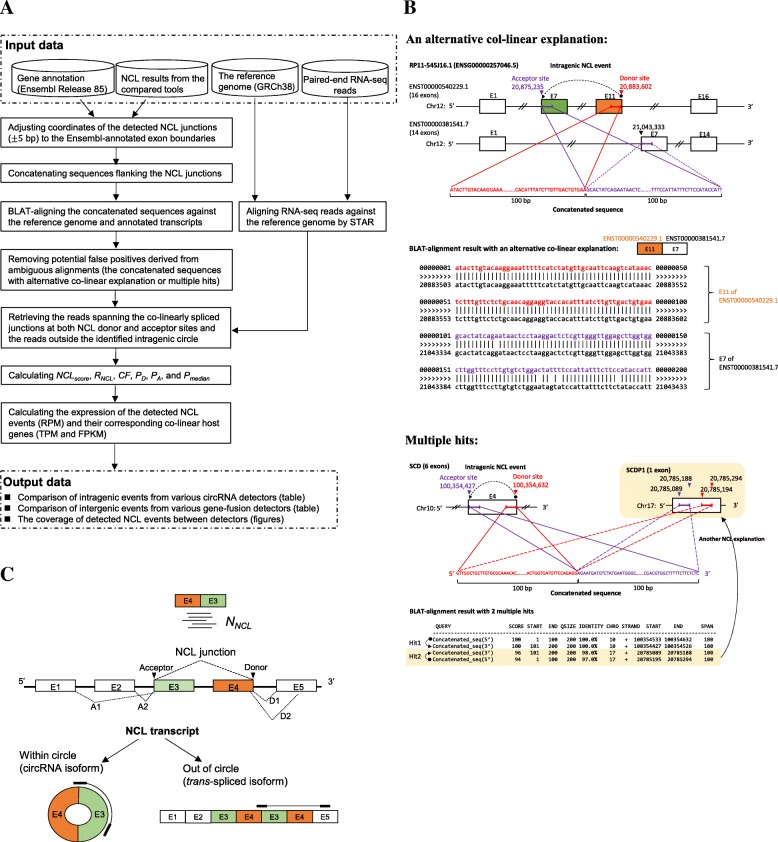
The flowchart of the NCLcomparator pipeline is listed in Fig. 1a. The input data include the identified NCL events from various detectors and the corresponding paired-end RNA-seq data. The input data for each tool should include the coordinates of the detected NCL donor/acceptor sites and the number of reads spanning the NCL junction (NNCL). NCLcomparator only considers the detected NCL events in which splice junctions agree to well-annotated junctions (co-linear) for comparisons for two reasons. First, such events were reported to be more reliable [2, 7, 40, 49] and second, some tools only detect NCL donor/acceptor sites at known co-linear exon boundaries (e.g., NCLscan [1] and UROBORUS [57]). To reduce possible alignment errors around the splice junctions among tools, an NCL event is recorded when the distance between the known co-linear junction and junction identified by the tested tool is equal to or less than 5 bp, in which the coordinates of the NCL junction is adjusted to those of the well-annotated boundary.
Fig. 1.
The NCLcomparator process. a Flowchart of NCLcomparator. b Examples of NCL events derived from ambiguous alignments with an alternative co-linear explanation (top) and multiple hits (bottom). For the top panel, the host gene (RP11-545 J16.1) includes two alternatively spliced transcripts (ENST00000540229.1 with 16 exons and ENST00000381541.7 with 14 exons). The concatenated sequence of the intragenic NCL event (E11-E7; ENST00000540229.1) has an alternative co-linear explanation (E11 of ENST00000540229.1 and E7 of ENST00000381541.7). For the bottom panel, the concatenated sequence of the intragenic NCL event (E4-E4; SCD) mapped to multiple positions (2 hits). c Schematic illustration of the NCL junction and the corresponding co-linear junctions at both NCL donor (D1 and D2) and acceptor (A1 and A2) sites. The mapped read spanning within (circRNA isoform) or outside (trans-spliced isoform) a detected intragenic circle is illustrated. E, exon. NNCL, number of reads spanning the NCL junction
Since repetitive sequences or paralogous genes often masquerade as NCL events due to ambiguous alignments of short RNA-seq reads [1, 25, 58–60], NCLcomparator checks the alignment ambiguity of the input NCL events and removes such potentially false positives. To this end, for each input NCL event, NCLcomparator concatenates the exonic sequence flanking the NCL junction (within − 100 nucleotides to + 100 nucleotides of each NCL junction) and then aligns the 200 bp concatenated sequence against the reference genome and well-annotated transcripts using BLAT [61]. Of note, the concatenated sequence may be shorter than 200 bp if the flanking exonic circRNA sequence is shorter than 200 bp. A concatenated sequence is regarded as false positives derived from ambiguous alignments, if it contains at least an alternative co-linear explanation (the sequence similarity of the alternative co-linear explanation is more than 80% identical to that of the non-co-linear one; Fig. 1b, top) or maps to multiple positions with similar BLAT mapping scores (difference of BLAT-mapping scores < 3; Fig. 1b, bottom).
To extract the reads spanning the co-linearly spliced junctions at both NCL donor and acceptor sites (Fig. 1c) according to the adjusted NCL junctions, the paired-end RNA-seq reads are aligned against the reference genome using STAR with the ‘chimeric alignment’ model [62]. The reads mapping outside the identified intragenic circle are also extracted, which is often employed to distinguish between trans-splicing and circRNA events [4] (Fig. 1c). In addition, to evaluate the variation in NNCL identified by the tools compared, we define τNCL as.
| 1 |
where n is the number of tools compared, NNCL(i) indicates NNCL of the NCL event of interest identified by Tool i, and Max(NNCL) is the highest NNCL of the NCL event across all examined tools. Of note, τNCL of an NCL event is defined as the heterogeneity of its NNCL value provided by the tools compared, which ranges from 0 to 1 with higher τNCL values indicating greater variation (or higher tool-specificity) in NNCL. The measurement of τNCL value is similar to that applied for evaluating sample specificity of DNA methylation level [63]. The NCL score, NCLscore, is then defined as.
| 2 |
where Median(NNCL) is the median NNCL of an NCL event across all examined tools and κ is a pseudocount arbitrarily set as 0.01 to avoid the occurrence of undefined values. A higher NCLscore of an NCL event indicates a greater median NNCL with a smaller variation (τNCL) in NNCL among tools compared, suggesting a higher level of confidence. To quantify the abundance of each detected NCL event as compared with that of its corresponding co-linear isoform(s), we calculate NCL ratio (RNCL) [59] and circular fraction (CF) [24] using NNCL and the number of reads spanning the co-linearly spliced junctions at both NCL donor (ND) and acceptor (NA) sites. RNCL and CF are defined as
| 3 |
respectively. Noteworthily, both RNCL and CF range from 0 to 1, with RNCL > 0.5 or CF > ~ 1/3 indicating a higher expression level in a NCL isoform than in its corresponding co-linear isoform. To quantify the usage of the co-linear junctions at both NCL donor and acceptor splice sites in the corresponding host gene, we also define PD and PA as
| 4 |
respectively. “all co-linear junction reads” means the sum of reads spanning the co-linearly spliced junctions at all well-annotated splice sites in the host gene. For comparison, the median frequency (Pmedian) of occurrence of all well-annotated splice sites (co-linear) in the host gene is also provided. The expression levels of NCL events are determined as the number of supporting reads per million raw reads (RPMraw) or per million uniquely mapped reads (RPMmapped) [64]; those of the corresponding co-linear host gene are estimated by transcripts per million (TPM) and fragments per kilobase of transcript per million mapped reads (FPKM) using RSEM [65]. Since synonymous constraint elements (SCEs) were suggested to be important in RNA secondary structures, RNA splicing, microRNA binding, and nucleosome positioning [66, 67], we also determine whether the NCL donor and acceptor junctions are located within SCEs. The union of the detected NCL events from the compared tools is exported into two tab-delimited text files (intragenic and intergenic results, respectively), in which the related information stated above is included. The figures representing coverage of identified NCL events among the compared tools and distribution of the number of supporting tools are also provided.
Results
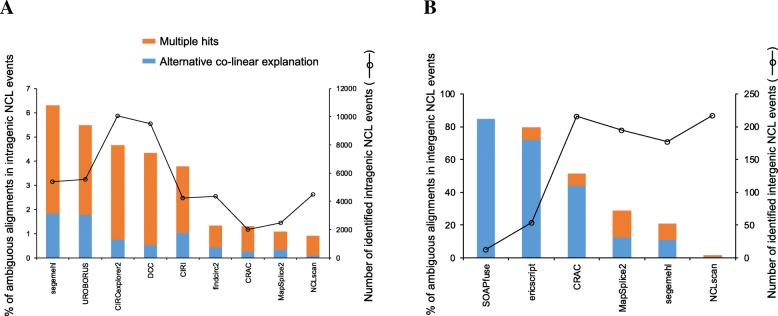
NCLcomparator is applied to an rRNA depleted RNA-seq of HeLa cells (SRR1637089). Nine intragenic and six intergenic NCL detectors are selected and run independently with default parameters or the parameters suggested by the authors (Additional file 1: Table S1). Some tools can simultaneously detect intragenic and intergenic NCL events (e.g., NCLscan, Segemehl [68], and MapSplice [69]). These tools totally identified 17,313 intragenic and 766 intergenic NCL events (i.e., the input NCL events; Additional file 2: Table S2). NCLcomparator first checks the alignment ambiguity of the input NCL events. Of the 17,313 intragenic NCL events, 269 events contain alternative co-linear explanations and 792 events map to multiple positions with similar BLAT mapping scores (Table 1). Of the 766 intergenic NCL events, 184 and 69 events have alternative co-linear explanations and multiple hits, respectively (Table 1). We can find that the proportions of false positives derived from ambiguous alignments vary among the compared tools and are generally higher in intergenic NCL events than in intragenic NCL events (Fig. 2a and b), reflecting previous reports that many intergenic NCL events may arise from false positives (sequencing/alignment errors or in vitro artifacts) [7, 42]. Particularly, more than 50% of intergenic events identified by CRAC, ericscript, and SOAPfuse are derived from ambiguous alignments. Of note, the NCLscan results have the lowest percentages of false positives derived from ambiguous alignments among the results of the compared tools; such percentages are consistently low (~ 1%) in both intragenic and intergenic NCL event detections (Fig. 2a and b). This result consists with previous reports that NCLscan has the highest precision compared with other tools [1, 51]. The NCL events that are determined to be derived from ambiguous alignments (designated as “ambiguous NCL events”) are removed. Therefore, a total of 16,252 intragenic and 513 intergenic events (designated as “non-ambiguous NCL events”) are retained for the following comparisons (Table 1).
Table 1.
The number of intragenic and intergenic NCL events before and after screening
| Number of NCL events | |
|---|---|
| Intragenic NCL events | |
| Before screening | 17,313 |
| Alignment ambiguity (ambiguous NCL events) | 1061 |
| Alternative co-linear explanation | 269 |
| Multiple hit | 792 |
| After screening (non-ambiguous NCL events) | 16,252 |
| Intergenic NCL events | |
| Before screening | 766 |
| Alignment ambiguity (ambiguous NCL events) | 253 |
| Alternative co-linear explanation | 184 |
| Multiple hit | 69 |
| After screening (non-ambiguous NCL events) | 513 |
Fig. 2.
Distributions of (a) intragenic and (b) intergenic NCL events derived from ambiguous alignments with an alternative co-linear explanation and multiple hit(s) among various tools on the rRNA depleted RNA-seq data of HeLa cells (SRR1637089)
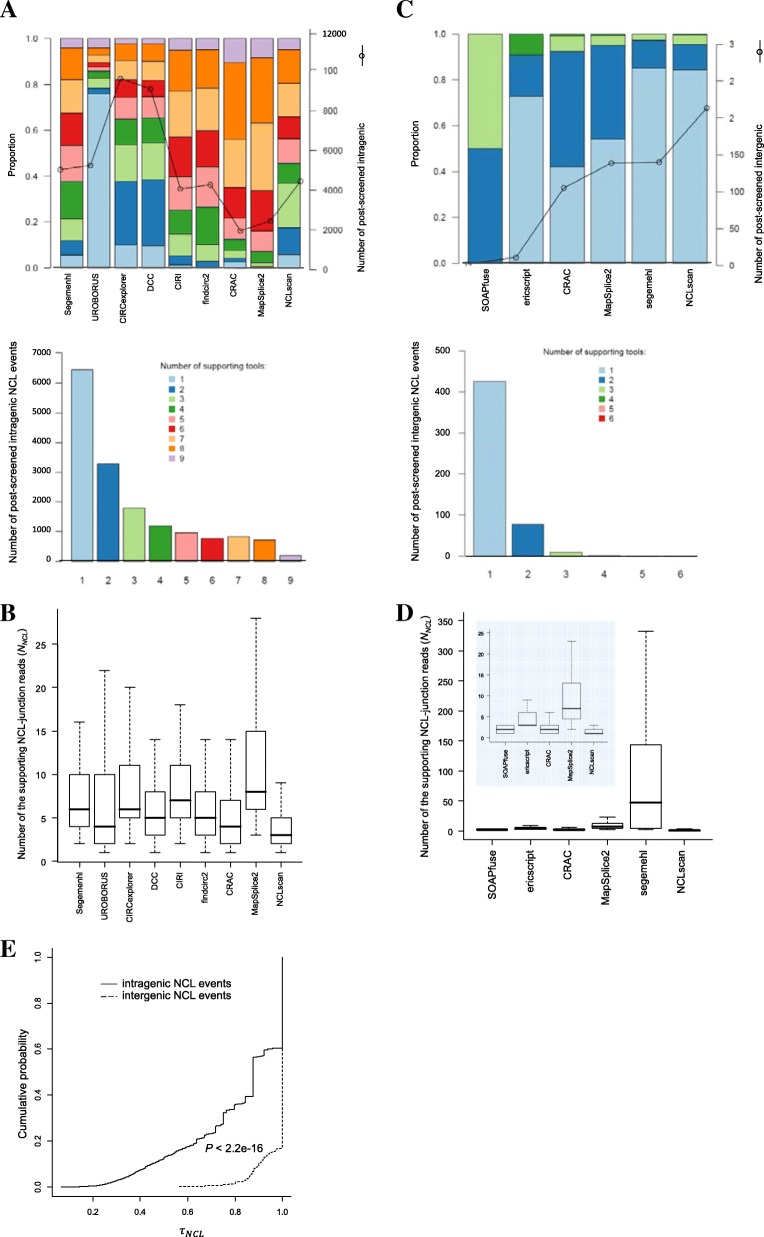
In addition to tab-delimited text files (e.g., Additional file 2: Table S2), NCLcomparator provides figures for comparison of identification results from different tools (see Fig. 3a and b for intragenic events and Fig. 3c and d for intergenic events). Here we take the result of intragenic NCL events as an example. We can find quite a large variation in numbers of detected events among tools; every tool identifies a considerable proportion of tool-specific intragenic NCL events (Fig. 3a, top). More than 40% (6450 events) of the intragenic NCL events are exclusively identified by a single tool (Fig. 3a, bottom). Particularly, even though the intragenic NCL events (205 events) detected by all the nine tools compared, the number of supporting NCL-junction reads (NNCL) vary among the compared tools (Fig. 3b). These observations reflect great discrepancies in the detection results (including the number and NNCL values of identified events) among tools. Such great discrepancies are much more remarkable in intergenic events than in intragenic ones. We can find that except for SOAPfuse and ericscript every tool identifies more than 30% of tool-specific intragenic NCL events (Fig. 3c, top), more than 83% (426 out of 513) of intergenic events are exclusively identified by a single tool (Fig. 3c, bottom), and the NNCL values highly vary among the compared tools (Fig. 3d). In addition, comparisons of the cumulative distribution of τNCLshow that τNCL values are significantly higher in intergenic events than in intragenic ones (P value < 2.2e-16 by the Kolmogorov-Smirnov test; Fig. 3e). These observations highlight the importance of a careful screen for the NCL events, especially for intergenic events, identified by currently available NCL event detectors.
Fig. 3.
Variation in number of detected events and NNCL among various tools. Of note, the analysis is based on the non-ambiguous NCL events. a The coverage of identified intragenic NCL events between the compared tools (top) and the distribution of the number of supporting tools (bottom). b Boxplot representing the number of the supporting NCL-junction reads (NNCL) of the intragenic NCL events (205 events) that are identified by all nine examined circRNA detectors. c and d representing the results of intergenic NCL events identified by 6 gene-fusion detectors, related to the intragenic cases in (a) and (b), respectively. For (d), boxplot represents the 87 intergenic NCL events identified by at least two gene-fusion detectors. A zoom-in view for NNCL of SOAPfuse, ericscript, CRAC, MapSplice2, and NCLscan is shown the middle panel of (d). The identified intragenic and intergenic NCL events by various tools are listed in Additional file 2: Table S2. e Comparisons of cumulative distribution of τNCL for the non-ambiguous intragenic and intergenic NCL events. P value is determined by the Kolmogorov-Smirnov test
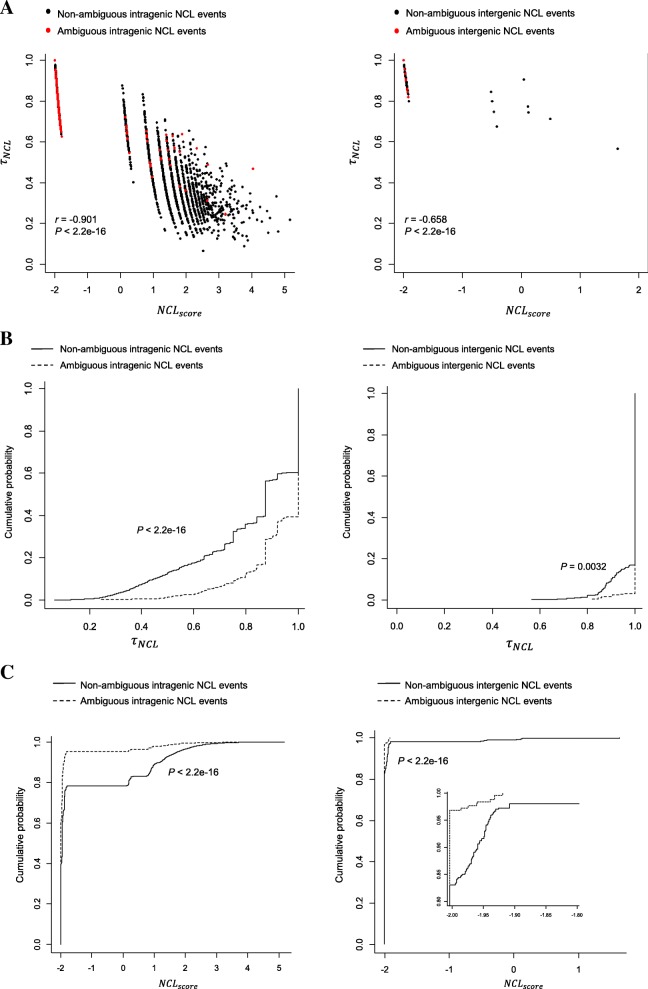
Importantly, NCLcomparator provides two measurements, τNCL and NCLscore (see Eqs. (1) and (2), respectively) to help users selecting potentially high- plausible NCL events. Since NNCL values vary remarkably between tools depending on the level of strictness of the filtering steps used [51, 60] (see also Fig. 3b and d), we speculate that high-confidence NCL events tend to have a large NCLscore (in other words, a large median NNCL with a small τNCL). Since ambiguous NCL events can be regarded as false positives, we examine τNCLand NCLscore values of ambiguous and non-ambiguous NCL events. Indeed, we find a significantly negative correlation between these two measurements, in which τNCL values are significantly higher in ambiguous NCL events than in non-ambiguous ones (Fig. 4a and b), whereas the reverse trends are observed for NCLscore values (Fig. 4a and c), regardless of whether the events are intragenic or intergenic (both P values < 2.2e-16 by the Kolmogorov-Smirnov test). For example, for τNCLτNCL, more than 95% of ambiguous events are filtered out, if we set the thresholds as < 0.6 and < 1 for intragenic and intergenic events, respectively (Fig. 4b). Meanwhile, for NCLscore, more than 95% of ambiguous events are removed, if we set the thresholds as > − 1 and > − 2 for intragenic and intergenic events, respectively (Fig. 4c). These results reveal that NCLscore is a good indicator for selecting NCL events with high level of confidence, suggesting that NCL events with a large NCLscore may be of high reliability and considered to take priority over the other for further experimental validation and functional analysis.
Fig. 4.
Comparisons of τNCL and NCLscore between ambiguous and non-ambiguous NCL events. a Correlation between τNCL and NCLscore for intragenic (left) and intergenic (right) NCL events. The black and red dots represent non-ambiguous and ambiguous NCL events, respectively. The correlation coefficient r and P values are determined by Pearson’s r test. b and c Comparisons of cumulative distribution of (b) τNCL and (c) NCLscore for the ambiguous and non-ambiguous NCL events in intragenic (left) and intergenic (right) detections. In (c), a zoom-in view is shown in the lower right panel. P values are determined by the Kolmogorov-Smirnov test
Discussion
There are several major challenges for detection of NCL events. In addition to false positives arising from alignment ambiguity and biased identification of NCL events from different bioinformatics approaches as stated above, identification of NCL events is often hampered by in vitro artifacts, particularly template switching during reverse transcription (RT) [2, 7, 42, 45, 59, 70, 71]. Actually, to minimize potential RT-artifacts, it would be better to confirm identified NCL events using both RT- and non-RT-based experiments (e.g., Northern blot or RNase protection assay [72]). However, it is required to develop a method for systematic identification of NCL events with controlling for experimental artifacts. While a study successfully detected a huge number of experimental artifacts based on Drosophila hybrid mRNAs (D. melanogaster females vs. D. sechellia males) and a mixed mRNA-negative control sample [42], this approach would not be applied to human studies. Alternatively, it has been demonstrated that RTase-dependent RNA products are likely to be RT artifacts [2, 4, 7, 73, 74]. RT-based artifacts can be detected by comparisons of different RTase products, which was shown to serve as effectively as RTase-free validation [2, 7]. On the basis of comparisons of Avian Myeloblastosis Virus- and Moloney Murine Leukemia Virus-derived RTase products, a recent study successfully applied this concept to human samples and systematically identified NCL events with controlling for experimental artifacts [59].
Moreover, NCL junctions can be generated during post-transcriptional processes (trans-spliced or circular RNAs) or by genetic rearrangements (fusion RNAs) at the DNA level. Thus, discrimination between post-transcriptional NCL events and genetic rearrangements presents another challenge to detection/analysis of NCL events. Since NCL events that are observed in multiple biological samples or conserved across multiple species are less likely to be formed by somatic recombination, post-transcriptional NCL events may be extracted by this simple rule [2, 7]. A more efficient approach is to analyze both RNA-seq data and whole genome sequencing (WGS) data from the same sample. Some systematic pipelines have been developed, which integrated WGS-based rearrangement detection with RNA-seq-based NCL detection to identify fusion RNAs, and successfully applied to analysis of functionally recurrent gene fusions in human diseases [75–80]. While many studies have focused on identification/analysis of fusion RNAs that consist of sequence fragments from different genes, transcribed rearrangements in an intragenic fashion is relatively less investigated.
With more and more NCL events are identified, the reliability and function of such a large number of NCL events remains an open question worthy of further investigation. To reduce the cost of subsequent validation and functional analysis, carefully evaluating the reliability of detected NCL events with considering all abovementioned challenges awaits further development.
Conclusion
Dozens of RNA-seq-based detectors have been developed and successfully identify thousands of NCL transcript candidates (circular, trans-spliced, or fusion RNA) in diverse species. However, there are great discrepancies in the identification results (including the number of NCL events and the number of the supporting NCL junction reads of the identified events) among tools, indicating a considerable proportion of potentially false positives in the results. NCLcomparator screens out potentially false positives originating from ambiguous alignments and provides a series of useful measurements, including NCL score (NCLscore), NCL ratio (RNCL), circular fraction (CF), the usage of the co-linear junctions at both NCL donor and acceptor splice sites in the corresponding host gene (PD, PA, and Pmedian), and the expression levels of NCL events (RPMraw and RPMmapped) and their corresponding co-linear host genes (FPKM and TPM), for users to screen the NCL events from various detectors. On the basis of the NCLcomparator-provided information, users can easily select potentially high-plausible NCL candidates with a high expression level and/or a low variation of supporting NCL junction reads from multiple NCL detectors. The software, a post-processing tool for screening identified NCL events from existing detectors, thus help to facilitate future studies into NCL events, shedding light on the fundamental biology of this important but understudied class of transcripts.
Availability and requirements
Project name: NCLcomparator.
Project home page: https://github.com/TreesLab/NCLcomparator
Operator system(s): Linux-like environment (Bio-Linux).
Programming language: shell script.
Other requirement: None.
License: None.
Any restrictions to use by non-academics: None.
Data: The tested RNA-seq data was derived from HeLa cells with rRNA depletion, which was downloaded from the NCBI Sequence Read Archive (SRR1637089) at https://trace.ddbj.nig.ac.jp/DRASearch/run?acc=SRR1637089. All parameter settings and identification results of intragenic/intergenic NCL detectors tested in this study are reported in Additional file 1: Table S1 and Additional file 2: Table S2, respectively.
Additional files
Table S1. Parameter settings of intragenic/intergenic NCL detectors tested in this study. (DOCX 34 kb)
Table S2. The totally identified 17,313 intragenic and 766 intergenic NCL events by the 9 intragenic and 6 intergenic NCL detectors on the RNA-seq data of HeLa cells (XLSX 6083 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by grants of the Genomics Research Center, Academia Sinica, Taiwan and the Ministry of Science and Technology (MOST), Taiwan (under the contract MOST 103–2628-B-001-001-MY4 and MOST 107–2311-B-001-046). The funding body did not play any role in the study design and collection, analysis and interpretation of the data and the write-up of the manuscript.
Availability of data and materials
The implementation of NCLcomparator software package, source code, and test data sets are available at https://github.com/TreesLab/NCLcomparator.
Abbreviations
- circRNA
Circular RNA
- FPKM
Fragments per kilobase of transcript per million mapped reads
- NCL
Non-co-linear
- RNA-seq
RNA sequencing
- SCEs
Synonymous constraint elements
- TPM
Transcripts per million
- WGS
Whole genome sequencing
Authors’ contributions
TJC designed the research and wrote the manuscript. CYC implemented the software and conducted the case studies. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chia-Ying Chen, Email: cychen0704@gate.sinica.edu.tw.
Trees-Juen Chuang, Email: trees@gate.sinica.edu.tw.
References
- 1.Chuang TJ, Wu CS, Chen CY, Hung LY, Chiang TW, Yang MY. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44(3):e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu CY, Liu HJ, Hung LY, Kuo HC, Chuang TJ. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res. 2014;42(14):9410–9423. doi: 10.1093/nar/gku643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gingeras TR. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461(7261):206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6(5):563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Wu CS, Yu CY, Chuang CY, Hsiao M, Kao CF, Kuo HC, Chuang TJ. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24(1):25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 9.Chwalenia K, Facemire L, Li H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip Rev RNA. 2017;8(6):e1427. [DOI] [PubMed]
- 10.Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315(6020):550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 11.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36(4):331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 12.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 13.Frohling S, Dohner H. Chromosomal abnormalities in cancer. N Engl J Med. 2008;359(7):722–734. doi: 10.1056/NEJMra0803109. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, Cornelissen JJ, Fischer T, Hochhaus A, Hughes T, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 15.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 16.Tkachuk DC, Westbrook CA, Andreeff M, Donlon TA, Cleary ML, Suryanarayan K, Homge M, Redner A, Gray J, Pinkel D. Detection of bcr-abl fusion in chronic myelogeneous leukemia by in situ hybridization. Science. 1990;250(4980):559–562. doi: 10.1126/science.2237408. [DOI] [PubMed] [Google Scholar]
- 17.Westbrook CA, Hooberman AL, Spino C, Dodge RK, Larson RA, Davey F, Wurster-Hill DH, Sobol RE, Schiffer C, Bloomfield CD. Clinical significance of the BCR-ABL fusion gene in adult acute lymphoblastic leukemia: a Cancer and leukemia group B study (8762) Blood. 1992;80(12):2983–2990. [PubMed] [Google Scholar]
- 18.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321(5894):1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 19.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20(2):127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69(7):2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu CY, Kuo HC. The trans-spliced long noncoding RNA tsRMST impedes human embryonic stem cell differentiation through WNT5A-mediated inhibition of the epithelial-to-mesenchymal transition. Stem Cells. 2016;34(8):2052–2062. doi: 10.1002/stem.2386. [DOI] [PubMed] [Google Scholar]
- 22.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9(3):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16(1):4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44(3):1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng J, Liu X, Xue Y, Gong W, Ma J, Xi Z, Que Z, Liu Y. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35(30):3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 32.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Non-coding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 37.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu M, Xu Y, Chen Y, Yan F, Circular BANP. An upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 40.Kim P, Yoon S, Kim N, Lee S, Ko M, Lee H, Kang H, Kim J. ChimerDB 2.0--a knowledgebase for fusion genes updated. Nucleic Acids Res. 2010;38(Database issue):D81–D85. doi: 10.1093/nar/gkp982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha KC, Lalonde E, Li L, Cavallone L, Natrajan R, Lambros MB, Mitsopoulos C, Hakas J, Kozarewa I, Fenwick K, et al. Identification of gene fusion transcripts by transcriptome sequencing in BRCA1-mutated breast cancers and cell lines. BMC Med Genet. 2011;4:75. doi: 10.1186/1755-8794-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McManus CJ, Duff MO, Eipper-Mains J, Graveley BR. Global analysis of trans-splicing in Drosophila. Proc Natl Acad Sci U S A. 2010;107(29):12975–12979. doi: 10.1073/pnas.1007586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20(5):646–654. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc Natl Acad Sci U S A. 2009;106(6):1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106(30):12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma L, Yang S, Zhao W, Tang Z, Zhang T, Li K. Identification and analysis of pig chimeric mRNAs using RNA sequencing data. BMC Genomics. 2012;13:429. doi: 10.1186/1471-2164-13-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inaki K, Hillmer AM, Ukil L, Yao F, Woo XY, Vardy LA, Zawack KF, Lee CW, Ariyaratne PN, Chan YS, et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011;21(5):676–687. doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Balool HH, Weber D, Liu Y, Wade M, Guleria K, Nam PL, Clayton J, Rowe W, Coxhead J, Irving J, et al. Post-transcriptional exon shuffling events in humans can be evolutionarily conserved and abundant. Genome Res. 2011;21(11):1788–1799. doi: 10.1101/gr.116442.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13(6):e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abate F, Acquaviva A, Paciello G, Foti C, Ficarra E, Ferrarini A, Delledonne M, Iacobucci I, Soverini S, Martinelli G, et al. Bellerophontes: an RNA-Seq data analysis framework for chimeric transcripts discovery based on accurate fusion model. Bioinformatics. 2012;28(16):2114–2121. doi: 10.1093/bioinformatics/bts334. [DOI] [PubMed] [Google Scholar]
- 53.Carrara M, Beccuti M, Cavallo F, Donatelli S, Lazzarato F, Cordero F, Calogero RA. State of art fusion-finder algorithms are suitable to detect transcription-induced chimeras in normal tissues? BMC Bioinformatics. 2013;14(Suppl 7):S2. doi: 10.1186/1471-2105-14-S7-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB Journal. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 55.Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44(6):e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci Rep. 2016;6:21597. doi: 10.1038/srep21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, Yu Y, Zhu D, Nickerson ML, Wan S, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14(2):R12. doi: 10.1186/gb-2013-14-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chuang TJ, Chen YJ, Chen CY, Mai TL, Wang YD, Yeh CS, Yang MY, Hsiao YT, Chang TH, Kuo TC, et al. Integrative transcriptome sequencing reveals extensive alternative trans-splicing and cis-backsplicing in human cells. Nucleic Acids Res. 2018;46(7):3671–3691. doi: 10.1093/nar/gky032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen C-Y, Chuang T-J. Comment on A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comp Biol. 2018; in press. [DOI] [PMC free article] [PubMed]
- 61.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chuang TJ, Chen FC, Chen YZ. Position-dependent correlations between DNA methylation and the evolutionary rates of mammalian coding exons. Proc Natl Acad Sci U S A. 2012;109(39):15841–15846. doi: 10.1073/pnas.1208214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478(7370):476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin MF, Kheradpour P, Washietl S, Parker BJ, Pedersen JS, Kellis M. Locating protein-coding sequences under selection for additional, overlapping functions in 29 mammalian genomes. Genome Res. 2011;21(11):1916–1928. doi: 10.1101/gr.108753.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermuller J, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15(2):R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shao X, Shepelev V, Fedorov A. Bioinformatic analysis of exon repetition, exon scrambling and trans-splicing in humans. Bioinformatics. 2006;22(6):692–698. doi: 10.1093/bioinformatics/bti795. [DOI] [PubMed] [Google Scholar]
- 71.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Djebali S, Lagarde J, Kapranov P, Lacroix V, Borel C, Mudge JM, Howald C, Foissac S, Ucla C, Chrast J, et al. Evidence for transcript networks composed of chimeric RNAs in human cells. PLoS One. 2012;7(1):e28213. doi: 10.1371/journal.pone.0028213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Houseley J, Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5(8):e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong Y, Zhou H, Yu Y, Chen L, Hao P, Li X. The evolutionary landscape of intergenic trans-splicing events in insects. Nat Commun. 2015;6:8734. doi: 10.1038/ncomms9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McPherson A, Wu C, Hajirasouliha I, Hormozdiari F, Hach F, Lapuk A, Volik S, Shah S, Collins C, Sahinalp SC. Comrad: detection of expressed rearrangements by integrated analysis of RNA-Seq and low coverage genome sequence data. Bioinformatics. 2011;27(11):1481–1488. doi: 10.1093/bioinformatics/btr184. [DOI] [PubMed] [Google Scholar]
- 76.Wang Q, Xia J, Jia P, Pao W, Zhao Z. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief Bioinform. 2013;14(4):506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McPherson A, Wu C, Wyatt AW, Shah S, Collins C, Sahinalp SC. nFuse: discovery of complex genomic rearrangements in cancer using high-throughput sequencing. Genome Res. 2012;22(11):2250–2261. doi: 10.1101/gr.136572.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen K, Navin NE, Wang Y, Schmidt HK, Wallis JW, Niu B, Fan X, Zhao H, McLellan MD, Hoadley KA, et al. BreakTrans: uncovering the genomic architecture of gene fusions. Genome Biol. 2013;14(8):R87. doi: 10.1186/gb-2013-14-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, White NM, Schmidt HK, Fulton RS, Tomlinson C, Warren WC, Wilson RK, Maher CA. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res. 2016;26(1):108–118. doi: 10.1101/gr.186114.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma C, Shao M, Kingsford C. SQUID: transcriptomic structural variation detection from RNA-seq. Genome Biol. 2018;19(1):52. doi: 10.1186/s13059-018-1421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Parameter settings of intragenic/intergenic NCL detectors tested in this study. (DOCX 34 kb)
Table S2. The totally identified 17,313 intragenic and 766 intergenic NCL events by the 9 intragenic and 6 intergenic NCL detectors on the RNA-seq data of HeLa cells (XLSX 6083 kb)
Data Availability Statement
The implementation of NCLcomparator software package, source code, and test data sets are available at https://github.com/TreesLab/NCLcomparator.