Abstract
Induction of interferons during viral infection is mediated by cellular proteins that recognise viral nucleic acids. MDA5 is one such sensor of virus presence and is activated by RNA. MDA5 is required for immunity against several classes of viruses, including picornaviruses. Recent work showed that mutations in the IFIH1 gene, encoding MDA5, lead to interferon-driven autoinflammatory diseases. Together with observations made in cancer cells, this suggests that MDA5 detects cellular RNAs in addition to viral RNAs. It is therefore important to understand the properties of the RNAs which activate MDA5. New data indicate that RNA length and secondary structure are features sensed by MDA5. We review these developments and discuss how MDA5 strikes a balance between antiviral immunity and autoinflammation.
Keywords: MDA5, IFIH1, RNA, type I interferon, antiviral defence, autoinflammation
MDA5, a Key RNA Sensor
Mammalian cells use pattern-recognition receptors (PRRs) to detect the presence of infectious microorganisms [1]. These receptors are activated by pathogen-associated molecular patterns (PAMPs). In the case of virus infection, PAMPs are often nucleic acids 2, 3. For example, viral RNAs trigger PRRs, including the endosomal toll-like receptors (TLRs) 3 and 7 and the cytosolic RIG-I-like receptors (RLRs) 2, 3. The RLR family comprises three members: retinoic acid inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) 2, 3. These proteins are ubiquitously expressed at low levels. All RLRs contain a DExD/H-box RNA helicase domain and a C terminal domain (CTD), both responsible for RNA binding [4]. In addition, RIG-I and MDA5 have two N terminal caspase activation and recruitment domains (CARDs). Upon activation of RLRs by RNA binding, the CARDs interact with the adaptor mitochondrial antiviral signalling protein (MAVS), ultimately leading to the transcription of the genes encoding type I interferons (IFNs) (see Glossary) 2, 3, 4 (Figure 1 ). Autocrine and paracrine IFN stimulation subsequently induces transcription of hundreds of IFN-stimulated genes (ISGs), several of which encode proteins with direct antiviral functions [5]. RLRs themselves are encoded by ISGs, constituting a feed-forward loop. Type I IFNs further coordinate cellular immune responses to virus infection and are thus essential for antiviral immunity [6]. In addition to inducing type I IFNs, RLRs and MAVS also activate apoptosis, leading to the elimination of the infected cell [7].
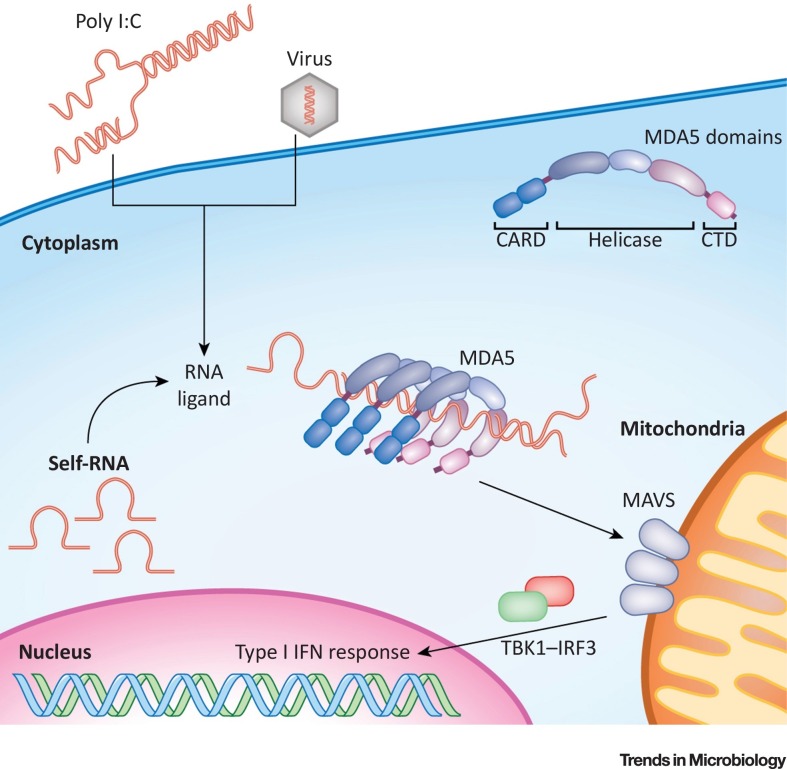
Figure 1.
Overview of the MDA5 Pathway. MDA5 comprises two N terminal CARD domains, a central helicase domain, and a C terminal domain (CTD). Both the helicase domain and the CTD are involved in RNA binding. MDA5 recognises synthetic RNAs, such as poly I:C, viral RNAs, and endogenous RNAs. Length and secondary structure are likely to be key determinants of MDA5 activation, which has been suggested to be mediated by multimerisation of the protein on RNA agonists. This results in the clustering of multiple CARD domains, which, in turn, allows the engagement of MAVS. Activation of this adaptor protein triggers a signalling cascade involving TBK1 and IRF3, leading to transcriptional induction of the genes encoding type I IFNs and other antiviral genes. CARD, caspase activation and recruitment domain; MAVS, mitochondrial antiviral signalling protein.
Given the abundance of nucleic acids in healthy cells, a key question is to understand the mechanisms by which nucleic acid-sensing PRRs become activated specifically following virus infection. The features of RIG-I-stimulatory RNAs are well understood and include the presence of two or three phosphate groups at the 5′-end, the absence of 5′-cap methylation, and base-pairing adjacent to the 5′-end 8, 9, 10, 11, 12, 13, 14, 15. These features are characteristic of viral RNAs produced by some viruses, such as influenza A virus or reovirus 8, 9, 12, but are not typically found in cellular RNAs present in the cytosol of healthy cells, explaining selective activation of RIG-I in virus-infected cells. It is also possible that, in some settings, mislocalised cellular RNAs activate RIG-I and other nucleic acid sensors of the innate immune system 16, 17. Although RIG-I and MDA5 share similar domains, they detect different viral infections [18]. For example, RIG-I detects infection with orthomyxoviruses, such as influenza A virus, while MDA5 senses picornavirus infection. In contrast to RIG-I, the mechanisms that allow MDA5 to recognise viral RNAs while avoiding cellular RNAs are less well understood. Biochemical and structural work using recombinant MDA5 protein demonstrated that MDA5 senses RNA length and secondary structure 19, 20, 21. Understanding molecular features of MDA5-stimulatory RNAs is important for another reason: in addition to its protective role in antiviral defence, MDA5 has been implicated in autoimmune and autoinflammatory diseases such as type 1 diabetes (T1D), systemic lupus erythematosus (SLE) and Aicardi–Goutières syndrome (AGS) 2, 3, 22. Moreover, new data indicate that MDA5 is activated during some forms of cancer treatment 23, 24 or in settings where mitochondrial RNA degradation is compromised [17]. These observations highlight that, in some circumstances, cellular RNAs trigger MDA5. Indeed, transcripts from repetitive genome segments, including endogenous retroelements, such as Alu elements, and mitochondrial RNA have been suggested to bind and activate MDA5 17, 23, 24, 25, 26. Here, we review RNA sensing by MDA5 in the context of antiviral immunity and autoinflammation. We discuss how important recent developments in this area set the stage for future exploration of MDA5-RNA interactions in living cell systems, including models of authentic virus infection and autoinflammation.
The Discovery of MDA5
MDA5 was first described in 2002 as a helicase protein in mouse and human cells 27, 28. Interestingly, these initial reports suggested that MDA5 is involved in the execution of apoptosis. In 2004, Rick Randall’s group found that overexpression of MDA5 alone induces the expression of IFN-β, a type I IFN [29]. Furthermore, this study showed that the IFN-β response of cells stimulated by transfection of polyriboinosinic:polyribocytidylic acid (poly I:C), a synthetic RNA, is greatly enhanced by MDA5 overexpression [29]. Shortly afterwards, knockouts of the mouse Ifih1 gene encoding MDA5 demonstrated that MDA5 is essential for the type I IFN response to poly I:C 18, 30. Together, these landmark studies established MDA5 as an RNA sensor inducing type I IFN (Figure 1).
Activation of MDA5 by Synthetic RNAs
Poly I:C is often used as a synthetic mimic of double-stranded RNA (dsRNA). This led to the hypothesis that the PAMP recognised by MDA5 is the double-stranded conformation of RNA. Indeed, structural studies of MDA5 show that the protein adopts a ring-like conformation around dsRNA [21]. Contacts between MDA5 and dsRNA are along the phosphodiester backbone, suggestive of sequence-nonspecific binding [21]. Experiments in the test tube using recombinant MDA5 and in vitro transcribed dsRNAs established that MDA5 forms filaments along dsRNA 19, 31. These filaments are particularly stable on long dsRNA molecules and are mediated by both protein–RNA and protein–protein interactions 20, 21. These biophysical studies used filament formation and ATP hydrolysis by the helicase domain of MDA5 as surrogates for its activation. Consistent with these data is the finding in cells that the IFN response to transfected poly I:C depends on length: long poly I:C molecules preferentially trigger MDA5, whereas short poly I:C activates RIG-I [32]. Together, these observations have led to the widely held notion that MDA5 detects long dsRNA. However, other observations suggest that dsRNA – that is, two complementary RNA molecules annealed to form a helical A-form duplex – may not always be sufficient to explain activation of MDA5. For example, IFN induction is particularly strong in response to poly I:C, while other dsRNAs such as poly A:U, trigger no response 33, 34. It should nevertheless be noted that, compared with G:C and I:C duplexes, A:U duplexes have a lower stability, which could explain their lower signalling activity. It is also noteworthy that poly I:C consists of annealed inosine and cytidine homopolymers. These are produced enzymatically from ribonucleoside diphosphates, using polynucleotide phosphorylase, and are heterogeneous in length [35]. Annealing thus results in double-stranded regions with single-stranded overhangs. These are available for base-pairing with other molecules, potentially resulting in the formation of more complex, branched RNA structures, which have been proposed to play a role in MDA5 activation [34].
RNA Sensing by MDA5 during Virus Infection
Early studies in MDA5-deficient mice reported that these animals are highly susceptible to infection with encephalomyocarditis virus (EMCV), failing to induce type I IFNs 18, 30. More recently, MDA5 deficiency in humans has been shown to increase susceptibility to viral infection 36, 37, 38. EMCV is a widely used model belonging to the Picornaviridae family that includes important human pathogens such as hepatitis A virus, coxsackie B virus, enterovirus, and rhinovirus. As summarised in Table 1 , subsequent work by many laboratories revealed that MDA5 is involved in type I IFN induction during infection with several other types of viruses. This includes virus families characterised by genomes consisting of single-stranded (ss), positive- or negative-sense RNA, dsRNA, and dsDNA. However, in contrast to EMCV infection, in which MDA5 is essential for type I IFN induction, MDA5 plays a partial role in other infections (Table 1). For example, dsDNA viruses are also detected by the cytosolic DNA sensing pathway, and some RNA viruses trigger both MDA5 and RIG-I.
Table 1.
Viral Infections Detected by MDA5
| Virus family | Genome | Examples | Role of MDA5 in type I IFN induction | Selected Refs |
|---|---|---|---|---|
| Picornaviridae | ssRNA (+) | Encephalomyocarditis virus; Rhinovirus; Coxsackie B virus | Essential | 18, 30, 43, 102, 103 |
| Flaviviridae | ssRNA (+) | West Nile virus; Hepatitis C virus; Zika virus | Partial | 44, 104, 105 |
| Togaviridae | ssRNA (+) | Sindbis virus | Partial | 34, 106, 107 |
| Coronaviridae | ssRNA (+) | SARS coronavirus | Partial | 57, 108, 109 |
| Paramyxoviridae | ssRNA (−) | Measles virus; human Metapneumovirus; Sendai virusa | Partial | 110, 111, 112 |
| Reoviridae | dsRNA | Rotavirus | Partial | 8, 32, 113 |
| Poxviridae | dsDNA | Vaccinia virus | Partial | 34, 114 |
| Herpesviridae | dsDNA | Herpes simplex virus 1 | Partial | [115] |
| Hepadnaviridae | dsDNA | Hepatitis B virusb | Partial | [116] |
Some Sendai virus stocks, particularly the Cantell strain, activate mostly RIG-I.
Hepatitis D virus, a satellite virus that only infects HBV-infected cells and has a circular, ssRNA(−) genome, is also sensed by MDA5 [117].
Immunofluorescence experiments using monoclonal antibodies, called J2 and K1, that recognise dsRNA [39] revealed strong staining in cells infected with many of the viruses that activate MDA5 34, 40, 41, 42. In the case of ssRNA viruses, this dsRNA is likely to be generated during replication of the viral genome, which involves the synthesis of a complementary RNA molecule. For dsDNA viruses, overlapping transcription of both the positive and negative DNA strand may generate dsRNA [42].
Since many viruses have evolved mechanisms to inhibit nucleic acid sensors, one protocol to study immunostimulatory RNAs is to isolate total RNA from virus-infected cells, followed by retransfection of this sample containing both cellular and viral RNAs into uninfected reporter cells. The transfected RNA then stimulates innate immune receptors in recipient cells without the presence of inhibitory viral mechanisms. For example, total RNA from cells infected with picornaviruses or Zika virus triggers MDA5-dependent IFN responses 34, 43, 44. Fractionation of the RNA isolated from infected cells can be used to further characterise MDA5 agonists. Using two picornaviruses, Mengo virus and Coxsackievirus B3, Feng at al. showed that RNA from a band containing long viral dsRNA, called the replicative form, triggers MDA5 [43], and similar results were reported by Triantafilou et al. [41]. In another study using EMCV, fractionation revealed that the activity stimulating MDA5 is not found in a dsRNA band specifically present in infected cells [34]. Instead, MDA5-stimulatory RNA was recovered from a high-molecular-weight fraction containing both ssRNA and dsRNA, and similar observations were made analysing RNA from vaccinia virus-infected cells [34].
Although a useful approach, transfecting total RNA from infected cells has limitations; for example, the role of RNA-binding proteins may be overlooked. In order to directly study MDA5-stimulatory RNAs generated during virus infection, a number of studies attempted to purify these RNAs from infected cells using different pull-down approaches 45, 46, 47. Indeed, immunoprecipitations (IPs) have been used successfully to identify RIG-I-associated RNAs 8, 9, 12, 16, 48. However, in contrast to RIG-I, we and others found that similar protocols using native conditions for IP of MDA5 do not copurify IFN-stimulatory RNAs 25, 47. One possible explanation for this observation is that MDA5 forms multimers on its natural RNA agonists, as has been reported for synthetic RNAs, and that such MDA5 filaments are unstable after cell lysis 19, 20, 25. To circumvent this technical obstacle, one study used photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation (PAR-CLIP) [46]. This method involves incubating cells with modified nucleosides, which are incorporated into newly synthesised RNA and allow UV-A-induced cross-linking of the RNA to nearby proteins [49]. Using antibodies against endogenous MDA5, Runge et al. isolated IFN-stimulatory RNAs from cells infected with measles virus (MV, a paramyxovirus) [46]. Sequencing of these RNAs revealed an enrichment of AU-rich sequences derived from mRNA of the MV L gene [46]. A later study using a streptavidin-based pull-down approach and native conditions, however, concluded that MDA5 binds to the MV N mRNA [45] and did not confirm the preference of MDA5 for AU-rich sequences observed with PAR-CLIP 45, 46. The reason for these different results is currently unclear and may relate to the different experimental methods used. Another approach to purify MDA5-stimulatory RNAs from virus-infected cells utilised IP of LGP2 [47]. This RLR lacks CARD domains and plays a dual role regulating both RNA interference by association with Dicer [50] as well as RIG-I and MDA5 signalling [51]. Indeed, LGP2 may enhance MDA5-dependent type I IFN responses by facilitating the access of MDA5 to RNA agonists 52, 53. IP of LGP2 from EMCV-infected cells under native conditions captures an RNA that activates MDA5 upon retransfection into reporter cells [47]. Deep sequencing of this RNA revealed an enrichment of negative-sense RNA corresponding to the L region of the EMCV genome [47]. Further experiments showed that in vitro transcribed L antisense RNA activates MDA5, and that the L region is required for type I IFN induction following EMCV infection in cells and mice [47].
Despite the differences in methodologies employed and results obtained, all of these studies find that MDA5-stimulatory RNAs are derived from one strand of the investigated viral sequences, namely, positive-sense RNA for MV 45, 46 and negative-sense RNA for EMCV [47]. Although a degree of intramolecular base-pairing is conceivable, these findings are consistent with the notion that physiological MDA5 agonists may not always simply correspond to viral dsRNA. Instead, dsRNA – as a product of infection – might be a correlate of MDA5 activation 34, 40, 42. We speculate that a more complex RNA motif or structure may activate MDA5 in some settings. Further studies are needed to identify and characterise this viral PAMP detected by MDA5 with precision. The recently described MDA5-RNase-protection assay [25], and advances in CLIP technologies [49], are likely to facilitate research into this question. Although CLIP analysis reveals both activating and nonactivating RNA ligands, it may be particularly informative as it allows interrogation of MDA5-associated RNAs in virus infected cells. This is important because viral RNAs are typically coated by viral proteins, and RLRs have been suggested to compete with viral RNA-binding proteins [54]. As such, the actual RNA agonist recognised by MDA5 in infected cells is likely to be determined by a combination of factors, including (i) the RNA binding preferences of MDA5, (ii) accessory host factors such as LGP2 [53], DHX29 [55], and LncITPRIP-1 [56] that facilitate RNA binding by MDA5, (iii) the abundance and properties of viral RNA-binding proteins [54], and (iv) viral evasion strategies such as RNA methylation [57] and RNA degradation [58]. Another interesting avenue for future research is to study at the subcellular level MDA5 activation, the formation of aggregates or filaments, and interaction with MAVS. For example, the subcellular localisation of MAVS suggests that active MDA5 signalling complexes may be formed on mitochondria and/or peroxisomes 59, 60, 61. Super-resolution and live-cell microscopy methods should be utilised to address these questions.
Activation of MDA5 in Autoimmune and Autoinflammatory Diseases
In addition to its role in the detection of viral infections, MDA5 has also been implicated in a number of sterile, inflammatory conditions. For example, single-nucleotide polymorphisms (SNPs) in IFIH1, the gene encoding MDA5, have been associated with T1D, psoriasis, rheumatoid arthritis, vitiligo, multiple sclerosis, and SLE 62, 63, 64, 65, 66, 67, 68. How IFIH1 risk alleles relate to the development of these diseases is not fully understood but it is likely that this involves chronic induction of type I IFNs, which then initiate or enhance autoinflammation and autoimmune responses. In the context of T1D, the IFIH1 risk alleles may result in increased MDA5 protein levels or increased responses to RNA derived from cellular sources or viral infection 69, 70, 71. Alternatively, it is possible that some IFIH1 SNPs change the conformation of MDA5 such that the protein becomes constitutively active, irrespective of RNA binding [72].
IFIH1 mutations can also cause a number of rare disorders characterised by aberrant type I IFN production, some of which are monogenic diseases and, as such, are good models for molecular studies [22]. This includes Singleton–Merten syndrome [73] and AGS 74, 75. AGS is characterised by severe neurological dysfunction and often results in death in childhood [76]. Type I IFNs are detectable in cerebrospinal fluid and in peripheral blood [77]. The AGS-associated IFIH1 mutations result in substitutions of single amino acids in the helicase domain of MDA5 74, 75. How do these missense mutations lead to chronic type I IFN production? In overexpression assays, mutated versions of MDA5 are more potent than wild-type MDA5 in activating the IFN-β promoter in the absence of an exogenous stimulus 74, 75. Two explanations have been proposed for this. In one, mutated MDA5 signals constitutively in an RNA-independent manner [75]. Indeed, Oda et al. find that type I IFN induction following EMCV infection is not enhanced by expression of mutated MDA5 [75], and similar observations were reported in a mouse model in which an Ifih1 mutation leads to type I IFN induction [72]. The other explanation suggests that MDA5 mutations alter the interaction between the protein and RNA, resulting in spontaneous signalling triggered by cellular RNAs. In line with the idea of enhanced RNA sensing, Rice et al. found that overexpression of mutated MDA5 in cell lines potentiates the type I IFN response to transfected synthetic RNA agonists [74]. Furthermore, coexpression of viral RNA-binding proteins prevents the response triggered by overexpression of MDA5 without additional stimulation [25]. This observation suggests that mutated MDA5 aberrantly detects cellular RNAs.
In order to identify cellular RNAs that stimulate mutant MDA5, Ahmad et al. recently described an RNase protection assay where total RNA extracted from cells is mixed in the test tube with recombinant MDA5 protein bearing a mutation in its helicase domain, which had been identified in an AGS patient 25, 74. RNA not bound by MDA5 is then degraded with RNase A and any remaining material is sequenced [25]. This analysis revealed that RNA derived from Alu elements arranged as inverted repeats (IRs) is bound by MDA5 [25]. Transcription of IR-Alu elements generates single-stranded RNA molecules with extensive self-complementarity, resulting in the formation of a long, base-paired stem. In cell-free systems, mutant MDA5 forms filaments on synthetic IR-Alu RNAs, which also stimulate its ATPase activity and MDA5-dependent dimerisation of a key downstream transcription factor, IRF3 [25]. Ahmad et al. further propose that the presence of mismatched bases in RNA stem structures formed by IR-Alu elements prevents recognition by wild-type MDA5 [25]. These elegant experiments provide a molecular explanation for type I IFN induction in cells from AGS patients: mutant MDA5 is less selective than wild-type MDA5 in terms of RNA binding and associates with base-paired RNA stems formed by IR-Alu elements containing bulges and mismatches [25]. Furthermore, these data lend support to the notion that nucleic acids derived from endogenous retroelements play important roles in autoinflammatory responses [78].
In addition to IFIH1, mutations in six other genes have been shown to cause AGS: TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1 and ADAR1 [22]. TREX1, RNASEH2A-C and SAMHD1 mutations all result in aberrant activation of the dsDNA sensing cGAS-STING pathway, driving chronic type I IFN production 79, 80, 81, 82, 83, 84. In contrast, the molecular consequence of ADAR1 mutations is the engagement of MDA5 85, 86, 87. ADAR1 (adenosine deaminase acting on RNA 1) is an RNA-editing enzyme that binds to dsRNA and converts adenosine to inosine (A-to-I). A-to-I edits are predicted to change RNA structure and stability, in addition to introducing mutations into proteins as the translational machinery reads ‘I’ bases as guanosines instead of adenosines [88]. ADAR1 has two isoforms: ADAR1-p110 is constitutively expressed, and localises to the cell nucleus, while ADAR1-p150 is type I IFN-inducible and additionally found in the cytoplasm. In AGS patients, mutations in the ADAR1 gene mostly map to regions of ADAR1 involved in A-to-I editing [89], and some mutations diminish editing activity 89, 90. In mice, ADAR1-deficiency triggers a type I IFN response and is embryonically lethal [91]. This phenotype is partially rescued by crossing Adar1–/– mice with either Ifnar–/– animals lacking the type I IFN receptor or Mavs–/– mice lacking the common adaptor protein for RLRs 87, 90. Similarly, IFN induction and embryonic lethality are rescued in Adar1–/– mice by additional knock-out of MDA5, but not RIG-I [87]. Interestingly, much like the situation in Adar1–/– mice (lacking both ADAR1-p110 and -p150), specific knock-out of ADAR1-p150 results in embryonic lethality that can be rescued by additional MAVS-deficiency [87]. Further evidence for the role of MDA5 and MAVS in inducing type I IFN downstream of ADAR1-deficiency was obtained in animals expressing an editing-deficient version of ADAR1 (E861A) [86]. Adar1E861A/E861A animals die during in utero development and recapitulate the type I IFN induction seen in Adar1–/– mice, and these effects are reversed in Adar1E861A/E861A; Ifih1–/– animals 86, 92.
Sites bound and edited by ADAR1 have been identified and, in human cells, are often found within or close to Alu elements 85, 93, 94, 95. Moreover, in the RNase protection assay described above, wild-type MDA5 protects IR-Alu RNAs amongst total RNA extracted from ADAR1-deficient cells, but not amongst RNA from wild-type cells [25]. Together with the MDA5-dependency of type I IFN induction in ADAR1-deficient mice and cells 85, 86, 87, these data suggest an attractive model: ADAR1 edits RNAs derived from IR-Alu repeats, leading to destabilization of their base-paired structure, which in turn prevents activation of MDA5 25, 96. This model is further consistent with the notion that IFIH1 mutations found in AGS change the RNA specificity of MDA5 such that it binds to IR-Alu elements containing mismatches and bulges [25].
Collectively, these observations highlight the importance of long base-paired stretches of RNA formed by IR-Alu elements in the activation of MDA5 in autoinflammatory disease. It is interesting, however, to note that Chung et al. recently reported that adenosines edited by ADAR1 tend to be found in single-stranded, rather than base-paired, regions of both single Alu and IR-Alu elements [85]. Moreover, A-to-I editing can in some circumstances stabilise RNA duplexes formed by Alu elements [96] and inosine-containing RNAs may in some settings inhibit RLRs [97]. Finally, RNAs derived from IR-Alu elements are bound by additional proteins such as DHX9 [98], which may compete with ADAR1 and MDA5. It will therefore be important to study RNAs bound by MDA5 in living cells harbouring ADAR1 or IFIH1 mutations.
A recent study found that mitochondrial dsRNA escapes into the cytosol when the mitochondrial enzymes SUV3 and PNPase, which are involved in RNA degradation, are depleted from cells or are dysfunctional due to mutations [17]. At the same time, type I IFN is induced in an MDA5-dependent manner. This observation further highlights the potential of MDA5 to detect cellular RNAs and parallels the finding that RIG-I can detect nuclear RNAs mislocalised to the cytosol [16].
The Role of MDA5 in Cancer
In addition to its myriad roles in viral and autoimmune diseases, MDA5 has recently been found to be activated during cancer treatment with DNA-demethylating agents 23, 24. Indeed, much like during viral infection, exposure of colorectal cancer cells to inhibitors of DNA methylation, such as 5-azacytidine-2-deoxycytidine, results in accumulation of RNAs detected by the dsRNA-specific antibody J2 [23]. Furthermore, these and similarly treated ovarian cancer cells contain extractable IFN-stimulatory RNAs, secrete type I and III IFNs, and upregulate ISGs 23, 24. These effects are MDA5- and MAVS-dependent and result in reduced cell growth and self-renewal 23, 24. Mechanistically, demethylation of normally repressed areas of the genome may result in transcription of endogenous retroviruses, and such RNAs have been suggested to activate MDA5 23, 24. In broad agreement with these data, a recent study, using acute myeloid leukemia cell lines, also shows that loss of epigenetic regulation can result in de-silencing of retroelements, dsRNA accumulation, and MDA5 activation [99]. It would be interesting to systematically investigate expression, subcellular localisation, and MDA5-association of RNAs derived from endogenous retroviruses and other endogenous retroelements such as Alu elements in cancer cells. Furthermore, IFIH1 genotype and MDA5 expression levels may be instructive in predicting the response of patients to drugs targeting epigenetic regulation. In another study, overexpression of MDA5 using adenoviral vectors induced cell death in cancer cells and also facilitated antitumour immunity via type I IFN production [100]. Taken together, these observations further support the notion that self-RNAs can trigger MDA5. In addition, synthetic MDA5-stimulatory RNAs, such as poly I:C, may induce cancer cell apoptosis and immune responses against tumours (reviewed in [101]).
Concluding Remarks
MDA5 is an important RNA sensor implicated in the detection of viral infections, in a range of autoimmune and autoinflammatory diseases, as well as in cancer. Recent efforts in identifying an RNA molecular pattern detected by MDA5 suggest that length and secondary structure of RNA are important determinants; however, we speculate here that activation of MDA5 in some settings may require more than simply dsRNA and that a yet-to-be identified consensus RNA PAMP detected by MDA5 may exist. We predict that recent technical advances, including an RNase protection assay [25] and, in particular, CLIP protocols [49], will facilitate research in this direction. The description of a defined RNA that selectively activates MDA5 would be of great value (see Outstanding Questions). MDA5 agonists could be used to boost antiviral and antitumoural immunity and may guide the developments of MDA5 antagonistic RNAs for use in autoinflammation.
Outstanding Questions.
What are the RNAs detected by MDA5? This question should be studied in the contexts of different virus infections, autoinflammation, and cancer. Models using living cells will be important given that other RNA-binding proteins are likely to compete with MDA5 for RNA binding. CLIP technologies may be particularly useful given that RNA–MDA5 complexes can be crosslinked in live cells.
Is there a common RNA structure or motif that is recognised by MDA5 in both virus infection and sterile conditions leading to MDA5 activation?
Is it possible to design synthetic RNAs that either activate or inhibit MDA5? Would such MDA5 agonists and antagonists have therapeutic value?
What is the relationship between endogenous retroelements and MDA5? At present, a systematic study of different retroelements in terms of their ability to generate MDA5-stimulatory RNAs has not been reported. Other interesting questions pertain to the stage in the life cycle of retroelements at which MDA5 is activated and to the role of MDA5 in suppressing these elements.
What are the molecular consequences of IFIH1 risk or pathologic alleles? Is mutated MDA5 spontaneously active or is RNA binding required? In which types of cells is MDA5 activated, and are there differences in the amount of MDA5-stimulatory RNA between cell types?
Does MDA5 form higher-order structures, such as filaments, in living cells? What is the spatiotemporal behaviour of MDA5 during virus infection? Advanced imaging technologies, such as super-resolution microscopy, are likely to provide insights into these questions.
What are the molecular mechanisms by which accessory proteins, such as LGP2 and DHX9, regulate the activity of MDA5?
Acknowledgments
The authors apologise to those colleagues whose work could not be cited due to space constraints. Work in the Rehwinkel group is supported by the UK Medical Research Council and Wellcome Trust. The authors thank Annemarthe G. Van der Veen and members of the Rehwinkel laboratory for critical discussion.
Glossary
- Aicardi–Goutières syndrome (AGS)
this progressive and severe brain disease is characterised by spontaneous production of type I IFNs that are detectable in the patients’ blood and cerebrospinal fluid [22]. AGS can manifest at birth and has some phenotypic overlap with congenital virus infection as well as with SLE. Mutations in any one of seven genes cause AGS. The proteins encoded by these genes are all enzymes involved in nucleic acid or nucleotide metabolism.
- Alu elements
endogenous retroelements of the short interspersed nuclear element (SINE) class that are ca. 300 nt in length. They are present in the human genome in more than one million copies, and some Alu elements are arranged as inverted repeats [118]. The vast majority of Alu elements do not complete retrotransposition, and Alu elements are absent in mice.
- Endogenous retroelements
a large proportion of mammalian genomes consists of endogenous retroelements, which are subdivided into endogenous retroviruses and retrotransposons [78]. These heritable DNA sequences resemble extant infectious retroviruses: retroelements can be transcribed into RNA that is reverse transcribed into a cDNA copy, which then reintegrates into the genome. These activities are usually silenced efficiently by cells to maintain genome integrity and to avoid innate immune activation.
- Interferons (IFNs)
secreted proteins that play pivotal roles in antiviral immune responses and are divided into types I, II, and III IFNs [6]. Humans have 17 different genes encoding type I IFNs (13 subtypes of IFN-α as well as IFN-β, -ε, -κ, and -ω). All type I IFNs signal via the ubiquitously expressed type I IFN receptor to induce the expression of hundreds of genes, several of which have antiviral functions. The single type II IFN, IFN-γ, plays important roles in cellular immune responses. Type III IFNs (also known as IFN-λs) have similar effects as type I IFNs but act primarily on mucosal tissues.
- Poly I:C
a synthetic RNA prepared by annealing inosine and cytidine homopolymers. Poly I:C is frequently used to stimulate type I IFN production and engages both TLR3 as well as RLRs.
- Single-nucleotide polymorphisms (SNPs)
this term describes substitutions of individual nucleotides relative to the reference genome. SNPs can occur within or outside of protein-coding sequences, and some SNPs affect the amino acid sequence of the encoded protein or gene expression, respectively.
- Systemic lupus erythematosus (SLE)
this common autoimmune disease is characterised by a range of symptoms, including rash, photo-sensitivity, arthritis, fatigue, and nephritis. IFN-α and autoantibodies play crucial roles in the development of SLE.
References
- 1.Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Barrat F.J., et al. Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 2016;67:323–336. doi: 10.1146/annurev-med-052814-023338. [DOI] [PubMed] [Google Scholar]
- 3.Hartmann G. Nucleic acid immunity. Adv. Immunol. 2017;133:121–169. doi: 10.1016/bs.ai.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goubau D., et al. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan N., Chen Z.J. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNab F., et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orzalli M.H., Kagan J.C. Apoptosis and necroptosis as host defense strategies to prevent viral infection. Trends Cell Biol. 2017;27:800–809. doi: 10.1016/j.tcb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goubau D., et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J., et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Pichlmair A., et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V., et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 12.Baum A., et al. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt A., et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuberth-Wagner C., et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlee M., et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang J.J., et al. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 2018;19:53–62. doi: 10.1038/s41590-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhir A., et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018;560:238–242. doi: 10.1038/s41586-018-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 19.Peisley A., et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peisley A., et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3340–E3349. doi: 10.1073/pnas.1208618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B., et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 22.Crow Y.J., Manel N. Aicardi–Goutieres syndrome and the type I interferonopathies. Nat. Rev. Immunol. 2015;15:429–440. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 23.Roulois D., et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiappinelli K.B., et al. Inhibiting DNA. methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmad S., et al. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell. 2018;172:797–810. doi: 10.1016/j.cell.2017.12.016. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K., et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 2018;90:105–115. doi: 10.1016/j.jaut.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 27.Kovacsovics M., et al. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr. Biol. 2002;12:838–843. doi: 10.1016/s0960-9822(02)00842-4. [DOI] [PubMed] [Google Scholar]
- 28.Kang D.C., et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. U. S. A. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrejeva J., et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gitlin L., et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berke I.C., Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J. 2012;31:1714–1726. doi: 10.1038/emboj.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H., et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colby C., Chamberlin M.J. The specificity of interferon induction in chick embryo cells by helical RNA. Proc. Natl. Acad. Sci. U. S. A. 1969;63:160–167. doi: 10.1073/pnas.63.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichlmair A., et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grunberg-Manago M., et al. Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of Azotobacter vinelandii. 1956. Biochim. Biophys. Acta. 1989;1000:65–81. [PubMed] [Google Scholar]
- 36.Zaki M., et al. Recurrent and prolonged infections in a child with a homozygous IFIH1 nonsense mutation. Front. Genet. 2017;8:130. doi: 10.3389/fgene.2017.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamborn I.T., et al. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 2017;214:1949–1972. doi: 10.1084/jem.20161759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asgari S., et al. Severe viral respiratory infections in children with IFIH1 loss-of-function mutations. Proc. Natl. Acad. Sci. U. S. A. 2017;114:8342–8347. doi: 10.1073/pnas.1704259114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schonborn J., et al. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 1991;19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber F., et al. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triantafilou K., et al. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J. Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen T.A., et al. SIDT2 Transports extracellular dsRNA into the cytoplasm for innate immune recognition. Immunity. 2017;47:498–509. doi: 10.1016/j.immuni.2017.08.007. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Q., et al. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertzog J., et al. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur. J. Immunol. 2018;48:1120–1136. doi: 10.1002/eji.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez David R.Y., et al. Comparative analysis of viral RNA signatures on different RIG-I-like receptors. eLife. 2016;5 doi: 10.7554/eLife.11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runge S., et al. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deddouche S., et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife. 2014;3 doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang M., et al. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. 2018;173 doi: 10.1016/j.cell.2018.03.064. 906–919.e13. [DOI] [PubMed] [Google Scholar]
- 49.Lee F.C.Y., Ule J. Advances in CLIP technologies for studies of protein–RNA interactions. Mol. Cell. 2018;69:354–369. doi: 10.1016/j.molcel.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 50.van der Veen A.G., et al. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018;37 doi: 10.15252/embj.201797479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez K.R., et al. MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J. Virol. 2014;88:8194–8200. doi: 10.1128/JVI.00640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchikawa E., et al. Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5. Mol. Cell. 2016;62:586–602. doi: 10.1016/j.molcel.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruns A.M., et al. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5–RNA interaction and filament assembly. Mol. Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao H., et al. ATP-dependent effector-like functions of RIG-I-like receptors. Mol. Cell. 2015;58:541–548. doi: 10.1016/j.molcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Q., et al. DHX29 functions as an RNA co-sensor for MDA5-mediated EMCV-specific antiviral immunity. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie Q., et al. LncITPRIP-1 positively regulates innate immune response through promoting oligomerization and activation of MDA5. J. Virol. 2018 doi: 10.1128/JVI.00507-18. Published online June 13, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zust R., et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kindler E., et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Odendall C., et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixit E., et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bender S., et al. Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smyth D.J., et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 63.Sheng Y., et al. Sequencing-based approach identified three new susceptibility loci for psoriasis. Nat. Commun. 2014;5:4331. doi: 10.1038/ncomms5331. [DOI] [PubMed] [Google Scholar]
- 64.Martinez A., et al. Association of the IFIH1-GCA-KCNH7 chromosomal region with rheumatoid arthritis. Ann. Rheum. Dis. 2008;67:137–138. doi: 10.1136/ard.2007.073213. [DOI] [PubMed] [Google Scholar]
- 65.Jin Y., et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enevold C., et al. Multiple sclerosis and polymorphisms of innate pattern recognition receptors TLR1-10, NOD1-2, DDX58, and IFIH1. J. Neuroimmunol. 2009;212:125–131. doi: 10.1016/j.jneuroim.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 67.Cunninghame Graham D.S., et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nejentsev S., et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lincez P.J., et al. Reduced expression of the MDA5 gene IFIH1 prevents autoimmune diabetes. Diabetes. 2015;64:2184–2193. doi: 10.2337/db14-1223. [DOI] [PubMed] [Google Scholar]
- 70.Gorman J.A., et al. The A946T variant of the RNA sensor IFIH1 mediates an interferon program that limits viral infection but increases the risk for autoimmunity. Nat. Immunol. 2017;18:744–752. doi: 10.1038/ni.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Downes K., et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Funabiki M., et al. Autoimmune disorders associated with gain of function of the intracellular sensor MDA5. Immunity. 2014;40:199–212. doi: 10.1016/j.immuni.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 73.Rutsch F., et al. A specific IFIH1 gain-of-function mutation causes Singleton–Merten syndrome. Am. J. Hum. Genet. 2015;96:275–282. doi: 10.1016/j.ajhg.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice G.I., et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oda H., et al. Aicardi–Goutières syndrome is caused by IFIH1 mutations. Am. J. Hum. Genet. 2014;95:121–125. doi: 10.1016/j.ajhg.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crow Y.J., Rehwinkel J. Aicardi–Goutières syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rice G.I., et al. Assessment of interferon-related biomarkers in Aicardi–Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol. 2013;12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Volkman H.E., Stetson D.B. The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maelfait J., et al. Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep. 2016;16:1492–1501. doi: 10.1016/j.celrep.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mackenzie K.J., et al. Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 2016;35:831–844. doi: 10.15252/embj.201593339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ablasser A., et al. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 82.Gray E.E., et al. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi–Goutières syndrome. J. Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao D., et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gall A., et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung H., et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172 doi: 10.1016/j.cell.2017.12.038. 811–824.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liddicoat B.J., et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pestal K., et al. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43:933–944. doi: 10.1016/j.immuni.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eisenberg E., Levanon E.Y. A-to-I RNA editing – immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018;19:473–490. doi: 10.1038/s41576-018-0006-1. [DOI] [PubMed] [Google Scholar]
- 89.Rice G.I., et al. Mutations in ADAR1 cause Aicardi–Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mannion N.M., et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hartner J.C., et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heraud-Farlow J.E., et al. Protein recoding by ADAR1-mediated RNA editing is not essential for normal development and homeostasis. Genome Biol. 2017;18:166. doi: 10.1186/s13059-017-1301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan M.H., et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550:249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramaswami G., Li J.B. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42(Database issue):D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bahn J.H., et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:6355. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Solomon O., et al. RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat. Commun. 2017;8:1440. doi: 10.1038/s41467-017-01458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vitali P., Scadden A.D. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aktas T., et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 99.Cuellar T.L., et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 2017;216:3535–3549. doi: 10.1083/jcb.201612160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu X., et al. Activation of the MDA-5-IPS-1 viral sensing pathway induces cancer cell death and type I IFN-dependent antitumor immunity. Cancer Res. 2016;76:2166–2176. doi: 10.1158/0008-5472.CAN-15-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu Y., et al. The anticancer functions of RIG-I-like receptors, RIG-I and MDA5, and their applications in cancer therapy. Transl. Res. 2017;190:51–60. doi: 10.1016/j.trsl.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 102.Wang J.P., et al. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J. Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Slater L., et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cao X., et al. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J. Hepatol. 2015;62:771–778. doi: 10.1016/j.jhep.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 105.Fredericksen B.L., et al. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burke C.W., et al. Characteristics of alpha/beta interferon induction after infection of murine fibroblasts with wild-type and mutant alphaviruses. Virology. 2009;395:121–132. doi: 10.1016/j.virol.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Akhrymuk I., et al. Both RIG-I and MDA5 detect alphavirus replication in concentration-dependent mode. Virology. 2016;487:230–241. doi: 10.1016/j.virol.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zalinger Z.B., et al. MDA5 is critical to host defense during infection with murine coronavirus. J. Virol. 2015;89:12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li J., et al. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ikegame S., et al. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 2010;84:372–379. doi: 10.1128/JVI.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gitlin L., et al. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banos-Lara Mdel R., et al. Critical role of MDA5 in the interferon response induced by human metapneumovirus infection in dendritic cells and in vivo. J. Virol. 2013;87:1242–1251. doi: 10.1128/JVI.01213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dou Y., et al. The innate immune receptor MDA5 limits rotavirus infection but promotes cell death and pancreatic inflammation. EMBO J. 2017;36 doi: 10.15252/embj.201696273. 274–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pham A.M., et al. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Melchjorsen J., et al. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 2010;84:11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu H.L., Liao F. Melanoma differentiation-associated gene 5 senses hepatitis B virus and activates innate immune signaling to suppress virus replication. J. Immunol. 2013;191:3264–3276. doi: 10.4049/jimmunol.1300512. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Z., et al. Hepatitis D virus replication is sensed by MDA5 and induces IFN-beta/lambda responses in hepatocytes. J. Hepatol. 2018;69:25–35. doi: 10.1016/j.jhep.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 118.Chen L.L., Yang L. ALUternative regulation for gene expression. Trends Cell Biol. 2017;27:480–490. doi: 10.1016/j.tcb.2017.01.002. [DOI] [PubMed] [Google Scholar]