Abstract
Background
For a long time, the Christie-Atkinson-Munch-Peterson (CAMP) test has been a standard test for the identification of Streptococcus agalactiae, and a positive result for S.agalactiae has been considered sensitive enough.
Methods
To confirm whether a positive CAMP test is a requirement for the identification of S.agalactiae, five suspected CAMP-negative S.agalactiae isolates from two hospitals, confirmed as Gram-positive and catalase-negative streptococci, were verified by the CAMP test in three batches of plates from two manufacturers and identified by the Phoenix system, MALDI-TOF MS, the PCR assay and the 16S rDNA gene sequencing.
Results
All five suspected strains were S.agalactiae, four of which were CAMP-negative and one of which was not S.agalactiae by the PCR assay.
Conclusions
A positive CAMP test was lacking sensitivity for the identification of S.agalactiae, and the question of whether the cfb gene is worthy of targeting should be further studied.
Keywords: CAMP test, Streptococcus agalactiae, PCR, 16S rDNA gene sequencing
Background
Streptococcus agalactiae, also known as Group B Streptococcus (GBS), causing intrapartum and postpartum infection, is the most common cause of neonatal sepsis, and has a high mortality rate [1–4]. In view of the dangers of GBS, prenatal GBS screening of pregnant women at 35 to 37 weeks’ gestation has been recommended by the competent departments of health in many countries [5–8]. The CAMP (Christie, Atkinson, Munch, Peterson) test is used in some laboratories to verify whether bacteria have enhanced staphylococcus beta-lysis activity test, which has long been considered as a key, confirmed test for the identification of GBS [9–12]. Some PCR-based assays for GBS have targeted the cfb gene, which encodes the CAMP factor [13, 14].
However, a positive CAMP test for GBS seemed to be lacking sensitivity. From the beginning of the twenty-first century, several studies have isolated the CAMP-negative GBS [15, 16], but it did not attract much attention from scholars in clinical medicine. Those academic achievements were generally derived from the same team; furthermore, all of those studies were performed on bovine mastitis. Until 2016, CAMP-negative GBS was only reported to be found in clinical laboratories [17]. In this paper we present evidence CAMP-negative GBS did exist, and not just in low proportions.
Methods
GBS isolates
Four isolates of suspected GBS and CAMP-negative strains, sequentially numbered 50, 51, 53 and 54, were obtained from the vaginal swabs of 174 patients admitted to the Shenzhen Maternity & Child Healthcare Hospital by using the GBS chromogenic agar plates and the blood agar plates (BAPs) (Autobio Diagnostics, Zhengzhou, China) in October 2016, where 22 samples of suspected GBS were isolated. Another isolate of suspected GBS and CAMP-negative origin, numbered 49, was obtained from 286 patients admitted to the Zhengzhou Maternity & Child Healthcare Hospital in November 2016, where 27 samples of suspected GBS were isolated. All five suspected strains were identified as Gram-positive and catalase-negative streptococci.
CAMP test
BAPs used in the study were derived from two manufacturers in three batches. Batch numbers 20161228B-Y, 20161104B and 20161117B came from Autobio Diagnostics (Zhengzhou, China), and batch numbers XWO1601811, XWO1601812 and XWO1601838 came from OXOID (Beijing, China). Staphylococcus aureus (ATCC25923) was streaked in the middle of the BAPs, and the lines of each of the suspected strains, as well as the QC strains, were streaked perpendicularly to S.aureus. GBS (ATCC13813) and Streptococcus pyogenes (ATCC19615) were used as positive and negative QC, respectively. The distance between two lines perpendicular to each other was approximately 1–2 mm. The plates were incubated aerobically at 35–37 °C for 18–24 h after inoculation. An arrow-head shaped zone of beta-haemolysis at the junction of perpendicular lines constituted a positive CAMP reaction.
Bacterial identification by four assays
To confirm whether the five strains were GBS, bacterial identification was carried out by the following four assays: the biochemical identification system, the matrix-assisted laser desorption ionisation time-of-flight mass spectrometry (MALDI-TOF MS), the PCR assay and the 16S rDNA gene sequencing.
The Phoenix™—100 ID/AST system (Becton Dickinson, Sparks, USA) was adopted as a biochemical identification system, and isolates activated on BAPs (Autobio Diagnostics, Zhengzhou, China) were inoculated in a 0.5 McFarland suspension into the SMIC/ID-2 cards.
MALDI-TOF MS analysis of activated isolates was performed on a Bruker Microflex (Bruker Daltonics, Bremen, Germany) using the Flexcontrol software (version 3.4). A colony was directly spotted on a 96-spot polished steel target plate in duplicates, and was pipetted in 1 μl of matrix solution (ultra pure water: 475 μl, formic acid: 25 μl, acetonitrile: 500 μl, α-cyano-4-hydroxycinnamic acid: 10 mg) was added after drying. Then, the target plate was dried again and loaded on the instrument, measured and analysed. An Escherichia coli DH5-α standard (Bruker Daltonics, Bremen, Germany) was used for external calibration, and the identification results were presented in the MALDI biotyper 3.0 as the log score values.
The PCR assay was performed using the Group B Streptococcus Nucleic Acid Detection Kit (Fluorescent PCR, Lot: 241611, Triplex, Fuzhou, China) that targets the cfb gene, in strict accordance with the manufacturer’s instruction. The concentration of activated isolates used for analysis was 104 CFU/ml.
The genomic DNA was isolated and purified from cells after activation by using SK 8255 (Sangon Biotech, Shanghai, China), a genomic extraction kit, as per the manufacturer’s instructions. DNA 16S region amplification was performed using the primer set 7F-1540R. The 16S rDNA gene (~ 1500 bp) was amplified employing universal primers (7F 5’-CAGAGTTTGATCCTGGCT-3′, 1540R 5’-AGGAGGTGATCCAGCCGCA-3′). Amplification was carried out on the Applied Biosystems 2720 thermal cycler (ThermoFisher, Foster City, USA) according to the manufacturer’s recommendations. The amplified fragment was purified from the agarose gel with SK8131 (Sangon Biotech, Shanghai, China), the gel extraction kit, as per the manufacturer’s instructions. The amplified product was sequenced by a standard procedure on the Applied Biosystems 3730 sequencer (ThermoFisher, Foster City, USA). The obtained 16S rDNA sequence data were aligned and compared with similar sequences from the GenBank database of NCBI using the BLAST program.
GBS (ATCC13813) was used as positive QC, S.pyogenes (ATCC19615) and S.aureus (ATCC25923) were included as negative QC as well in these four assays.
Results
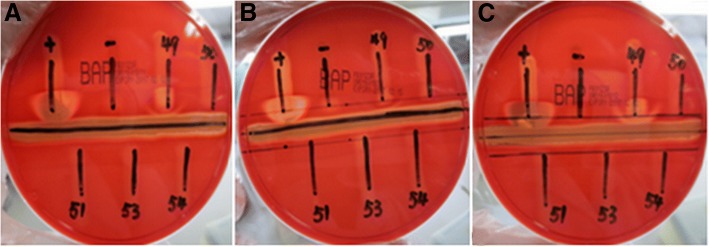
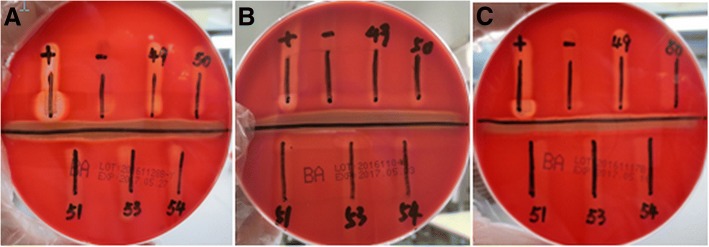
Four suspected strains numbered 50, 51, 53 and 54, were clearly demonstrated to be CAMP-negative on six different batches of BAPs from two different manufacturers, but the results of strain No.49 differed (Fig. 1 and Fig. 2). This isolate was CAMP-negative on two batches of BAPs from Autobio and CAMP-positive on the third batch (20161128B-Y), while all three batches of OXOID, demonstrated an extremely weak or obvious arrow-head shape of the zone of enhanced haemolytic activity.
Fig. 1.
The CAMP test of five suspected strains on three batches of BPAs from OXOID (A: XWO1601811, only No.49 was weakly positive, B: XWO1601838, only No.49 was almost no positive, C: XWO1601812, only No.49 was positive)
Fig. 2.
The CAMP test of five suspected strains on three batches of BPAs from Autobio (A: 20161228B-Y, B: 20161104B, C: 20161117B, only No.49 was positive on BPAs of batch number 20161228B-Y)
Strain No.51 could not be identified as GBS by the PCR assay only, however, all other four suspected strains were GBS by all four methods. Strain No.51 was identified as GBS by all the other three methods (Table 1). Compared with six similar 16S rDNA sequences (Accessions numbers: CP016501, CP010875, CP010874, CP010319, CP011327 and CP011326) from GenBank, the sequence of No.51 had the ident of 99% with all six sequences. However, all the other four strains had the ident of 100%.
Table 1.
ID results of five suspected strains by four methods
| Strain numbers | Biochemical identification | Fluorescent PCR | MALDI-TOF MS | 16S rDNA identification | ||||
|---|---|---|---|---|---|---|---|---|
| Confidence (%) | ID results | Ct values* | ID results | Log score values | ID results | Identity (%) | ID results | |
| 49 | 99 | GBS | 23 (positive) | GBS | 2.221 | GBS | 100(6)** | GBS |
| 50 | 98 | GBS | 28 (grey zone) | GBS | 2.478 | GBS | 100(6) | GBS |
| 51 | 98 | GBS | No Ct | No GBS | 2.317 | GBS | 99(6) | GBS |
| 53 | 99 | GBS | 27 (grey zone) | GBS | 2.236 | GBS | 100(6) | GBS |
| 54 | 99 | GBS | 27 (grey zone) | GBS | 2.169 | GBS | 100(6) | GBS |
| ATCC 13813 | 99 | GBS | 23 (positive) | GBS | 2.436 | GBS | 100(5) & 99(1) | GBS |
| ATCC 19615 | 99 | S.pyogenes | No Ct | No GBS | 2.382 | S.pyogenes | 100(6) | S.pyogenes |
| ATCC 25923 | 0 | UNIORG*** | No Ct | No GBS | 2.357 | S.aureus | 100(6) | S.aureus |
*Positive control: < 23, Negative control: =30 or No Ct
**Figures in brackets denote the number of similar sequences
***SMIC/ID-2 cards are dedicated to the identification of streptococcus and cannot identify staphylococci
Discussion
Many diagnostic guidelines still recommend using a positive CAMP test as a significant condition for identification of GBS, so the resulting problem of failing to detect some GBS is given insufficient attention. The result of the CAMP test may be affected by many factors, such as culture conditions, culture time, and culture temperature [18, 19], but once these conditions are standardised, the most likely factor is the quality of the BPAs. Ensuring strict quality control and using multiple batches of products from different manufacturers to eliminate the effects of medium and inoculation, the detection rate of 18.18% (4/22) in Shenzhen suggest that CAMP-negative GBS not only existed in some parts of China but also were detected in large numbers. This proportion was higher than that reported in the literature [20]. To confirm these findings, additional studies with expanded sample sizes are needed.
The PCR kit used in this study targeted the cfb gene. To eliminate the false negatives due to insufficient sensitivity of the kit, we increased the concentration of bacterial suspensions to four McFarland; however, the sample No.51 was still negative in these conditions. These results suggest that at least some CAMP-negative GBS may not carry the cfb gene, which is different from the traditional view that the cfb gene is present in every GBS isolate [21, 22]. Strains with the cfb gene may also produce the CAMP-negative phenotype because the cfb gene is transcriptionally defective [23], gene expression is low, or the activity of the CAMP factor expression is low. The CAMP factor of GBS is a virulent protein with a complete sequence of 226 amino acids [24], but it is not essential for the systemic virulence of GBS [25], and some studies propose the existence of a different CAMP factor, named CAMP factor II [26].
In recent years, the culture-free GBS detection techniques have developed rapidly [27–30]. Since the CAMP factor can bind in a non-immune reaction to the IgG and IgM antibody classes in various mammalian species [31], it is not a suitable antigen for immunoassays. However, the cfb gene is the most commonly used target for PCR assays. Our research suggest that targeting the cfb gene or the CAMP factor results in missing CAMP-negative GBS. Since CAMP-negative GBS do exist, the question of whether the cfb gene is worthy of targeting should be further studied, especially if the number of these GBS is relatively high.
Conclusions
A positive CAMP test was lacking sensitivity for the identification of GBS, and the question of whether the cfb gene is worthy of targeting should be further studied.
Acknowledgements
The authors would like to thank Autobio Diagnostics Co. Ltd. for its convenience.
Funding
This research was not specifically funded by grants and another funding agency. The study was conducted using the existing facilities of Autobio Diagnostics Co. Ltd.
Availability of data and materials
The datasets analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- BAPs
blood agar plates
- CAMP
Christie-Atkinson-Munch-Peterson
- GBS
Group B Streptococcus
- MALDI-TOF MS
matrix-assisted laser desorption ionisation time-of-flight mass spectrometry
Authors’ contributions
DCG conceived of the study, designed all the experiments and was a major contributor in writing the manuscript. SMW provided the isolates and the information of the isolates. DCG, YX and SMW perfomed the experiments, analysed and interpreted data. ZYW provided valuable insight for designing the study and revising the manuscript. All authors read final version of the manuscript and approved its submission.
Ethics approval and consent to participate
No formal ethics approval was required in the study and informed written consent was waived by the Ethics Committee of Henan Provincial Center for Disease Control and Prevention, because the study is only about five clinical isolates of in-depth study, which does not involve in the human body, human specimens, or personal information.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dacheng Guo, Email: guodc@hncdc.com.cn.
Yu Xi, Email: microxy@126.com.
Shanmei Wang, Email: wsm1997@sina.com.
Zeyu Wang, Email: pine8844@126.com.
References
- 1.Schuchat A. Group B streptococcus. Lancet. 1999;353(9146):51–56. doi: 10.1016/S0140-6736(98)07128-1. [DOI] [PubMed] [Google Scholar]
- 2.Heyderman RS, Madhi SA, Slobod K, Clemens SAC. Variation in reported neonatal group B streptococcal disease incidence in developing countries. Clin Infect Dis. 2012;55(1):91–102. doi: 10.1093/cid/cis395. [DOI] [PubMed] [Google Scholar]
- 3.Camacho-Gonzalez A, Spearman PW, Stoll BJ. Neonatal infectious diseases. evaluation of neonatal sepsis Pediatr Clin North Am. 2013;60(2):367–389. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker CJ, Barrett FF. Group B streptococcal infections in infants. The importance of the various serotypes. JAMA. 1974;230(8):1158–1160. doi: 10.1001/jama.1974.03240080040025. [DOI] [PubMed] [Google Scholar]
- 5.The Canadian Task Force on Preventive Health Care Prevention of group B streptococcal infection in newborns: recommendation statement from the Canadian task force on preventive health care. CMAJ. 2002;166(7):928–930. [PMC free article] [PubMed] [Google Scholar]
- 6.Verani JR, Mc Gee L, Schrag SJ. Division of bacterial diseases National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention (CDC). Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recomm Rep. 2010;59(RR-10):1–36. [PubMed] [Google Scholar]
- 7.Mahieu L, Langhendries JP, Cossey V, De Praeter C, Lepage P, Melin P. Management of the neonate at risk.for early-onset Group B streptococcal disease (GBS EOD): new paediatric guidelines in Belgium. Acta Clin Belg. 2014;69(5):313–319. doi: 10.1179/2295333714Y.0000000054. [DOI] [PubMed] [Google Scholar]
- 8.Di Renzo GC, Melin P, Berardi A, Blennow M, Carbonell-Estrany X, Donzelli GP, Hakansson S, Hod M, Hughes R, Kurtzer M, et al. Intrapartum GBS screening and antibiotic prophylaxis: a European consensus conference. J Matern Fetal Neonatal Med. 2015;28(7):766–82. [DOI] [PubMed]
- 9.Wilkinson HW. CAMP-disk test for presumptive identification of group B streptococci. J Clin Microbiol. 1977;6(1):42–45. doi: 10.1128/jcm.6.1.42-45.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae BH, Bottone EJ. Modified Christie-Atkins-munch-Petersen (CAMP) test for direct identification of hemolytic and nonhemolytic group B streptococci on primary plating. Can J Microbiol. 1980;26(4):539–542. doi: 10.1139/m80-091. [DOI] [PubMed] [Google Scholar]
- 11.El Aila NA, Tency I, Claeys G, Saerens B, De Backer E, Temmerman M, Verhelst R, Vaneechoutte M. Genotyping of Streptococcus agalactiae (group B streptococci) isolated from vaginal and rectal swabs of women at 35-37 weeks of pregnancy. BMC Infect Dis. 2009;9:153. doi: 10.1186/1471-2334-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Aila NA, Tency I, Claeys G, Saerens B, Cools P, Verstraelen H, Temmerman M, Verhelst R, Vaneechoutte M. Comparison of different sampling techniques and of different culture methods for detection of group B streptococcus carriage in pregnant women. BMC Infect Dis. 2010;10:285. doi: 10.1186/1471-2334-10-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke D, Menard C, Picard FJ, Boissinot M, Ouellette M, Roy PH, Bergeron MG. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46(3):324–331. [PubMed] [Google Scholar]
- 14.Buchan BW, Faron ML, Fuller D, Davis TE, Mayne D, Ledeboer NA. Multicenter clinical evaluation of the Xpert GBS LB assay for detection of group B Streptococcus in prenatal screening specimens. J Clin Microbiol. 2015;6(1):42–45. doi: 10.1128/JCM.02598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan AA, Abdulmawjood A, Yildirim AO, Fink K, Lämmler C, Schlenstedt R. Identification of streptococci isolated from various sources by determination of cfb gene and other CAMP-factor genes. Can J Microbiol. 2000;46(10):946–51. [PubMed]
- 16.Hassan AA, Akineden O, Lämmler C, Huber-Schlenstedt R. Molecular characterization of phenotypically CAMP-negative Streptococcus agalactiae isolated from bovine mastitis. J Vet Med B Infect Dis VetPublic Health. 2002;49(5):257–9. [DOI] [PubMed]
- 17.Savini V, Marrollo R, De Filippis R, DaIncecco E, Imperi M, Pataracchia M, Alfarone G, Fusilli P, Coclite E, DrIncecco C, et al. CAMP-negative group B Streptococcus went unrecognized with Cepheid GeneXpert but was detected by Lioflchem® chromatic StrepB. Int J Clin Exp Pathol. 2016;9(11):12017–12020. [Google Scholar]
- 18.Darling CL. Standardization and evaluation of the CAMP reaction for the prompt, presumptive identification of Streptococcus agalactiae (Lancefield group B) in clinical material. J Clin Microbiol. 1975;1(2):171–4. [DOI] [PMC free article] [PubMed]
- 19.Phillips EA, Tapsall JW, Smith DD. Rapid tube CAMP test for identification of Streptococcus agalactiae (Lancefield group B). J Clin Microbiol. 1980l;12(2):135–7. [DOI] [PMC free article] [PubMed]
- 20.Hansen SM, Sørensen UB. Method for quantitative detection and presumptive identification of group B streptococci on primary plating. J Clin Microbiol. 2003;41(4):1399–403. [DOI] [PMC free article] [PubMed]
- 21.Ke D, Ménard C, Picard FJ, Boissinot M, Ouellette M, Roy PH, Bergeron MG. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46(3):324–31. [PubMed]
- 22.Gosiewski T, Brzychczy-Włoch M, Heczko PB. The application of multiplex PCR to detect seven different DNA targets in group B streptococci. Folia Microbiol (Praha). 2012;57(3):163–7. [DOI] [PMC free article] [PubMed]
- 23.Podbielski A, Blankenstein O, Lütticken R. Molecular characterization of the cfb gene encoding group B.streptococcal CAMP-factor. Med Microbiol Immunol. 1994;183(5):239–256. doi: 10.1007/BF00198458. [DOI] [PubMed] [Google Scholar]
- 24.Ruhlmann J, Wittmann-Liebold B, Jurgens D, Fehrenbach FJ. Complete amino acid sequence of protein B. FEBS Lett. 1988;235(1–2):262–266. doi: 10.1016/0014-5793(88)81275-4. [DOI] [PubMed] [Google Scholar]
- 25.Hensler ME, Quach D, Hsieh C, Doran KS, Nizet V. CAMP factor is not essential for systemic virulence of group B Streptococcus. Microb Pathogenesis. 2008;44(1):84–88. doi: 10.1016/j.micpath.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuzeville S, Puymège A, Madec JY, Haenni M, Payot S. Characterization of a new CAMP factor carried by an integrative and conjugative element in Streptococcus agalactiae and spreading in streptococci. PLoS One. 2012;7(11):e48918. [DOI] [PMC free article] [PubMed]
- 27.Das A, Ray P, Sharma M, Gopalan S. Rapid diagnosis of vaginal carriage of group B beta haemolytic streptococcus by an enrichment cum antigen detection test. Indian J Med Res. 2003;117:247–52. [PubMed]
- 28.Wernecke M, Mullen C, Sharma V, Morrison J, Barry T, Maher M, Smith T. Evaluation of a novel real-time PCR test based on the ssrA gene for the identification of group B streptococci in vaginal swabs. BMC Infect Dis. 2009;9:148. 10.1186/1471-2334-9-148. [DOI] [PMC free article] [PubMed]
- 29.Jamal W, Albert MJ, Rotimi VO. Real-time comparative evaluation of bioMerieux VITEK MS versus Bruker microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol. 2014;14:289. doi: 10.1186/s12866-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konikkara KP, Baliga S, Shenoy S, Bharati B. Evaluation of culture, antigen detection and polymerase chain reaction for detection of vaginal colonization of group B Streptococcus (GBS) in pregnant women. J Clin Diagn Res. 2014. 8(2):47–9. [DOI] [PMC free article] [PubMed]
- 31.Jürgens D, Sterzik B, Fehrenbach FJ. Unspecific binding of group B streptococcal cocytolysin (CAMP factor) to immunoglobulins and its possible role in pathogenicity. J Exp Med. 1987;165(3):720–732. doi: 10.1084/jem.165.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study available from the corresponding author on reasonable request.