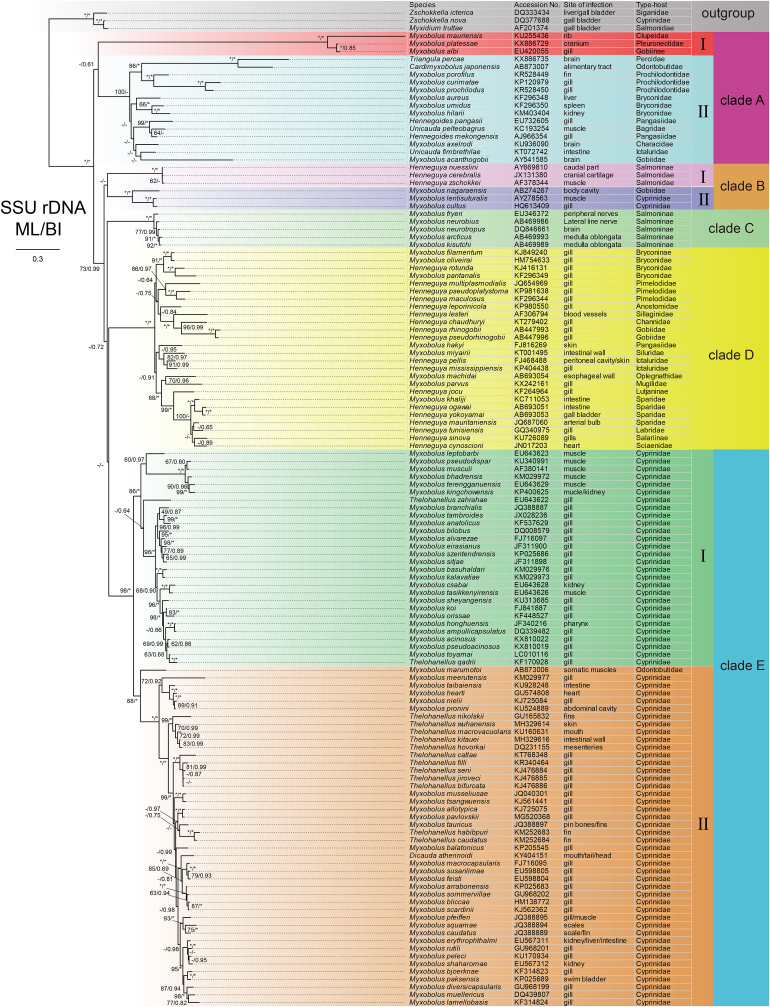
Abstract
Thelohanellus Kudo, 1933 is a species rich genus of Myxosporea, sharing many morphological similarities with species of Myxobolus but the former possesses a single polar capsule, and the latter has two. Based on molecular phylogenetic analyses, this single distinguishing feature is not monophyletic, and members of Thelohanellus are intermixed with Myxobolus species, calling into question the validity of genus Thelohanellus. The occurrence of two polar capsules in a small proportion of Thelohanellus spores as observed in this study suggests that these species have the capacity to express this Myxobolus-like trait, clouding the distinction of these two genera further. Herein, using the most comprehensive data set to date, we explored the phylogenetic relationships of Thelohanellus to other myxobolids, to investigate the evolutionary history of the genus Thelohanellus and the origins of single polar capsule in this group. The phylogenetic analyses and statistical tests of topology revealed Thelohanellus as a strongly supported polyphyletic lineage, clustering in five distinct branches within Myxobolus clade. Ancestral state reconstruction for polar capsule number showed that Thelohanellus species have evolved from myxosporean species with two polar capsules at least four times, which could be classified in three possible evolutionary pathways. The polyphyly of Thelohanellus and the multiple evolutionary origins of single polar capsule of Thelohanellus demonstrate that the distinction of this genus from Myxobolus is largely for convenience, and does not reflect their evolutionary history.
Keywords: Thelohanellus, Myxobolus, Single polar capsule, Phylogeny, Evolution, SSU rDNA
Graphical abstract
Highlights
-
•
Atypical Thelohanellus spores with two polar capsules were firstly observed.
-
•
Most myxobolid genera involved including Thelohanellus were poly- or paraphyletic.
-
•
Thelohanellus species have evolved at least four times which could be classified in three different evolutionary pathways.
1. Introduction
Myxosporeans are a group of diverse and widely distributed metazoan endoparasites with extremely reduced body size and structure (Okamura et al., 2015). Under the recent advance of multi-gene analyses, myxosporeans were found to diverge from free-living cnidarian ancestors in ancient times (Jiménez-Guri et al., 2007; Feng et al., 2014; Chang et al., 2015; Holzer et al., 2018) and then became parasitic to a wide range of animals, typically fishes, and less frequently amphibians, reptiles, birds, and mammals (Kent et al., 2001; Fiala et al., 2015b).
To date, approximately 2600 myxosporean species have been described and classified in 2 orders, 16 families and 62 genera based mainly on the structure and shape of myxospores (Lom and Dyková, 2006; Fiala et al., 2015a; Okamura et al., 2018). However, this classical spore-morphology-based taxonomic system is often in conflict with DNA sequence-based phylogenies (Fiala, 2006; Lom and Dyková, 2006; Bartošová et al., 2013). Specifically, there are a number of poly- or paraphyletic genera, such as Henneguya, Sphaerospora, Myxidium, Chloromyxum, Kudoa, and Ceratomyxa, to name a few (Fiala, 2006; Lom and Dyková, 2006; Fiala et al., 2015a). In fact, most myxosporean genera where molecular sequences are available were resolved as non-monophyletic. Such great discrepancies between traditional classification and DNA sequence-based phylogenies have resulted in a number of taxonomic revisions (Whipps et al., 2004; Gleeson and Adlard, 2012; Bartošová et al., 2013) and have motivated taxonomists to consider both morphological and genetic data when classifying myxosporeans.
The characters used to establish many myxosporean genera centered around morphological features of the myxospore (polar capsule number and orientation, overall shape, number of valves, etc.). For example, what we now classify as Thelohanellus species were originally placed in the genus Myxobolus Bütschli, 1882 based on the presence of iodinophilous vacuole in the sporoplasm, and other general features of the spore (pyriform, two valves, no tails). Subsequently, Kudo (1933) established genus Thelohanellus to delineate species with spores containing one polar capsule from those of Myxobolus which possess two polar capsules. Currently, Thelohanellus comprises 108 nominal species, most of which are plasmodia-forming parasites infecting various tissues of freshwater fishes, and some of which are pathogenic, causing economical losses in aquaculture (Zhang et al., 2013). The monophyly of genus Thelohanellus is not supported by phylogenetic analyses, with Thelohanellus species clustered in several Myxobolus clades (Griffin and Goodwin, 2011; Shin et al., 2014; Yuan et al., 2015; Liu et al., 2016; Ye et al., 2017). This suggests that the difference in polar capsule number between Thelohanellus and Myxobolus is not a character that reflects a single evolutionary divergence, but has evolved several times. To investigate this further, we explored the phylogenetic relationships of Thelohanellus species and traced the evolutionary origins of the single polar capsule within the larger myxobolid clade.
2. Materials and methods
2.1. Myxospore samples and morphological observation
Fresh spores of two Thelohanellus species were collected from their hosts in Hubei Province in China during 2015–2016 (Table 1). Spores were photographed using an Olympus BH2 microscope equipped with the Olympus DP73 camera and measured following the guidelines of Lom and Arthur (1989). All measurements are given in micrometers (μm).
Table 1.
Myxosporean species collected and identified in present study.
| Myxosporean species | Host species | Dates of collection | Location | Site of infection | Accession No. |
|---|---|---|---|---|---|
| Thelohanellus kitauei | Cyprinus carpio | March 2015 | Baishazhou Fish Market, Wuhan City, Hubei Province, China | Intestine | MH329616 |
| Thelohanellus wuhanensis | Carassius auratus gibelio | Jun 2016 | Honghu Lake, Hubei Province, China | Skin | MH329614 |
2.2. DNA extraction, amplification and sequencing
Total genomic DNA was extracted from fresh spores using General All Gen Kit (animal tissue protocol; CoWin Biosciences, China) following manufacturer's protocol. Small subunit ribosomal DNA (SSU rDNA) was amplified using primers pair 18e of Hillis and Dixon (1991) and 18R of Whipps et al. (2003). Polymerase chain reactions (PCRs) were performed in a 50-μL reaction solution comprising approximately 200 ng of extracted genomic DNA (2 μl), 25 μl 2×Es Taq MasterMix (CWBIO, China), and 10 pmol each primer. The reactions were run on an Eppendorf Mastercycler® nexus GX2 gradient thermocycler (Germany) with cycling parameters as follows: an initial denaturation at 94 °C for 7 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 60 s, and a terminal extension at 72 °C for 10 min. The PCR products were purified using the Gel Extraction Kit (CWBIO, China) and sequenced in both directions on the ABIPRISM 3730XL DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA). The SeqMan utility of the Lasergene software package (DNAStar, USA) was used to assemble contiguous sequences which were subsequently uploaded to NCBI with accession numbers MH329614 and MH329616.
2.3. Phylogenetic analyses
Only species with molecular sequence and morphological records provided by the same peer reviewed paper were included in the present study. In addition, most sequences used were selected with a length threshold set above 1300 bp. These included mostly Myxobolus, Henneguya and Thelohanellus species, but also Cardimyxobolus, Dicauda, Triangula, Unicauda and Hennegoides. Three species from the family Myxidiidae were used as outgroup. Alignments of SSU rDNA sequences were performed in MAFFT v7.306b (Katoh and Standley, 2013) with L—INS—I method which is optimized for sequence with multiple conserved domains and long gaps, such as rDNA genes. Gblocks 0.91b (Castresana, 2000) was used to remove ambiguous regions and highly variable sections (parameters were: t = d, -b1 = 65, -b2 = 67, -b3 = 10, -b4 = 5, -b5 = a), 71% positions of the original data set were saved. Substitution saturation was evaluated with DAMBE 5 (Xia and Xie, 2001). Maximum likelihood (ML) analysis was performed using RAxML 8.2.9 (Stamatakis, 2014) with a GTR GAMMA model as recommended by the author of the program. One thousand bootstrap replicates were performed to assess clade support. The above analyses were performed again after removing ambiguous species identified by RogueNaRok algorithm (Aberer et al., 2012). Bayesian Inference (BI) analysis was done in MrBayes v3.2.6 (Ronquist et al., 2012) with GTR + G + I model indicated by evaluation with jModelTest 2.1.10 (Darriba et al., 2012) under Akaike Information Criterion (AIC). Four parallel MCMC runs of 1 million generations were conducted with a burn-in of 25% of the samples and sampled every 100 generations. A 50% majority-rule consensus tree, with mean branch lengths, and posterior probabilities of clades was generated to summarize the results of the post burn-in posterior distribution of trees. Convergence of the Markov chains was confirmed in Tracer v.1.6 (Rambaut and Drummond, 2007) by a combined effective sample size (ESS) > 200.
2.4. Testing alternative phylogenetic hypotheses
Phylogenetic trees that constrained the involved genera to be monophyletic and per-site log likelihood scores for unconstrained and constrained ML trees were created using RAxML v8.2.9 with the GTR GAMMA model mentioned above. We calculated approximately unbiased (AU) values using CONSEL v1.20 (Shimodaira and Hasegawa, 2001) to determine statistical significance of differences in the likelihood scores. The alternative tree topologies could be rejected at the 5% confidence level.
2.5. Ancestral state reconstruction
In order to investigate the origin of Thelohanellus species, we traced the evolutionary lineage of the polar capsule number with the following possible three states: 1) single polar capsule; 2) two unequal polar capsules; 3) two equal polar capsules. In some cases of Myxobolus species, it is difficult to determine whether the two polar capsules are equal or not (Lom and Dyková, 2006) because this relies largely on prior authors’ opinions. In these cases, where polar capsules were reported as slightly different, almost equal, similar, etc., we classified them as equal (Supplementary Table 1). In addition, to avoid mismatches of molecular data and morphology brought by misidentification of species, morphological descriptions in present study were referred to articles that provided the molecular data rather than original records. The SSU-based ML trees were chosen as basis for reconstruction of ancestral states. A standard categorical matrix of polar capsule number (2 - two equal polar capsules, 1 - single polar capsule, 0 - two unequal polar capsules) was created to map on the tree. Mesquite v3.20 (Maddison, 2008) was used to trace the evolutionary history of this character. Reconstruction of character states at ancestor nodes was done by likelihood method with Markov k-state 1 parameter model. Any change in character state is equally probable within this model.
3. Results
3.1. Atypical spore morphology
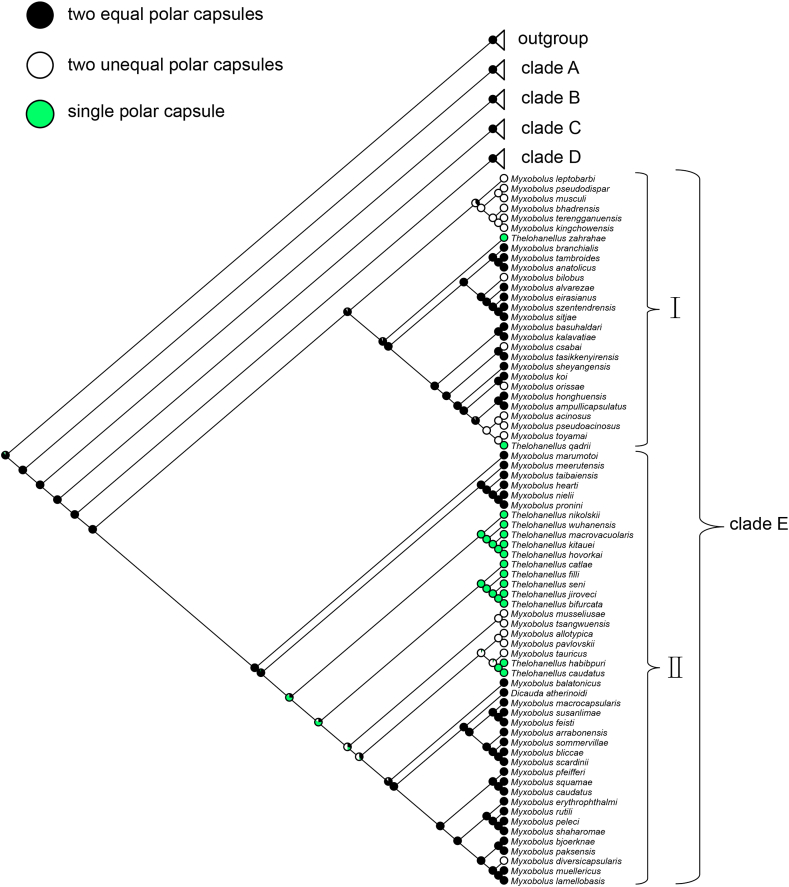
Two Thelohanellus species (Thelohanellus kitauei and Thelohanellus wuhanensis) showed two morphotypes (Fig. 1) in a plasmodium: typical spores with one polar capsule and atypical spores with two polar capsules. Besides the polar capsule number, they were different in spore shape, spore length and spore width. The atypical spore and typical spore of T. kitauei (Fig. 1 A) are contrasted as follows: oval versus prolonged pyriform shape, 22.62 versus 29.34 ± 1.34 (26.49–32.08) (n = 30) in spore length, and 16.00 versus 9.52 ± 0.88 (8.34–11.42) (n = 30) in spore width. The atypical spore and typical spore of T. wuhanensis (Fig. 1 B, C) are contrasted as follows: oval versus prolonged pyriform shape, 19.22 versus 22.86 ± 0.77 (21.63–24.69) (n = 30) in spore length, and 19.73 versus 12.05 ± 0.71 (11.01–13.58) (n = 30) in spore width. Despite the differences in overall spore shape, the polar capsules of the atypical spore and typical spore of T. kitauei (Fig. 1 A) and T. wuhanensis (Fig. 1 B, C) were similar in shape whether there was one or two. The proportion of atypical spores in the plasmodia of both present Thelohanellus species was exceedingly low, approximately 0.1% (n = 1000).
Fig. 1.
Photomicrograph of fresh spores of Thelohanellus kitauei and Thelohanellus wuhanensis. A - the spore of T. kitauei with two polar capsules (arrow); B, C - the spores of T. wuhanensis with two polar capsules (arrows). Scale bars indicate 20 μm for A, B and C.
3.2. Phylogenetic relationships among myxobolid species
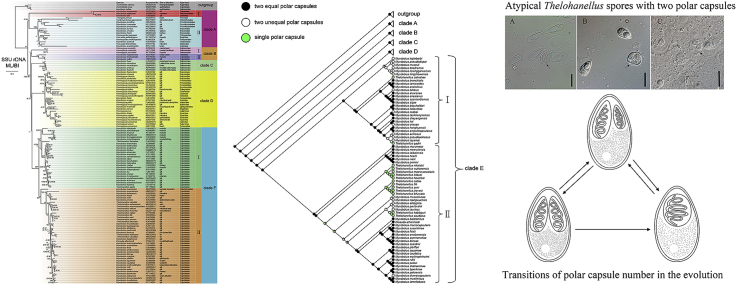
Identification of rogue taxa was implemented on the complete data set (138 taxa) using RogueNaRok, showed that 9 taxa (Supplementary Table 2), significantly increased the nodal support and did not affect the topology of the tree when removed from the analysis. The subsequent phylogenetic analyses were performed on the pruned data set, with the ingroup consisting of 126 myxobolid species covering 8 genera (Myxobolus, Thelohanellus, Henneguya, Unicauda, Dicauda, Hennegoides, Cardimyxobolus, Triangula). Tree topologies retrieved from ML and BI analyses were identical among main clades, both resulting in 5 clades though the branching order of the central part of the tree was weakly supported (Fig. 2).
Fig. 2.
SSU rDNA based maximum likelihood tree of selected myxobolid species. The table containing detail information of taxa is shown behind taxa names; Bootstrap supports and posterior probabilities are given beside the nodes, respectively. Asterisks represent values = 100/1.00. Dashes represent values < 60/0.60.
Apart from Dicauda, Cardimyxobolus and Triangula, which are represented by single species, none of other five genera included were monophyletic. Myxobolus species intermixing with species from other genera were found in every clade. Two species of the genus Hennegoides (Hennegoides pangasii and Hennegoides mekongensis) were closely related but not a monophyletic lineage. All Henneguya species clustered in clades B and D. Thelohanellus was polyphyletic, appearing in 5 different locations in clade E. Unicauda pelteobagrus showed close genetic relationship with Hennegoides species rather than Unicauda fimbrethilae (Fig. 2).
Clade A branched at the base of the tree and split into two major subclades (subclade I and subclade II). Subclade I included species (Myxobolus mauriensis, Myxobolus albi, Myxobolus platessae) with the extremely long branches in the phylogenetic tree, which might be consequence of their uncommon host environment (marine or brackish water, rare in Myxobolidae). In contrast, subclade II consisted of numerous species which infect freshwater fishes and exhibited much shorter branches length than subclade I species (Fig. 2).
The position of clade E as one of the most diverged branches was stable regardless of any variable phylogenetic methods involved (model selection, add/delete species, Gblock parameters). Apart from Myxobolus marumotoi, species within clade E differed from other clades by exclusively infecting cyprinid fishes. All members of Thelohanellus clustered in clade E, which split in two main subclades (subclade I, subclade II) with strong support. Within subclade II, Thelohanellus species clustered in three distinct clades. Five Thelohanellus species including the type species Thelohanellus hovokai formed an assemblage with high support. The clade with Thelohanellus catlae at the base was made up of Thelohanellus species that infect the gills. Two fin-infecting Thelohanellus species (Thelohanellus caudatus and Thelohanellus habibpuri) clustered outside the main Thelohanellus clade and were sister to Myxobolus tauricus which also infects fins. Within clade I, there was significant genetic divergence between Thelohanellus zahrahae and Thelohanellus qadrii. (Fig. 2).
In addition, four alternative topologies (Supplementary Fig. 1 topology 2, topology 3, topology 4, topology 5) were obtained from the phylogenetic analyses of SSU rDNA data set using different MAFFT strategies, Gblocks parameters and outgroups (Supplementary Table 3). We also presented another topology (Supplementary Fig. 1 topology 6) derived from Karlsbakk et al. (2017). The alternative topologies indicated the instability of the central part of the tree. However, clade E, comprising all of Thelohanellus species, clustered stably as one of the most diverged branches free from the impact of different phylogenetic methods.
3.3. Hypothesis testing
We performed the AU tests to further test the hypothesis that most of the genera are non-monophyletic group (Table 2). The monophyly of Thelohanellus, Myxobolus, Unicauda and Henneguya was significantly rejected (p ≪ 0.05). However, the monophyly of Hennegoides was not rejected (p = 0.541).
Table 2.
Results of approximately unbiased tests comparing the best maximum likelihood tree with the best constrained trees obtained under alternative hypotheses of monophyletic genera. Results in which P-values < 0.05 are shown in bold.
| Alternative hypothesis | P-value |
|---|---|
| unconstrained tree | 0.459 |
| monophyly of Thelohanellus | 5×10−120 |
| monophyly of Myxobolus | 6×10−60 |
| monophyly of Henneguya | 8×10−05 |
| monophyly of Dicauda | 2×10−12 |
| monophyly of Hennegoides | 0.541 |
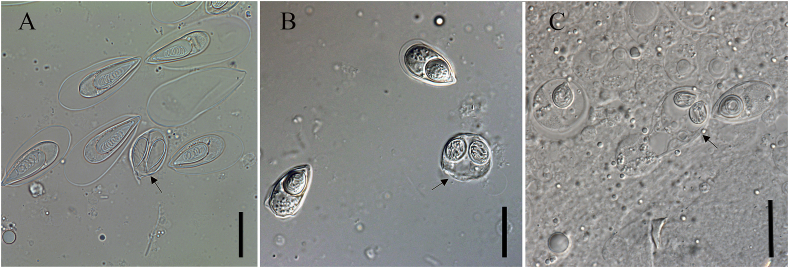
3.4. Reconstruction of ancestral polar capsule number
We conducted an ancestral state reconstruction analysis based on the preferred ML topology (Fig. 2) to investigate myxobolid evolution, particularly focused on the transitional events of the polar capsule number (Fig. 3). The analysis assigned high probability for most nodes. The ancestor of all myxobolid species was reconstructed to have two equal polar capsules with a probability 89%. The polar capsule number of the most recent common ancestors of clade E and clade A, B, C, D had not shifted, and were suggested to be two equal polar capsules with a probability >95%. Within clade E, there were evidences for six different evolutionary lineages of the polar capsule morphology (number and relative size): 1) Species with two equal polar capsules evolved to species with two unequal polar capsules, which was the most common shift, forming species like Myxobolus orissae, Myxobolus kingchowensis, and Myxobolus csabai. 2) Species with two equal polar capsules transferred to species with single polar capsule, resulting in species like T. wuhanensis, T. kitauei, and T. zahrahae. 3) Species with two equal polar capsules evolved to species with two unequal polar capsules, and then transformed to species with single polar capsule, leading to T. qadrii. 4) Species with two equal polar capsules evolved to species with single capsule, transformed to species with two unequal polar capsules subsequently, transformed to species with single capsule finally, resulting in T. caudatus and T. habibpuri. 5) Species with two equal polar capsules converted into species with single polar capsule, and then became species with two unequal polar capsules, evolved to species with two equal polar capsules eventually, resulting in species like Myxobolus macrocapsularis, Myxobolus bliccae, and Myxobolus shaharomae. 6) Species with two equal polar capsules evolved to species with single polar capsule, then transferred to species with two unequal polar capsules, became species with two equal polar capsules subsequently, transformed to species with two unequal polar capsules finally, leading to Myxobolus diversicapsularis.
Fig. 3.
Ancestral polar capsule number reconstruction for myxobolid species based on the preferred ML tree (Fig. 2). Pie charts show the marginal probability of the ancestral state at each node. Green circle represents species with single polar capsule, black circle represents species with two equal polar capsules, white circle represents species with two unequal polar capsules. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To further evaluate the effect of topological instability on the results of reconstruction, we also reconstructed the ancestral state based on the five alternative tree topologies (Supplementary Fig. 2). There is no difference from the evolutionary lineages documented above according to the ancestral state reconstruction based on topology 2, topology 4, topology 5 and topology 6. However, the phylogenetic position of T. caudatus and T. habibpuri altered in topology 3, making the origins of T. caudatus and T. habibpuri obscured.
4. Discussion
Myxosporean classification was established based mainly on spore morphology, and currently there are 16 established families (Fiala et al., 2015a). Of them, the family Myxobolidae has the most nominal species and is split into 14 genera based largely on the number of polar capsules and projections, and shape of sutural line. However, the monophyly of some genera of Myxobolidae was not supported by phylogenetic analyses (Kent et al., 2001; Fiala, 2006; Fiala and Bartošová, 2010; Moreira et al., 2014; Rocha et al., 2015; Capodifoglio et al., 2016). Our phylogenetic analyses included eight myxosporean genera, which were also shown as paraphyletic or polyphyletic groups except Dicauda, Cardimyxobolus and Triangula, for which there is only one SSU rDNA sequence included. Members of Myxobolus were distributed throughout the phylogenetic tree, indicating the close interrelatedness with the species of other genera, such as Henneguya, Dicauda, Thelohanellus, Unicauda. In fact, spore features that distinguish these genera from Myxobolus, such as the spore projections of Henneguya or single polar capsule of Thelohanellus, likely arose from Myxobolus ancestors (Fiala and Bartošová, 2010). The genus Hennegoides was paraphyletic but both species are within the same clade including a Unicauda species. Still, the monophyly of Hennegoides was not rejected by AU tests. Consistent with the results of Rosser (2016), the genus Unicauda was resolved as non-monophyletic group in present study. Henneguya species nested with Myxobolus species in clade B and D, but formed a monophyletic assemblage in clade BⅠ in the present study. The polyphyly of Henneguya is well noted as early as the first broad phylogenetic analysis of Myxosporea (Kent et al., 2001). All members of Thelohanellus clustered in clade E, but didn't form a monophyletic group, consistent with previous studies (Griffin and Goodwin, 2011; Yuan et al., 2015; Liu et al., 2016). The present study highlights the general non-monophyly of genera within Myxobolidae, indicating that taxonomic features used to establish different genera may be plastic and unstable. Gain or loss of these features probably occurs several times in their evolutionary history.
According to polar capsule number, the family Myxobolidae was divided into genera with two polar capsules (ten genera) and genera with a single polar capsule (four genera) (Lom and Dykova, 2006; Sarkar, 2009). Among these two groups, Myxobolus and Thelohanellus were the first established genera characterized by 2-PCs and one-PC, respectively. Based on the reconstruction of myxozoan morphotypes, Fiala and Bartošová (2010) proposed that the ancestor of myxobolids had two polar capsules, which was supported by our results. Moreover, we found that the two polar capsules of the ancestor of myxobolids were equal in size. Consideration of polyphyly of genus Thelohanellus and the placement of Thelohanellus species within several Myxobolus clades, Yuan et al. (2015) speculated that ancestors of Thelohanellus underwent several times of loss or gain of polar capsule in the evolutionary history. The ancestral state reconstruction in present study revealed that the formation of single polar capsule of Thelohanellus happened four times after the most recent common ancestor of Thelohanellus infecting cyprinid fish. However, the possibility of ancestral state of the clade rooted by T. catlae to be single polar capsule (0.61) is not very high. The situation that species within this clade evolved with an independent origin of a single polar capsule could not be excluded, which makes us conclude that Thelohanellus evolved at least four times. These transitional events could be classified in three different ways according to the evolutionary process: (1) the direct type: single polar capsule formed by two equal polar capsules losing one polar capsule directly; (2) the gradual type: single polar capsule formed by one of the two polar capsules gradually degenerating and finally missing; and (3) the devious type: single polar capsule formed by losing one of the two equal polar capsules firstly, gaining one polar capsule subsequently, and losing one polar capsule eventually. However, this evolutionary process of the devious type might be changeable due to the unstable phylogenetic position of T. caudatus and T. habibpuri.
Surprisingly, the Thelohanellus spores observed in the present study sometimes possessed two polar capsules, which indicated Thelohanellus species have the genetic capacity to express two polar capsules. In addition, Desser et al. (1983) observed two capsulogenic cells in the young sporoblast of Thelohanellus nikolskii and its single polar capsule was formed by abortion of the second capsulogenic cell, supporting the idea that ancestor of Thelohanellus species had two polar capsules.
A single polar capsule is the only feature that discriminates Thelohanellus from Myxobolus, yet this character is of limited taxonomic significance as its multiple evolutionary routes and the polyphyletic character of Thelohanellus. A possible course of action would be to synonymize Thelohanellus with Myxobolus. This may be the ultimate course of action, but we recognize that there is utility in maintaining separate genera for the ease of identification, particularly with pathogenic species.
Funding
This work was supported by the Nature Science Foundation of China (projects Nos. 31572233 and 31501848), Research and Demonstration of Key Techniques for High Quality Aquatic Products (project No. 2016620000001046).
Declarations of interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2018.11.006.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Schematic drawings of tree topologies obtained from phylogenetic analyses.
Ancestral polar capsule number reconstructions based on five alternative phylogenetic trees. The representation of pie charts is exactly same as that of Fig. 3.
References
- Aberer A.J., Krompass D., Stamatakis A. Pruning rogue taxa improves phylogenetic accuracy: an efficient algorithm and webservice. Syst. Biol. 2012;62:162–166. doi: 10.1093/sysbio/sys078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartošová P., Fiala I., Jirků M., Cinková M., Caffara M., Fioravanti M.L., Atkinson S.D., Bartholomew J.L., Holzer A.S. Sphaerospora sensu stricto: taxonomy, diversity and evolution of a unique lineage of myxosporeans (Myxozoa) Mol. Phylogenet. Evol. 2013;68:93–105. doi: 10.1016/j.ympev.2013.02.026. [DOI] [PubMed] [Google Scholar]
- Capodifoglio K.R., Adriano E.A., Milanin T., Silva M.R., Maia A.A. Morphological, ultrastructural and phylogenetic analyses of Myxobolus hilarii n. sp. (Myxozoa, Myxosporea), a renal parasite of farmed Brycon hilarii in Brazil. Parasitol. Int. 2016;65:184–190. doi: 10.1016/j.parint.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chang E.S., Neuhof M., Rubinstein N.D., Diamant A., Philippe H., Huchon D., Cartwright P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. 2015;112:14912–14917. doi: 10.1073/pnas.1511468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desser S.S., Molnar K., Weller I. Ultrastructure of sporogenesis of Thelohanellus nikolskii akhmerov, 1955 (Myxozoa: Myxosporea) from the common carp, Cyprinus carpio. J. Parasitol. 1983:504–518. [Google Scholar]
- Feng J., Xiong J., Zhang J., Yang Y., Yao B., Zhou Z., Miao W. New phylogenomic and comparative analyses provide corroborating evidence that Myxozoa is Cnidaria. Mol. Phylogenet. Evol. 2014;81:10–18. doi: 10.1016/j.ympev.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Fiala I. The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int. J. Parasitol. 2006;36:1521–1534. doi: 10.1016/j.ijpara.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Fiala I., Bartošová P. History of myxozoan character evolution on the basis of rDNA and EF-2 data. BMC Evol. Biol. 2010;10:228. doi: 10.1186/1471-2148-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala I., Bartošová-Sojková P., Whipps C.M. Classification and phylogenetics of Myxozoa. In: Okamura B., Gruhl A., Bartholomew J.L., editors. Myxozoan Evolution, Ecology and Development. Springer International Publishing; Cham, Switzerland: 2015. pp. 85–110. [Google Scholar]
- Fiala I., Hlavničková M., Kodádková A., Freeman M.A., Bartošová-Sojková P., Atkinson S.D. Evolutionary origin of Ceratonova shasta and phylogeny of the marine myxosporean lineage. Mol. Phylogenet. Evol. 2015;86:75–89. doi: 10.1016/j.ympev.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Gleeson R.J., Adlard R.D. Phylogenetic relationships amongst Chloromyxum Mingazzini, 1890 (Myxozoa: Myxosporea), and the description of six novel species from Australian elasmobranchs. Parasitol. Int. 2012;61:267–274. doi: 10.1016/j.parint.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Griffin M.J., Goodwin A.E. Thelohanellus toyamai (Syn. Myxobolus toyamai) infecting the gills of koi Cyprinus carpio in the eastern United States. J. Parasitol. 2011;97:493–502. doi: 10.1645/GE-2674.1. [DOI] [PubMed] [Google Scholar]
- Hillis D.M., Dixon M.T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Holzer A.S., Bartošová-Sojková P., Born-Torrijos A., Lövy A., Hartigan A., Fiala I. The joint evolution of the Myxozoa and their alternate hosts: a cnidarian recipe for success and vast biodiversity. Mol. Ecol. 2018;27:1651–1666. doi: 10.1111/mec.14558. [DOI] [PubMed] [Google Scholar]
- Jiménez-Guri E., Philippe H., Okamura B., Holland P.W. Buddenbrockia is a cnidarian worm. Science. 2007;317:116–118. doi: 10.1126/science.1142024. [DOI] [PubMed] [Google Scholar]
- Karlsbakk E., Kristmundsson Á., Albano M., Brown P., Freeman M.A. Redescription and phylogenetic position of Myxobolus aeglefini and Myxobolus platessae n. comb. (Myxosporea), parasites in the cartilage of some North Atlantic marine fishes, with notes on the phylogeny and classification of the Platysporina. Parasitol. Int. 2017;66:952–959. doi: 10.1016/j.parint.2016.10.014. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent M.L., Andree K.B., Bartholomew J.L., El-matbouli M., Desser S.S., Devlin R.H., Feist S.W., Hedrick R.P., Hoffmann R.W., Khattra J. Recent advances in our knowledge of the Myxozoa. J. Eukaryot. Microbiol. 2001;48:395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Kudo R. A taxonomic consideration of Myxosporidia. Trans. Am. Microsc. Soc. 1933;52:195. [Google Scholar]
- Liu Y., Zhai Y., Gu Z. Morphological and molecular characterization of Thelohanellus macrovacuolaris n. sp. (Myxosporea: Bivalvulida) infecting the palate in the mouth of common carp Cyprinus carpio L. in China. Parasitol. Int. 2016;65:303–307. doi: 10.1016/j.parint.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Lom J., Arthur J.R. A guideline for the preparation of species descriptions in Myxosporea. J. Fish. Dis. 1989;12:151–156. [Google Scholar]
- Lom J., Dyková I. Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol. (Praha) 2006;53:1. [PubMed] [Google Scholar]
- Maddison W.P. Mesquite: a modular system for evolutionary analysis. Evolution. 2008;62:1103–1118. [Google Scholar]
- Moreira G.S., Adriano E.A., Silva M.R., Ceccarelli P.S., Maia A.A. The morphological and molecular characterization of Henneguya rotunda n. sp., a parasite of the gill arch and fins of Salminus brasiliensis from the Mogi Guaçu River, Brazil. Parasitol. Res. 2014;113:1703–1711. doi: 10.1007/s00436-014-3815-y. [DOI] [PubMed] [Google Scholar]
- Okamura B., Gruhl A., Bartholomew J.L. An introduction to myxozoan evolution, ecology and development. In: Okamura B., Gruhl A., Bartholomew J.L., editors. Myxozoan Evolution, Ecology and Development. Springer International Publishing; Cham, Switzerland: 2015. pp. 1–20. [Google Scholar]
- Okamura B., Hartigan A., Naldoni J. Extensive uncharted biodiversity: the parasite dimension. Integr. Comp. Biol. 2018;58:1132–1145. doi: 10.1093/icb/icy039. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J. Tracer v1. 6: MCMC trace analyses tool. 2007. http://tree.bio.ed.ac.uk/software/tracer/ Software available via.
- Rocha S., Matos E., Velasco M., Casal G., Alves A., Azevedo C. Ultrastructure and phylogeny of Thelohanellus sp. (Myxozoa: Myxosporea) infecting the gills of Hypophthalmus marginatus (Actinopterygii: Pimelodidae), a fish from the amazon river. Microsc. Microanal. 2015;21:46. [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser T.G., Alberson N.R., Baumgartner W.A., Mauel M.J., Pote L.M., Griffin M.J. Morphological, histological, and molecular description of Unicauda fimbrethilae n. sp. (Cnidaria: Myxosporea: Myxobolidae) from the intestinal tract of channel catfish Ictalurus punctatus. J. Parasitol. 2016;102:105–113. doi: 10.1645/15-810. [DOI] [PubMed] [Google Scholar]
- Sarkar N.K. Thelohanelloides bengakensis gen. and sp. nov. (Myxosporea: Thelohanellidae) from the gall bladder of marine catfish of the Bay of Bengal, India. Uttar Pradesh J. Zool. 2009:251–254. [Google Scholar]
- Shimodaira H., Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shin S.P., Nguyen V.G., Jeong J.M., Jun J.W., Kim J.H., Han J.E., Baeck G.W., Park S.C. The phylogenetic study on Thelohanellus species (Myxosporea) in relation to host specificity and infection site tropism. Mol. Phylogenet. Evol. 2014;72:31–34. doi: 10.1016/j.ympev.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipps C.M., Adlard R.D., Bryant M.S., Lester R.J., Findlav V., Kent M.L. First report of three Kudoa species from eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus) J. Eukaryot. Microbiol. 2003;50:215–219. doi: 10.1111/j.1550-7408.2003.tb00120.x. [DOI] [PubMed] [Google Scholar]
- Whipps C.M., Grossel G., Adlard R.D., Yokoyama H., Bryant M.S., Munday B.L., Kent M.L. Phylogeny of the Multivalvulidae (Myxozoa: Myxosporea) based on comparative ribosomal DNA sequence analysis. J. Parasitol. 2004;90:618–622. doi: 10.1645/GE-153R. [DOI] [PubMed] [Google Scholar]
- Xia X., Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Ye L., Lu M., Quan K., Li W., Zou H., Wu S., Wang J., Wang G. Intestinal disease of scattered mirror carp Cyprinus carpio caused by Thelohanellus kitauei and notes on the morphology and phylogeny of the myxosporean from Sichuan Province, southwest China. Chin. J. Oceanol. Limnol. 2017;35:587–596. [Google Scholar]
- Yuan S., Xi B., Wang J., Xie J., Zhang J. Thelohanellus wangi n. sp. (Myxozoa, Myxosporea), a new gill parasite of allogynogenetic gibel carp (Carassius auratus gibelio Bloch) in China, causing severe gill myxosporidiosis. Parasitol. Res. 2015;114:37–45. doi: 10.1007/s00436-014-4157-5. [DOI] [PubMed] [Google Scholar]
- Zhang J., Gu Z., Kalavati C., Costa Eiras J., Liu Y., Guo Q., Molnár K. Synopsis of the species of Thelohanellus Kudo, 1933 (Myxozoa: Myxosporea: Bivalvulida) Syst. Parasitol. 2013;86:235–256. doi: 10.1007/s11230-013-9449-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic drawings of tree topologies obtained from phylogenetic analyses.
Ancestral polar capsule number reconstructions based on five alternative phylogenetic trees. The representation of pie charts is exactly same as that of Fig. 3.