Abstract
MicroRNAs (miRNAs) may regulate diverse biological processes and play an important role in cancer. And MiRNAs have been proposed as a useful tool for lung cancer diagnosis and therapeutics in cancer. The purpose of the present study was to investigate the association among the expression level of mature miR-200b-5p in peripheral blood and the risk of lung cancer and clinic pathological characteristics. This case-control study included 24 patients with lung cancer and 12 healthy controls. MiR-200b expression was deleted using real-time PCR. and the miR-200b expression of normal controls was significantly higher than that in lung cancer patients (1732.13 pg/mL vs 881.67 pg/mL, P < 0.05), no difference with age, sex, tissue type and clinical stage of lung cancer patients (P > 0.05). Furthermore, miR-200b expression level fluctuated with tumor progression in lung cancer, and there was highly significant for clinical stage II compared with the clinical stage III (P < 0.05). In addition, the down-regulation of miR-200b showed a highly discriminative receiver operating characteristic (ROC) curve profile, clearly distinguishing cancer patients from cancer-free subjects with an area under the ROC curve (AUROC) of 0.87. The detection of miR-200b expression yielded 83.30% sensitivity and 100.00% specificity in the diagnosis of lung cancer. Therefore, these findings suggested that miR-200b may be used as a marker for the detection and diagnosis of lung cancer in peripheral blood.
Keywords: MiR-200b, Lung cancer, Prognosis, Molecular markers
1. Introduction
Lung cancer is a malignant tumor that occurs in the epithelium of bronchial mucosa (Torre et al., 2015), which is one of most commonly diagnosed cancers, with the highest incidence and mortality malignant tumors in the world and abroad (Siegel et al., 2018). It is significantly higher in developed countries than that in developing countries recently (Pratap et al., 2018, Melino et al., 2004). According to the World Health Organization statistics, China will become the largest country with lung cancer, and the new lung cancer cases will increase by more than 1 million per year. Lung cancer is a complex pathogenesis involving activation of many oncogenes and inactivation of tumor suppressor genes (Croce, 2009).
MicroRNAs (miRNAs) are a class of small RNAs with non-protein-coding, endogenous, highly conservative, approximately 18–25 nt in length, which also participate in various biological signaling pathways (Croce, 2009). Although biological functions of miRNAs remain largely unclear, many studies show that the miRNAs may play a great important role in regulating tumor formation and many physiological and pathological processes such as cell proliferation, apoptosis, tumor development as well (Yu et al., 2015, Pratap et al., 2018, Wozniak et al., 2015, Boeri et al., 2015, Ivey et al., 2008).
Altered expression of miR-200 are association with various the progress process of cancers (Muralidhar and Barbolina, 2015, Zhang et al., 2016, Zhang et al., 2014, Xie et al., 2010, Yu et al., 2013, Feng et al., 2014), MiR-200b is the one of miR-200 family with significantly highly expressive in the gastric cancer, breast cancer, ovarian cancer and other malignant tumors (Senfter et al., 2016, Chang et al., 2015, Chen et al., 2015, Liu et al., 2015, Tang et al., 2013). Tumor-suppressive effects of miR-200 family has been repeatedly tested in the recently years (Zhang et al., 2016, Zhang et al., 2014, Xie et al., 2010, Yu et al., 2013, Feng et al., 2014), which supports the role of these small molecules in the tumorigenic process and their potential use as a biomarker providing a new approach to the diagnosis of oncogenesis (Senfter et al., 2016, Chang et al., 2015, Chen et al., 2015, Liu et al., 2015, Tang et al., 2013). However, the application of miRNA expression pattern biomarkers remain in their infancy in cancers diagnosis and prognosis (Tang et al., 2013), the relationship between serum miR-200b levels and the clinical parameters and prognosis of lung cancer remains to be clarified in southern-central Chinese population. Therefore, the purpose of this study was to perform the possible association between the serum miR-200b levels and the clinical significance of lung cancer.
2. Materials and methods
2.1. Patients and samples
A case control was made up of 24 patients with lung cancer and 12 normal controls in peripheral blood. All 36 samples were from the Hunan Provincial Tumor Hospital during September 2014 to December 2015. At recruitment, the written informed consent was obtained from each subject. The methods were carried out in according to the approved guidelines. The research agreement was approved by the Institutional Review Board of the hospital, and the research was approved by the Ethics Committee of Hunan Cancer Institute.
2.2. RNA isolation and real-time PCR
MiRNAs of blood samples were extracted by using BioTeKe kit (BioTeKe, China), in accordance with the manufacturer’s protocol. The RNA samples were reversely transcribed using the AMV first strand cDNA synthesis Kit(Sangon Biotech (Shanghai) Co., Ltd.) at 42 °C for 60 min and 85 °C for 5 min. Real-time PCR was performed to measure expressions of target miRNAs by using MicroRNAs qPCR Kit (SYBR Green Method) (Sangon Biotech (Shanghai) Co., Ltd.) on a Bio-Red IQ2 Real-time PCR Detection System (Bio-Red, Hercules, CA) (Li et al., 2011). The PCR conditions were set as follows: an initial denaturation at 95 °C for 7 min; 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold. Expression of target has-miR-200b-5p and corresponding primers were designed and synthesized by RiBo Biotech (Guangzhou) Co., Ltd. All assays were carried out in triplicates.
2.3. Statistical analysis
All data analysis was handled by using SPSS19.0 software (SPSS Inc. Chicago, IL, USA). The receiver operating characteristic curve (ROC) was applied to describe the sensitivity and specificity, and the area under the ROC curve was used as a standard to judge the diagnostic value. All tests of statistical significance were two-sided (Li et al., 2011, Hanley and McNeil, 1982). P-values < 0.05 were defined as statistical significance.
3. Results
3.1. MicroRNA-200b expression profile
RNA was extracted from peripheral blood and the mean ratio of absorbance at 280 and 260 nm was 1.84 (range 1.72–2.15 ± 0.14). Synthesized has-miR-200b-5p mimics were diluted by 10 orders of magnitude in DEPC-treated H2O to generate a standard curve, and the qRT-PCR assay revealed excellent linearity between the log of target input and the Ct value, which demonstrated that the assay had a strong range of at least 4 orders of magnitude (R2 = 0.9990) [Fig. 1(A and B)] and the concentration of miR-200b in all samples can be derived by the threshold cycle (Ct) in the standard curve. The 12 normal controls had high-level expression of miR-200b (1732.13 ± 618.68 pg/mL), while 24 lung cancer patients had low-level expression of miR-200b (881.67 ± 288.19 pg/mL) (Fig. 2), and significantly a higher difference existed in normal controls than lung cancer (P < 0.05).
Fig.1.
Real-time fluorescence quantitative PCR detection of miR-200b Standard. (A: Real-time fluorescence amplification curve; B: Standard curve).
Fig. 2.
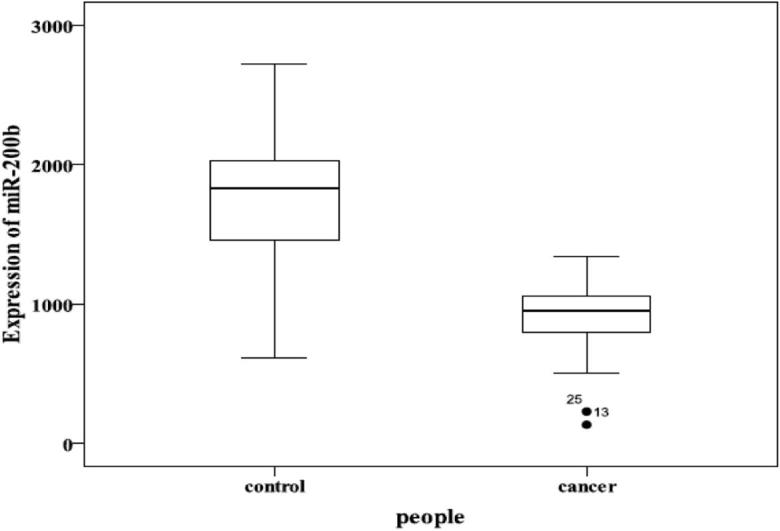
Comparison of miR-200b expression levels in normal and lung cancer blood samples.
3.2. Sensitivity, specificity and dynamic range of miRNA200b quantification in peripheral blood
Receiver-operator characteristic (ROC) curve analysis and areas under the ROC curve (AUROCs) were used to determine sample size in cancer patients and normal controls by SPSS19.0 (Li et al., 2011). And it was performed to analyze and evaluate the classification effect. In this way, AUROCs identified optimal sensitivity and specificity levels at which could be used to differentiate normal individuals from cancer patients (Li et al., 2011). ROC curves together with AUROCs for miR-200b expressions in blood between lung cancer and control cases were shown in Fig. 3. The figure showed highly discriminative receiver-operator characteristic (ROC) curve profile, clearly distinguishing cancer patients from cancer-free subjects with areas under the ROC curve at 0.87 ± 0.09. The detection of expression level of miR200b generated a 83.30% sensitivity and 100.00% specificity in the diagnosis of lung cancer. It was indicated that miR-200b could be used as a potential molecular biomarker to the diagnosis of oncogenesis (Fig. 3).
Fig. 3.
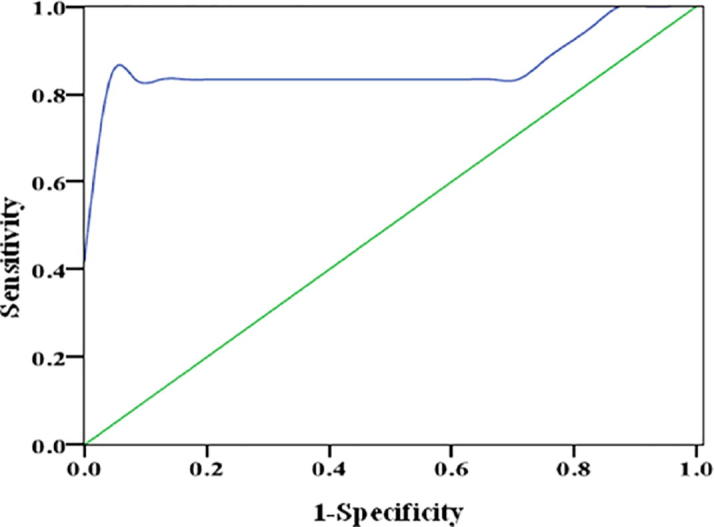
ROC curve analysis of miR-200b quantitative results.
3.3. Comparison analysis of miR-200b expression level and clinic pathologic characteristics
Real-time PCR method was used to detect the expressions of miR-200b in the peripheral blood samples between 24 patients and 12 normal subjects. And the results were shown in Table 1. The expression level of miR-200b was significantly higher difference in normal controls than lung cancer (P < 0.05). No association was found between the expression levels of miR-200b and clinicopathological characteristics (clinical stage, age, gender and cell type) (Hanley and McNeil, 1982).
Table 1.
Analysis of clinicopathological characteristics for miR-200b in lung cancer.
| Characteristics | Patients |
Mean ± SD(pg/ml) | P-value | |
|---|---|---|---|---|
| No. | % | |||
| Total | 24 | 100 | ||
| Age | ||||
| ≥60 | 16 | 66.67 | 843.17 ± 320.98 | 0.37 |
| <60 | 8 | 33.33 | 958.67 ± 204.83 | |
| Gender | ||||
| Male | 18 | 75.00 | 838.84 ± 307.73 | 0.21 |
| Female | 6 | 25.00 | 1010.17 ± 183.44 | |
| Cell type | ||||
| Squamous cell carcinoma | 13 | 54.17 | 881.50 ± 301.35 | 0.92 |
| Adenocarcinoma | 8 | 33.33 | 856.98 ± 320.01 | |
| Other carcinomas | 3 | 12.50 | 948.26 ± 206.73 | |
| Clinical stage | ||||
| I | 6 | 25.00 | 818.66 ± 341.26 | 0.13 |
| II | 6 | 25.00 | 1050.11 ± 208.07 | |
| III | 6 | 25.00 | 948.22 ± 238.50 | |
| IV | 6 | 25.00 | 709.69 ± 292.48 | |
3.4. The multiple comparison of miR-200b expression level with clinical stage
The correlation between miR-200b expression level and different clinical stage was investigated as a pathway to understand the role of miR-200b in lung cancer. The multiple comparison of miR-200b expression level with clinical stage was used by Chisquare partitioning method. The results were listed in Table 2. And the level of miR-200b expression was highly significant for clinical stage Ⅱ comparing with the clinical stage III (P < 0.05), and there were no association among each other comparisons with the level of miR-200b expression (P > 0.05).
Table 2.
The multiple comparison of miR-200b expression with clinical stage in lung cancer.
| I | II | III | IV | |
|---|---|---|---|---|
| I | \ | 0.19 | 0.46 | 0.57 |
| II | \ | \ | 0.45 | 0.04 |
| III | \ | \ | \ | 0.15 |
4. Discussion
The generation and development of lung cancer is a complex mechanism of multi-step and multi-gene combination. More and more studies have shown that the abnormal expression of miRNAs is closely related to the occurrence, development, invasion and metastasis of malignant tumors (Muralidhar and Barbolina, 2015, Yu et al., 2013, Chen et al., 2015, Weber et al., 2010). Therefore, it is necessary to identify and employ novel biomarkers as possible therapeutic targets or prognostic predictors, which will be used as an adjunct to the staging system and contribute to the optimization to cure lung cancer patients (Men et al., 2014).
The miRNA-200 family is involved in the regulation of tumor epithelial-mesenchymal transition (EMT) (Shang et al., 2017, Zubakov et al., 2010). And miR-200b is a member of the miR-200 family, which is located on chromosome 1p36 and is closely associated with carcinogenesis and disease progression in a wide range of cancer types (Yu et al., 2013, Feng et al., 2014, Men et al., 2014, Shang et al., 2017, Zubakov et al., 2010). And miR-200 may play an important role in tumorgenesis (Yu et al., 2013, Feng et al., 2014, Men et al., 2014, Shang et al., 2017, Zubakov et al., 2010, Hanson et al., 2009). And miR-200b is the one of miR-200 family with significantly highly expressive in the gastric cancer, breast cancer, ovarian cancer and other malignant tumors (Senfter et al., 2016, Chang et al., 2015, Chen et al., 2015, Liu et al., 2015). However, some reporters were shown that the miR-200b expression level was markedly declined in gastric cancer and breast cancer (Li et al., 2011). At present, miR-200b expression profiles were detected by qRT-PCR assay in blood with lung cancer. The expression level of miR-200b was 1732.13 pg/ml in controls and 881.67 pg/ml in lung cancer patients. A significant difference in the expression levels of miR-200b was detected between lung cancer patients and controls (P < 0.05). This process activates miR-200b down-regulation which is involved in many pathways, especially in tumourogenesis, such as cell proliferation, apoptosis, glucose metabolism and energy metabolism, and the microenvironment of tumor was also changed (Hanson et al., 2009, Davey et al., 2011), The assessment of miR-200b expression may play a significant effect on the development and serve as a potential diagnostic biomarker for lung cancer patients.
The results were shown that no association was between the expression levels of miR-200b in peripheral blood of non-small cell lung cancer patients and the various clinic pathological characteristics (clinical stage, age, gender and cell type), suggesting that the change of expression level was not specific to histologic type (P = 0.92) and clinical stage (P = 0.13). Similar results were obtained from age and gender. However, the expression level of miR-200b was actively correlated with the clinical stage and cell type. The expression level of clinical stage Ⅱ was higher than that of other three stages, and there was greatly significant for clinical stage Ⅱ comparing with the clinical stage IV (1050.11 vs 709.69, P < 0.05), and the expression level of adenocarcinoma was lower than that of other two cell types. Further studies would be necessary and important to prove our findings and to find the role of miR-200b as a creditable clinical molecular marker for the outcome of lung cancer patients because of the small sample size of patients in our investigation (Li et al., 2010).
Many scientific researchers have shown that the microRNA expression is used to diagnose different tumors as a powerful biomarker in the peripheral blood (Zuberi et al., 2015, Halvorsen et al., 2016), and 83.30% sensitivity rate is not sufficiently efficient for routine clinical application. Less sensitive may be considered because of lower expression in serum concentrations, and therefore more clinical specimens and experiments are needed to further confirm. Once confirmed, the expression level of miR-200b could be used as a potential molecular biomarker for lung cancer in peripheral blood.
All in all, although this study only used a small sample size, the miR-200b expression levels have significant differences between lung cancer patients and normal subjects with a high specificity in Southern-central Chinese population.
Peripheral blood miR-200b is expected to use as a marker for the detection and diagnosis of lung cancer and provide a new method in lung cancer diagnosis.
Acknowledgements
This work was supported by the National Key Technology R&D Program (2015BAD05B02), the National Natural Science Foundation of China (31501538, 51774128 and 21605046), the Natural Science Foundation of Hunan Province of China (2015JJ2049, 2015JJ3062, 2016JJ3053, 2017JJ4032, 2018JJ4061, 2018JJ2090 and 2018JJ4009), the Key Program of Hunan Provincial Department of Science and Technology (2016NK2096), Huxiang Youth Talent Support Program (2015RS4051),the Scientific Research Fund of Hunan Provincial Education Department (16C0470, YB2016B034, 17A055 and 17C0487), the China of Postdoctoral Science Foundation (2016T90769, 2016M592456, 2015M580707), Zhu zhou Key Science & Technology Program of Hunan Province (2017 and 2018), the China of Undergraduate Innovative Experiment Program (201511535003), Hunan Province Undergraduate Innovative Experiment Program (2015) and Green Packaging and Security Special Research Fund of China Packaging Federation (2017ZBLY14).
Footnotes
Peer review under responsibility of King Saud University.
References
- Boeri M., Sestini S., Fortunato O., Verri C., Suatoni P., Pastorino U., Sozzi G. Recent advances of microRNA-based molecular diagnostics to reduce false-positive lung cancer imaging. Expert Rev. Mol. Diagn. 2015;15(6):801–811. doi: 10.1586/14737159.2015.1041377. [DOI] [PubMed] [Google Scholar]
- Chang L., Guo F., Huo B., Lv Y., Wang Y., Liu W. Expression and clinical significance of the microRNA-200 family in gastric cancer. Oncol Lett. 2015;9(5):2317–2324. doi: 10.3892/ol.2015.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Yang D., Wang Q., Wang X. Expression and clinical pathological significance of miR-200a in concurrent cholangiocarcinoma associated with hepatolithiasis. Med. Sci. Monit. 2015;20(21):3585–3590. doi: 10.12659/MSM.895013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C.M. Causes and consequences of microRNA dys-regulation in cancer. Nat. Rev. Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J.W., Hohenlohe P.A., Etter P.D., Boone J.Q., Catchen J.M., Blaxter M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011;12(7):499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Feng X., Wang Z., Fillmore R., Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344(2):166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen A.R., Bjaanæs M., Leblanc M., Holm A.M., Bolstad N., Rubio L., Peñalver J.C., Cervera J., Mojarrieta J.C., López-Guerrero J.A., Brustugun O.T., Helland Å. A unique set of 6 circulating micrornas for early detection of non-small cell lung cancer. Oncotarget. 2016;7(24):37250–37259. doi: 10.18632/oncotarget.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley J.A., McNeil B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- Hanson E.K., Lubenow H., Ballantyne J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal. Biochem. 2009;387(2):303–314. doi: 10.1016/j.ab.2009.01.037. [DOI] [PubMed] [Google Scholar]
- Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E., Hsiao E.C., Schwartz R.J., Conklin B.R., Bernstein H.S., Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Deng J., Jiang P., Tang J. Association of 5'-CpG island hypermethylation of the FHIT gene with lung cancer in southern-central Chinese population. Cancer Biol. Ther. 2010;10(10):997–1000. doi: 10.4161/cbt.10.10.13231. [DOI] [PubMed] [Google Scholar]
- Li Y., Wen L., Ouyang Q., Hu S., Tang J. Detection of lung cancer with blood microrna-21 expression levels in chinese population. Oncol. Lett. 2011;2(5):991–994. doi: 10.3892/ol.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhong L., Zeng J., Zhang X., Yang Q., Liao D., Wang Y., Chen G., Wang Y. Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer. Neoplasma. 2015;62(4):550–559. doi: 10.4149/neo_2015_066. [DOI] [PubMed] [Google Scholar]
- Melino G., Bernassola F., Ranalli M., Yee K., Zong W.X., Corazzari M., Knight R.A., Green D.R., Thompson C., Vousden K.H. p73 induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 2004;279(9):8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- Men D., Liang Y., Chen L. Decreased expression of microRNA-200b is an independent unfavorable prognostic factor for glioma patients. Cancer Epidemiol. 2014;38(2):152–156. doi: 10.1016/j.canep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Muralidhar G.G., Barbolina M.V. The miR-200 family: versatile players in epithelial ovarian cancer. Int. J. Mol. Sci. 2015;16(8):16833–16847. doi: 10.3390/ijms160816833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap P., Raza S.T., Abbas S., Mahdi F. MicroRNA-associated carcinogenesis in lung carcinoma. J. Cancer Res. Ther. 2018;14(2):249–254. doi: 10.4103/0973-1482.187283. [DOI] [PubMed] [Google Scholar]
- Senfter D., Madlener S., Krupitza G., Mader R.M. The microRNA- 200 family: still much to discover. Biomol. Concepts. 2016;7(5–6):311–319. doi: 10.1515/bmc-2016-0020. [DOI] [PubMed] [Google Scholar]
- Shang Y., Chen H., Ye J., Wei X., Liu S., Wang R. HIF-1α/Ascl2/miR-200b regulatory feedback circuit modulated the (EMT) in colorectal cancer cells. Exp. Cell Res. 2017;360(2):243–256. doi: 10.1016/j.yexcr.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA: Cancer J. Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Tang H., Deng M., Tang Y., Xie X., Guo J., Kong Y., Ye F., Su Q., Xie X. miR-200B and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin. Cancer Res. 2013;19(20):5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA: Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak M.B., Scelo G., Muller D.C., Mukeria A2., Zaridze D., Brennan P. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0125026. e0125026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Todd N.W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S.A., Jiang F. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tan Q., Deng B., Fang C., Qi D., Wang R. The microRNA-520a-3p inhibits proliferation, apoptosis and metastasis by targeting MAP3K2 in non-small cell lung cancer. Am. J. Cancer Res. 2015;5(2):802–811. [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wu J., Guan L., Ma B., Zhan J., Wang Y., Fang W., Zhang H. Kindlin 2 promotes breast cancer invasion via epigenetic silencing of the microRNA200 gene family. Int. J. Cancer. 2013;133(6):1368–1379. doi: 10.1002/ijc.28151. [DOI] [PubMed] [Google Scholar]
- Zhang H.F., Alshareef A., Wu C., Jiao J.W., Sorensen P.H., Lai R., Xu L.Y., Li E.M. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37(9):858–869. doi: 10.1093/carcin/bgw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.F., Zhang K., Liao L.D., Li L.Y., Du Z.P., Wu B.L., Wu J.Y., Xu X.E., Zeng F.M., Chen B., Cao H.H., Zhu M.X., Dai L.H., Long L., Wu Z.Y., Lai R., Xu L.Y., Li E.M. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis. 2014;35(2):292–301. doi: 10.1093/carcin/bgt320. [DOI] [PubMed] [Google Scholar]
- Zubakov D., Boersma A.W., Choi Y., Kuijk P.F., Wiemer E.A., Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Legal Med. 2010;124(3):217–226. doi: 10.1007/s00414-009-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi M., Mir R., Das J., Ahmad I., Javid J., Yadav P., Masroor M., Ahmad S., Ray P.C., Saxena A. Erratum to: expression of serum mir-200a, mir-200b and mir-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 2015;17(10):779–787. doi: 10.1007/s12094-015-1303-1. [DOI] [PubMed] [Google Scholar]



