Abstract
Introduction/Background:
Cancer registries in the US collect timely and systematic data on new cancer cases, extent of disease, staging, biomarker status, treatment, survival, and mortality of cancer cases. Existing methodologies for accessing local cancer registry data for research are time-consuming and often rely on the manual merging of data by staff registrars. In addition, existing registries do not provide direct access to these data nor do they routinely provide linkage to discrete electronic health record (EHR) data, reports, or imaging data. Automation of such linkage can provide an impressive data resource and make valuable data available for translational cancer research.
Methods:
The UPMC Network Cancer Registry collects highly structured, longitudinal data on all reportable cancer patients, from the point of the diagnosis throughout treatment and follow-up/outcomes. Using commercial registry software, we collect data in compliance with standards governed by the North American Association of Central Cancer Registries. This standardization ensures that the data are highly structured with standard coding and collection methods, which support data exchange among central cancer registries and the Centers for Disease Control and Prevention.
Results:
At the UPMC Hillman Cancer Center and University of Pittsburgh, we explored the feasibility of linking this well-curated, structured cancer registry data with unstructured text (i.e., pathology and radiology reports), using the Text Information Extraction System (TIES). We used the TIES platform to integrate breast cancer cases from the UPMC Network Cancer Registry system and then combine these data with other EHR data as a pilot use case that can be replicated for other cancers.
Conclusions:
As a result of this integration, we now have a single searchable repository of information for breast cancer patients from the UPMC registry, combined with their pathology and radiology reports. The system that we developed is easily scalable to other health systems and cancer centers.
Keywords: Breast cancer, cancer registry, concept recognition, imaging data, pathology record
INTRODUCTION
Importance of cancer registry data for cancer research
The National Program of Cancer Registries (NPCR), established by US Congress in 1992,[1] aims to collect timely and systematic data on new cancer cases, extent of disease, staging, biomarker status, treatment, survival, and mortality of cancer patients in the US.[2] Since 1994, the NPCR has funded state cancer registries to collect population-based cancer incidence data, covering 96% of the US population.[2,3] Starting in 2001, the NPCR began receiving data annually from funded programs with the goals of ascertaining the quality of the data and eventually releasing the data for the use in public health planning.[3] The National Cancer Institute (NCI)-funded Surveillance, Epidemiology, and End Results Program (SEER)[4] is a population-based surveillance system from various cancer registries, established in 1973, which collects data on cancer for 28% of the US population, which has been used to conduct multiple research studies.[5,6,7] Over the past few decades, registry data have become a rich source of information for cancer researchers, especially for those working in the area of breast cancer and those seeking to analyze Medicare/SEER-linked databases.[8]
Cancer registries in breast cancer research
While registry data access has been very useful in conducting high-impact cancer research, registry research is often difficult due to the need to manually integrate cancer registry data with other data. In addition, traditional sources of cancer registry data have not been linked to useful sources of information such as full pathology and radiology reports, which are essential data sources for a large number of breast cancer studies. Breast cancer, because of its high prevalence, is a logical malignancy to develop and test a structured data import tool to link multiple data types into a single repository for research and clinical use. Breast cancer is the most common malignancy in US women, except for skin cancer. Among US women in 2018, there will be an estimated 266,120 new cases of invasive breast cancer, 63,960 new cases of breast carcinoma in situ, and 40,920 breast cancer deaths.[9] A recent SEER-based article reported widening racial disparities in breast cancer mortality, which is likely to continue in light of the increasing incidence of breast cancer in African American women.[10] US SEER data have been used to evaluate the large-scale effects of breast cancer radiotherapy on patient survival.[11] Registry data are useful for evaluating the course and survival from rare conditions, such as male breast cancer.[12] In addition, registry data have been used for the characterization of rare histologic types of breast cancer, including mucinous, tubular, comedo, inflammatory, medullary, and papillary carcinomas, which together account for about 10% of all breast cancer cases.[13]
Text Information Extraction System software and the future directions in registry research
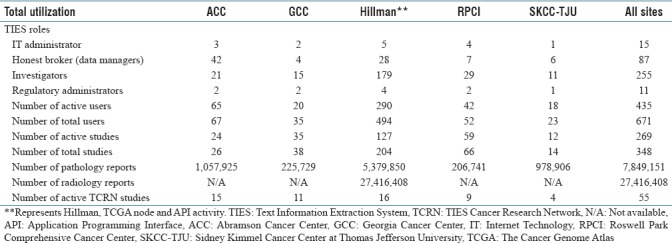
Over the past 15 years, the Department of Biomedical Informatics (DBMI) at the University of Pittsburgh developed a novel software system called Text Information Extraction System (TIES),[14] which uses natural language processing to create a unique platform for research cohort selection. The software currently encompasses ~33 million deidentified pathology and radiology clinical documents, and other data sources including ~50,000 deidentified whole slide images. One unique feature of TIES is that it has also been developed to operate a federated network, with the trust agreements necessary to share data and biospecimens. With funding from NCI, the TIES software has been deployed at several cancer centers, and we are currently operating a network of five cancer centers that share data and tissue through the TIES Cancer Research Network (TCRN).[15] This network was described in detail in a recent Cancer Research publication.[16] Current participants include UPMC Hillman Cancer Center, Roswell Park Cancer Center, University of Pennsylvania Abramson Cancer Center, Augusta University Cancer Center, Stony Brook University, and Thomas Jefferson University Kimmel Cancer Center. Across these six cancer centers, we currently share deidentified data for over 2.5 million cancer patients. Several pilot projects across the network have shown that TIES reduces the barriers to safe sharing of data and biospecimens among institutions.[17,18] Outside the TCRN network, using TIES stand alone, a recent study at Kaiser Permanente demonstrated that the TIES system could effectively identify potential breast cancer cases.[17] Summary of TCRN activities is highlighted in Table 1. The TIES open source tool is dual licensed (individual/educational non-profits and other users) and available through a SourceForge repository for download and use at other centers.
Table 1.
Text Information Extraction System Text Information Extraction System Cancer Research Network all sites summary as of May, 2018
This paper describes the integration of breast cancer registry data into the TIES system at the UPMC Hillman Cancer Center/University of Pittsburgh resulting in the development of invaluable data resource for clinicians and researchers. The aim of this manuscript is to describe the methodology that we used for integrating TIES and cancer registry data and to describe the usage of the TIES structure data import tool to present this data for use in translational cancer research.
METHODS
The focus of this work was to develop an open source structured data import tool (also called structured data loader in the technical documentation) for the University of Pittsburgh TIES data repository (i.e., pathology and radiology reports) to allow research access to the UPMC Network Cancer Registry structured data (i.e., Staging/Treatment/Outcomes) with proper approvals. This involved registry data collection in compliance with North American Association of Central Cancer Registries (NAACCR) formatting,[19] filtering, and reformatting data, followed by uploading the structured data extracted from cancer registry into the TIES repository. The work was guided by a working group of subject matter experts from the University of Pittsburgh, UPMC Hillman Cancer Center, and the UPMC Health System and covered as an Institutional Review Board approved project (PRO12050326 and PRO07050292). The working group's expertise in data, research, and clinical domains was essential to our success in securing access to and using the cancer registry data for research purposes. These experts ensured the appropriate interpretation of these data, given the complex access and clinical rules governing cancer registry data collection.
The working group included representation from (1) UPMC Network Cancer Registry (a) Director, Supervisor, and cancer-specific Research Registrar), (2) DBMI (a faculty member and programmer), (3) UPMC Hillman Cancer Center Informatics (a Database Administrator), and (4) Institute of Precision Medicine (a breast cancer researcher). The first task for the working group was to choose the set of common data elements (CDEs) to use from the Cancer Registry, on the basis of importance of these data elements for conducting and reporting research data.[20] Our cancer registry a data management system coordinats the entire cancer registry process, from data collection and follow-up, through reporting and analysis; it also provides standards compliance which satisfies current regulatory reporting and accreditation requirements. We exported the cancer registry data into NAACCR compliant simple data storage. This provided us with a method to quickly identify a set of over 130 CDEs [Supplement Table 1 (1.9MB, tif) ] with established coding standards (e.g., ICD-O, ICD-9/10, SEER) and ensure that other institutions can leverage this information and process for extracting data. Since the data are represented using a NAACCR data model, its data fields are normalized to machine computable values. This representation was ideal for the internal TIES structured data model, which supports an arbitrary set of document and patient level hierarchical property/value pairs. This data model allows TIES to represent any conceivable data model including those consistent with the NAACCR data dictionary.[21] Once the CDE list was established, the working group began to vet the entire list to further refine the elements into a smaller working set and then categorized these elements into several groupings: patient demographics, the extent of disease, treatment, outcomes, and disease-specific data. The demographic data consisted largely of identification data (e.g., MRN, name) to provide a method to link to the current patients in TIES. However, this data also included a family history of cancer, ethnicity, and race. The extent of disease as defined by the NCI, is a description of how far the tumor has spread from organ or site of origin (the primary site).[22] The extent of disease is an anatomic categorization using descriptors to group individual cases about the human body.” This allowed us to capture structured pathological and clinical staging related to the primary diagnosis. There are a number of data elements collected by cancer registries related to first-course treatment and radiation options; however, the working group decided not to include these at this time because TCRNs principle use is cohort discovery.
Common elements list
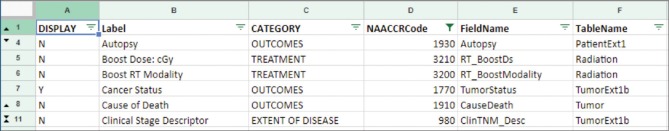
The outcomes and survival data present a mixture of computed and hand-curated fields. The computed fields are derived from variables such as date of diagnosis, recurrence, or date of metastatic disease diagnosis. These are used by the system to compute overall survival (i.e., months from the date of diagnosis to date of last contact), disease-free survival, and days from the diagnosis to the first recurrence. Various other data are entered by the registrar, such as tumor status, vital status, and cause of death gathered from the Electronic Medical Record records. Our cancer registry at the UPMC Hillman Cancer Center collects a number of disease-specific data elements using a combination of system user-defined fields and other fields coded using the standard Collaborative Stage Data Collection System for both staging and site-specific factors (SSFs).[23] We selected a number of SSF initially for breast cancer, from estrogen receptor, progesterone receptor, HER2 results to distant metastasis indicators. As we defined this list of CDEs, we also recorded other meta-data [Figure 1] about each CDE such as a mapping of the database table/column names, in a machine-readable format, for our SQL data extraction and load scripts. The associated NAACCR item number was used to acquire the preferred name and definitions using the SEER API service,[24] which was used with the data pipeline to provide consistent labeling and definitions of the CDEs when loaded into TIES.
Figure 1.
Common data element meta-data list
In the second stage of this process, we established a set of data extraction criteria working with our cancer registry team. These criteria included rules identifying when certain fields were collected between certain times and which were transitioned to other fields for collection. For example, one such rule involved tumor size which was collected under collaborative staging rules through 2015, then tumor size summary fields from 2016 onward; such rules were taken into account in our extraction and merge scripts. For our initial extraction, we were focused on only breast cancer cases and therefore limited our extraction to the primary site of breast cancer (i.e., ICD-O C500-C509). We developed our extract, transform, and load (ETL) SQL scripts for TIES against a staging environment [Figure 2].
Figure 2.
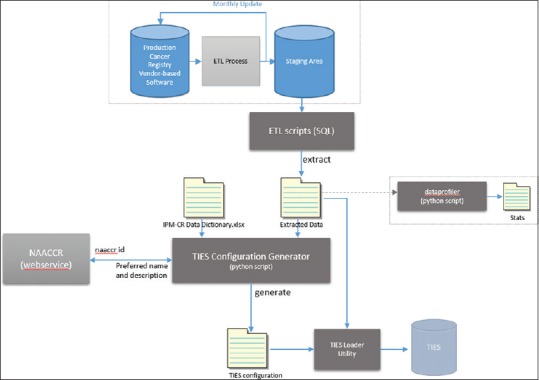
Data transfer pipeline
A set of automated extract scripts were developed to extract and populate data on a set basis to this staging area. This process was done to ensure that we would not affect the data integrity nor degrade the performance of the production server when performing the extracts and to meet the NAACCR standards [see the staging area in the data transfer pipeline in Figure 2]. With the staging area populated, we created SQL extract scripts based on the defined CDE categories and extraction criteria, which were then extracted to comma separated value (CSV) files. The remaining portion of the pipeline uses a python script to read the extracted data (i.e., CSV) and build a TIES configuration file (CFG) [Figure 3] for ingesting this data directly into TIES by the StructureDataLoader service.
Figure 3.
Sample text information extraction system configuration file
The construction of the CFGs by hand would be time prohibitive. To allow for a feasible time to iterate between modifying the extraction of SQL scripts and loading them into TIES, especially for testing, automation scripts are essential. The TIES configuration generator (i.e., python script) reads the CSV file and uses the CDE meta-data file to obtain the correct mapping for each field. SEER API is used to get descriptive labels, used for labeling and tooltips within the TIES user interface (UI). Data types, namespace grouping, and Protected Health Information indicators are also referred to generate all required files for the TIES CFG. Once this process is complete, a text file is generated containing the configuration for each field per the TIES configuration specification. This file, along with the data file, is used to ingest the data into the TIES repository. Linkage to the patient and document in TIES occurs through a common patient identifier supplied in the extracted data and identified by the CFG file.
After the data have been loaded, they become available as structured data in the TIES UI, linked to both the patient identifier and each clinical report. The data are tagged by the importer as data from the cancer registry. This tagging allows a user to limit their search results to only documents with available cancer registry data. Registry CDEs are also available in a faceted search, using the NAACCR standard values. On searching the structured data, it is displayed in the TIES UI and available for review [Figure 4]. This provides a grouped layout of all the data and allows tooltip hovering to provide an extensive definition. As CDEs may change in the process of updating the system, these two steps need to be implemented to add additional CDE to already existing data stream:[1] additional CDEs would need to be added to the TIES ETL tool for all data going forward [Figure 2 ETL level] and (2) all data that was already loaded from cancer registry would have to be reloaded to develop a richer data set.
Figure 4.
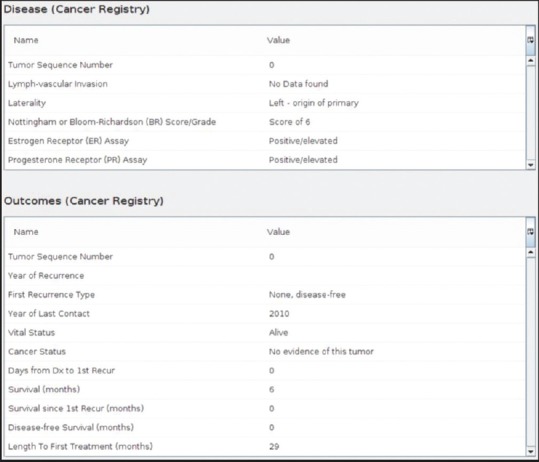
Cancer registry data in text information extraction system
The structured cancer registry data stored in TIES alongside the unstructured report text allows the users to choose from a richer, well-defined set of selection criteria and combine structured and unstructured data elements in search of better cohort identification. Successful accomplishment of the deliverables of the project required a number of modifications and extensions to TIES, development of new ETL scripts, and some alterations to agreements, policies, procedures, and processes, both locally and across the network. It is important to point out that this model is applicable and scalable to any structured data, not necessary that contained in TIES.
DISCUSSION
This paper describes the complex process of establishing a standard ingestion pipeline for integrating structured clinical data into the TIES system and represents a scalable model for intake of structured data from other sources (e.g., Electronic Health Record (EHR)). TIES uses a hierarchical property value data model to represent discrete data elements associated with clinical documents and patients, representing a paradigm that can ingest a wide variety of structured data in a simple and consistent format. At present, TIES uses this mechanism to store cancer registry data (as described in this publication), the Cancer Genome Atlas phenotypic information, and whole slide image metadata that is used to associate virtual slides with clinical reports. In the future, any discrete data element from EHR or manual human abstraction can be integrated into TIES including pharmaceutical and clinical trials data, vital signs, billing codes, results of manual abstraction, results of automatic information extraction or classification, results of whole slide image analysis, and manual whole slide image annotations.
While the focus of this project was breast cancer registry data, the methodologies outlined in this manuscript are applicable to other disease sites. The strength of this approach includes the use of national standards (e.g., NAACCR), validated systems (TIES), and new automated methods for combining various data types. Breast cancer was an optimal disease site to initiate this effort because of high disease prevalence. Thousands of cases are available for research on both common breast cancer types and rare breast cancer histologies. Once breast cancer registry data access for research is approved across our research community, we will work with the rest of the TCRN to deploy it at partnering institutions. Greater availability of this data to the personalized medicine research community across partnering institutions will create new opportunities for research programs that currently rely on the manual integration of cancer registry data. Ideally, users of this data across the network will be able to share deidentified cancer registry data linked with biospecimens and other data, including whole slide images. The strength of the model that we described in this paper is that it can be easily applied to other projects and data types, without being limited to the formats dictated by NAACCR. Any structured data from any source systems can be represented in this format. In the future, the systems we developed as a part of this project will help to resolve some of the key challenges associated with cancer registry data usage. For example, because a patient may have multiple primary tumors or same cancer may be reported to the registry by more than one provider, the same person can appear more than once in a registry database depending on how many UPMC facilities cared for that patient and their primary disease.[25] Because knowing the number of patients is critical for effective resource allocation and the development of innovative research studies, TIES system may further evolve to develop procedures to reliably estimate the number of patients from these joint data sources. Managing the numbers of patients and the number of cases, each distinctly and clearly for the user is in our plans for TCRN developing going forward.
Deep neural networks have surged in popularity and proven to be powerful tools for various artificial intelligence applications in computer vision, speech recognition, and natural language processing.[26] Recent publication by Gao et al.[27] demonstrated that deep learning approaches based on hierarchical attention networks (HANs) could improve model performance for multiple information extraction tasks from unstructured cancer pathology reports compared to traditional methods. As any supervised machine learning method, including HANs, requires a large set of labeled data for training purposes, manually curated structured data from cancer registries linked to unstructured document text within TIES become invaluable resources for the training of deep learning models. Furthermore, these datasets can potentially be used for the development of novel methods, including HANs, in a variety of research and clinical applications, including document classification, information extraction, and predictive modeling. Transfer learning may provide a boost to deep learning approaches, but their use for automatic abstraction of pathology reports needs to be further explored.[28] One of the future challenges that our group is planning to work with and address is the automation of cancer registry data acquisition. Typically, registries have large numbers of data elements, with many of them being difficult to interpret, especially for researchers not typically working with registry data. In addition, not all fields that are present in cancer registries are useful for cancer researches. While Nguyen et al. published one of the first reports on the developed of an automated medical text analysis system for registry type data,[29] more research is needed in this important area. Thus, one of the challenges that we will be solving in the multidisciplinary workgroup setting is figuring out how much data are sufficient for research and should be made available.
CONCLUSION
We used the TIES platform to extract breast cancer cases from the UPMC Network Cancer Registry system and integrate these entries with other EHR data as a pilot use case that can be replicated for other malignancies. Through this integration, we now have a single searchable repository of information for breast cancer patients from the UPMC registry, combined with their pathology and radiology reports. These data are available to scientists at the Hillman Cancer Center, with the TIES team providing a structured data import tool and implementation model useful for other TCRN sites. This open source tool is dual licensed (individual/educational nonprofits and commercial users)[30] and available through SourceForge repository[31] for download and use at other cancer and research centers.
Financial support and sponsorship
This project has been sponsored by the Institute for Precision Medicine, University of Pittsburgh School of Medicine and UPMC; UPMC Network Cancer Registry. The National Cancer Institute Grants UL1 TR001857 U24 CA180921 (and CA180921 - 0451 supplement) and R01CA132672 have supported the TIES system development.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2018/9/1/47/248451
REFERENCES
- 1.CDC. Cancer Registries Amendment Act 2015. [Last accessed on 2018 Jun 10]. Available from: https://www.cdc.gov/cancer/npcr/amendmentact.htm .
- 2.White MC, Babcock F, Hayes NS, Mariotto AB, Wong FL, Kohler BA, et al. The history and use of cancer registry data by public health cancer control programs in the United States. Cancer. 2017;123(Suppl 24):4969–76. doi: 10.1002/cncr.30905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. United States Cancer Statistics. 2016. [Last accessed on 2018 Jun 10]. Available from: https://www.cdc.gov/rdc/B1DataType/Dt131.htm .
- 4.NCI. Surveillance, Epidemiology, and End Results (SEER) Program. 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.seer.cancer.gov/
- 5.Felix AS, Linkov F, Maxwell GL, Ragin C, Taioli E. Racial disparities in risk of second primary cancers in endometrial cancer patients: Analysis of SEER data. Int J Gynecol Cancer. 2011;21:309–15. doi: 10.1097/IGC.0b013e318206a098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll R, Lawson AB, Jackson CL, Zhao S. Assessment of spatial variation in breast cancer-specific mortality using louisiana SEER data. Soc Sci Med. 2017;193:1–7. doi: 10.1016/j.socscimed.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder MC, Rastogi P, Geyer CE, Jr, Miller LD, Thomas A. Early and locally advanced metaplastic breast cancer: Presentation and survival by receptor status in surveillance, epidemiology, and end results (SEER) 2010-2014. Oncologist. 2018;23:481–8. doi: 10.1634/theoncologist.2017-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Chubak J, Boudreau DM, Barlow WE, Weiss NS, Li CI, et al. Use of antihypertensive medications and risk of adverse breast cancer outcomes in a SEER-medicare population. Cancer Epidemiol Biomarkers Prev. 2017;26:1603–10. doi: 10.1158/1055-9965.EPI-17-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ACS. How Common is Breast Cancer? 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.cancer.org/cancer/breast-cancer/about/how-common-isbreast-cancer.html .
- 10.DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A, et al. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 11.Henson KE, Jagsi R, Cutter D, McGale P, Taylor C, Darby SC, et al. Inferring the effects of cancer treatment: Divergent results from early breast cancer trialists’ collaborative group meta-analyses of randomized trials and observational data from SEER registries. J Clin Oncol. 2016;34:803–9. doi: 10.1200/JCO.2015.62.0294. [DOI] [PubMed] [Google Scholar]
- 12.Giordano SH, Cohen DS, Buzdar AU, Perkins G, Hortobagyi GN. Breast carcinoma in men: A population-based study. Cancer. 2004;101:51–7. doi: 10.1002/cncr.20312. [DOI] [PubMed] [Google Scholar]
- 13.Li CI, Uribe DJ, Daling JR. Clinical characteristics of different histologic types of breast cancer. Br J Cancer. 2005;93:1046–52. doi: 10.1038/sj.bjc.6602787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Text Information Extraction System (TIES) Software. [Last accessed on 2018 Jun 10]. Available from: http://www.ties.dbmi.pitt.edu/
- 15.Text Information Extraction System (TIES) Software. [Last accessed on 2018 Jun 10]. Available from: http://www.ties.dbmi.pitt.edu/
- 16.Jacobson RS, Becich MJ, Bollag RJ, Chavan G, Corrigan J, Dhir R, et al. Afederated network for translational cancer research using clinical data and biospecimens. Cancer Res. 2015;75:5194–201. doi: 10.1158/0008-5472.CAN-15-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie F, Lee J, Munoz-Plaza CE, Hahn EE, Chen W. Application of text information extraction system for real-time cancer case identification in an integrated healthcare organization. J Pathol Inform. 2017;8:48. doi: 10.4103/jpi.jpi_55_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.London WJ. Philadelphia, PA: IEEE; 2010. Using the Semantically Interoperable Biospecimen Repository Application, caTissue. 10th IEEE International Conference on Bioinformatics and Engineering. [Google Scholar]
- 19.NAACCR. North American Association of Central Cancer Registries. 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.naaccr.org/
- 20.Elekta. METRIQ Cancer Registry Data Management. 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.elekta.com/software-solutions/knowledge-management/registries/metriq/
- 21.NAACCR. Data Standards and Data Dictionary. [Last accessed on 2018 Jun 10];NAACCR. 2018 2 Available from: https://www.naaccr.org/data-standards-data-dictionary/ [Google Scholar]
- 22.NIH. SEER Training Modules. Introduction to Cancer Staging. [Last accessed on 2018 Jun 10]. Available from: https://www.training.seer.cancer.gov/staging/intro/
- 23.Collaborative Statge Data Collection System. 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.cancerstaging.org/cstage/about/Pages/default.aspx .
- 24.NCI. SEER API. 2018. [Last accessed on 2018 Jun 10]. Available from: https://www.api.seer.cancer.gov/
- 25.Izquierdo JN, Schoenbach VJ. The potential and limitations of data from population-based state cancer registries. Am J Public Health. 2000;90:695–8. doi: 10.2105/ajph.90.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alawad M, Yun HJ, Tourassi G. Energy Efficient Stochastic-Based Deep Spiking Neural Networks for Sparse Datasets. Paper Presented at: IEEE Explore. 2018 Mar 14; [Google Scholar]
- 27.Gao S, Young MT, Qiu JX, Yoon HJ, Christian JB, Fearn PA, et al. Hierarchical attention networks for information extraction from cancer pathology reports. J Am Med Inform Assoc. 2017;25:321–30. doi: 10.1093/jamia/ocx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu JX, Yoon HJ, Fearn PA, Tourassi GD. Deep learning for automated extraction of primary sites from cancer pathology reports. IEEE J Biomed Health Inform. 2018;22:244–51. doi: 10.1109/JBHI.2017.2700722. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen AN, Moore J, O’Dwyer J, Philpot S. Assessing the utility of automatic cancer registry notifications data extraction from free-text pathology reports. AMIA Annu Symp Proc 2015. 2015:953–62. [PMC free article] [PubMed] [Google Scholar]
- 30.TIES. License Types. 2018. [Last accessed on 2018 Oct 20]. Available from: http://www.ties.dbmi.pitt.edu/license/
- 31.TIES. SourceForge. 2018. [Last accessed on 2018 Oct 20]. Available from: https://www.sourceforge.net/projects/caties/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Common elements list