Abstract
SEC14 and Spectrin domain-1 (Sestd1) is a synapse protein that exhibits a striking shift from the presynaptic to postsynaptic space as neurons mature postnatally in the mouse hippocampus. Hippocampal pyramidal neurons from mice with global genetic deletion of Sestd1 have reduced dendrite arbors, spines, and excitatory synapses. Electrophysiologically this correlates with cell-autonomous reductions in both AMPA- and NMDA-excitatory postsynaptic currents in individual hippocampal neurons from which Sestd1 has been deleted in vivo. These neurodevelopmental and functional deficits are associated with increased activation of the Rho family GTPases Rac1 and RhoA. Co-immunoprecipitation and mass spectrometry reveal that the Breakpoint Cluster Region protein, a Rho GTPase activating protein (GAP), forms complexes with Sestd1 in brain tissue. This complements earlier findings that Sestd1 can also partner with other Rho family GAPs and guanine nucleotide exchange factors. Our findings demonstrate that Sestd1 is a developmentally dynamic synaptic regulator of Rho GTPases that contributes to dendrite and excitatory synapse formation within differentiating pyramidal neurons of the forebrain.
Keywords: glutamate, GTPase, hippocampus, projection, pyramidal
Introduction
Dendrites of pyramidal glutamatergic neurons in the cerebral cortex and hippocampus are decorated with protrusions called spines, specialized sites for excitatory postsynaptic terminals (Sekino et al. 2007; Arguello et al. 2013). Pathogenic disruption of dendrites, spines, and synapses is a contributor to cognitive disability and mental illness (Zoghbi and Bear 2012; Konopaske et al. 2014; Phillips and Pozzo-Miller 2015). Elucidating the molecular regulation of these structures is a central goal of basic neuroscience research.
The SEC14 and Spectrin domain-1 (Sestd1) protein has previously been characterized in endothelial cells as participating in phospholipid signaling (Miehe et al. 2010), and in developing neurons as an antagonist of TRIO8, a guanine nucleotide exchange factor (GEF) for the Rho superfamily GTPase, Rac1 (Lee et al. 2015). We previously found that during gastrulation Sestd1 is also a functional partner of the Dapper antagonist of catenin (Dact1) protein involved in Wnt signaling (Yang and Cheyette 2013). Sestd1 knockout (KO) mice die within a day of birth (Yang and Cheyette 2013) phenocopying Dact1KO mice and reflecting close cooperation of the 2 proteins in the regulation of cell signaling during germ layer formation at the primitive streak (Suriben et al. 2009; Wen et al. 2010). Nonetheless, both Dact1 and Sestd1 are also widely expressed later during development within neurons of the developing and adult brain (Fisher et al. 2006; Miehe et al. 2010). Moreover, forebrain neurons from Dact1KO animals exhibit reduced dendrite arbors, spines, and excitatory synapses correlating with reduced Rac1 activity (Okerlund et al. 2010). A recent human genetic study found an association between a common SNP in an intron of the human SESTD1 gene and lithium-responsive bipolar disorder (Song et al. 2015); although this association was later revised to below the threshold for genome-wide significance (Song et al. 2017), the finding does suggest potential biomedical significance for Sestd1 function in the forebrain. Furthermore, given the well-established importance of small GTPase regulation during formation and plasticity of dendritic spines and synapses (Um et al. 2014; Spence and Soderling 2015; Woolfrey and Srivastava 2016; Hedrick and Yasuda 2017), and given the fundamental importance of these processes to complex behavior and neuropsychiatry (Oh et al. 2010; Durand et al. 2012; Pavlowsky et al. 2012; Konopaske et al. 2014; Aceti et al. 2015), we conducted the present study to further explore the role of Sestd1 in these events.
Similar to Dact1KO mice, Sestd1KO mice have grossly normal brain morphology at birth, but Sestd1KO HCNs have reduced dendrite complexity, spines, and excitatory synapse density compared with those from wild-type (WT) littermates. Intracellular recordings in the CA3-CA1 hippocampal circuit demonstrate electrophysiological deficits consistent with decreased excitatory inputs to postsynaptic neurons lacking Sestd1. Loss of Sestd1 in glutamatergic neurons leads to increases in activity of 2 Rho GTPase family members, Rac1 and RhoA, within both the developing and mature hippocampus. Co-immunoprecipitation (co-IP) and mass spectrometry (MS) reveal that an endogenous partner of Sestd1 in the brain is the breakpoint cluster region (BCR) protein, a GTPase activating protein (GAP) previously shown to inhibit Rac1 in neurons (Um et al. 2014). Finally, neuronal fractionation experiments demonstrate an intriguing dynamic relocalization of Sestd1 as the brain matures: although predominantly presynaptic in the hippocampus at birth, Sestd1 becomes enriched postsynaptically by adulthood, suggesting that it may play distinct roles on each side of synapses as these structures form and stabilize.
Materials and Methods
Animals
All animal studies were examined and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California at San Francisco. All mouse lines were maintained and studied in a C57Bl/6J (JAX) genetic background, back-crossed a minimum of 6 times. The Sestd1 targeted mutant alleles (Sestd1flox and its genetic derivatives) were generated in the Cheyette lab (Yang and Cheyette 2013). “Sestd1KO” designates mice homozygous for the constitutive null allele; “WT” designates littermates homozygous for the wild type locus. Thy1-GFP-M transgenic mice (JAX) express GFP (green fluorescent protein) in a subset of pyramidal neurons within the hippocampus and neocortex (as well as other brain regions), producing a “pseudo-Golgi” fluorescence staining pattern useful for morphometric studies of individual neurons (Feng et al. 2000), including measurements of spine density along primary dendrites in situ (Martin et al. 2016). Emx1-IRES-Cre knockin mice (JAX) express Cre recombinase in glutamatergic neurons of the forebrain and their precursors starting in mid-embryonic stages (Gorski et al. 2002); in combination with a flox allele the Emx1-Cre transgene thereby enables assessment of phenotypes arising from selective deletion of a gene product in this neuron population. The BCRKO allele was created in the laboratory of Dr Nora Heisterkamp (Voncken et al. 1995) and graciously provided at her suggestion by the laboratory of Dr Masaaki Murakami at the Institute of Genetic Medicine, Hokkaido University, Japan. Wild type CD-1 mice (used for preparation of synaptosome fractions) were obtained commercially from Charles River Laboratories.
In every experiment, littermates were compared, and except where otherwise noted a 1:1 ratio of males and females were utilized within each cohort. Briefly, for generation of Sestd1 +/+ versus −/− animals (Figs 1 and 2) parents were an intercross of heterozygous (+/−) mice, producing equal numbers of “KO” (−/−) and “WT” (+/+) littermates, as well as heterozygotes that were not experimentally compared. For conditional mutant experiments involving Emx1-Cre (Figs 3 and 4), a Sestd1flox/flox; Cre+ mouse was crossed to Sestd1flox/+ (or Sestd1flox/+; Thy1-GFP+ for experiments involving the Thy1-GFP marker). These crosses produced experimental (Sestd1flox/flox; Cre+) and control (Sestd1flox/flox) littermates which were compared experimentally. For double mutant experiments involving the Sestd1KO and BCRKO alleles (Fig. 5C–E), all comparator genotypes were littermates obtained from intercrosses of double heterozygous parents (i.e., Sestd1−/+; BCR−/+ × Sestd1−/+; BCR−/+).
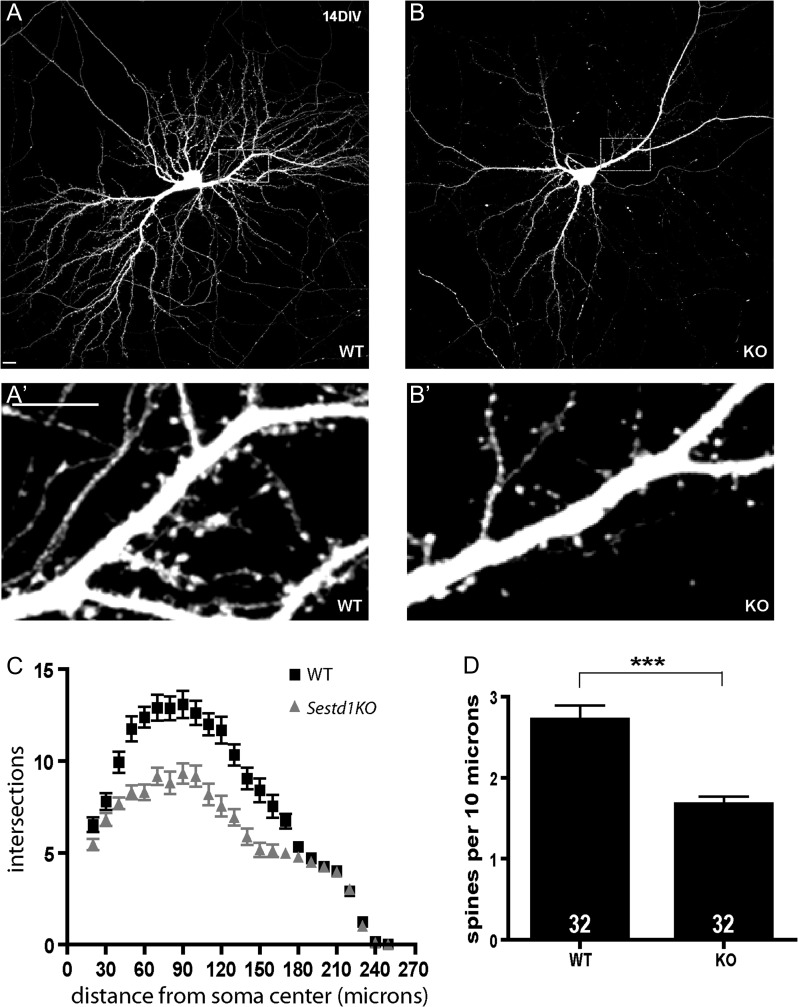
Figure 1.
Sestd1 mutant neurons have reduced dendrite arbors and spines. (A,B) Cultured hippocampal neurons (HCNs) transfected with GFP and imaged at 14 DIV. (A) WT (B) Sestd1KO. (A′,B′) Corresponding boxed regions from A to B at higher magnification. (C) Sholl analysis of WT (black squares) and Sestd1KO (gray triangles) HCNs. (D) Spine density is reduced in Sestd1KO HCNs. Scale bars: 10 μm. The number within each bar = n, 32 neurons/condition. ***P ≤ 0.001.
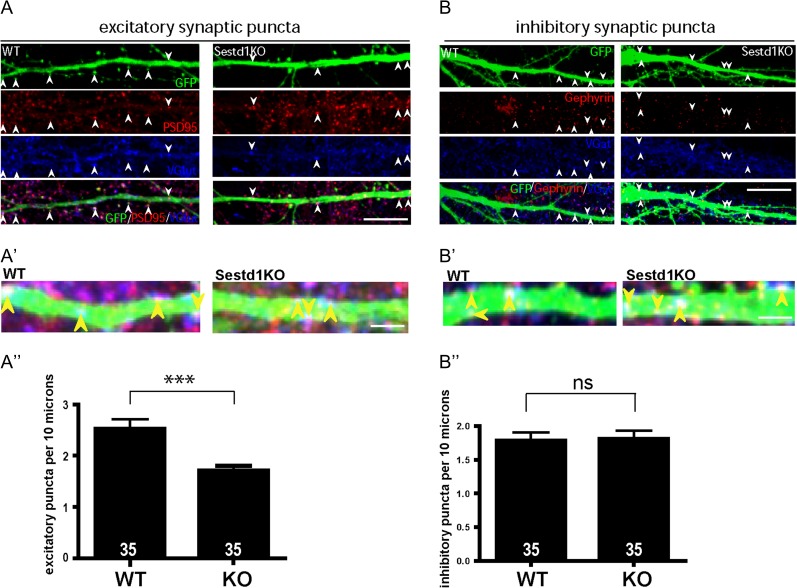
Figure 2.
Cultured HCNs from Sestd1KO mice have fewer excitatory synapses; inhibitory synapses are spared. (A) Comparison of excitatory synaptic puncta (colocalized Vglut1 and PSD95) between HCNs cultured from WT and Sestd1KO littermates. (B) Comparison of inhibitory synaptic puncta (colocalized VGAT and Gephyrin) between WT and Sestd1KO. (A′,B′) Further magnification of excitatory and inhibitory puncta, respectively. (A″) Excitatory synapse density is reduced in Sestd1KO compared with WT HCNs. (B″) Inhibitory synapse density is no different between genotypes. Scale bar: 10 μm in (A,B); 3 μm in (A′,B′). The number within each bar = n, 35 neurons/condition. ns, P > 0.05; ***P ≤ 0.001
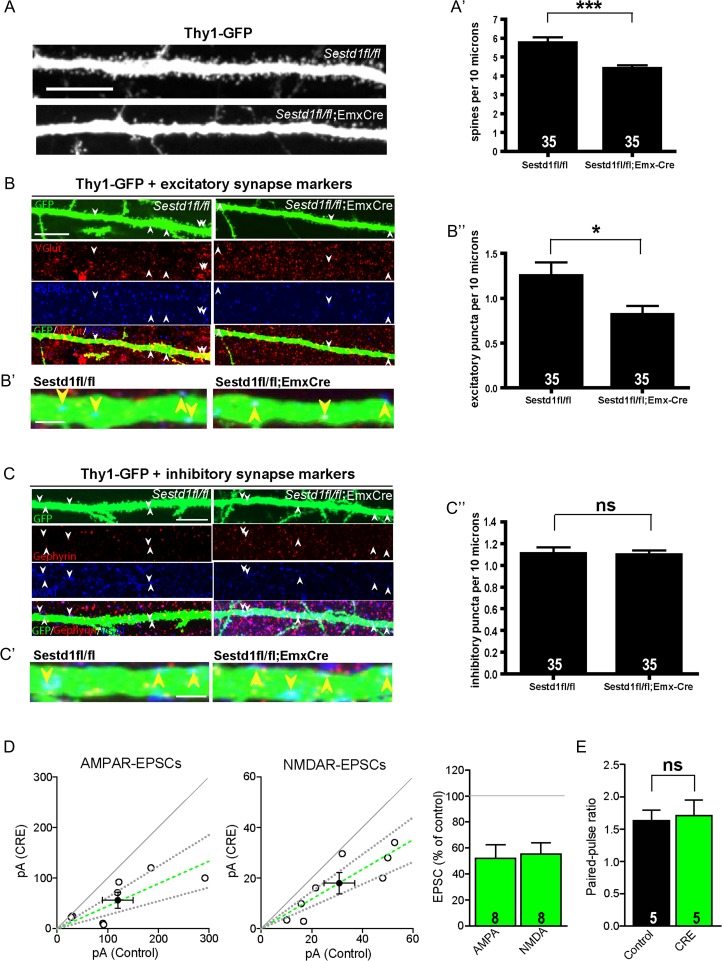
Figure 3.
Sestd1 mutant neurons within the hippocampus have fewer spines and excitatory synapses. (A–A′) Dendritic spine density in hippocampal tissues of Sestd1fl/fl; EmxCre+;Thy1-GFP mice is lower than in Sestd1fl/fl;Thy1-GFP littermate controls.(B–B″) Excitatory synapse density (colocalized Vglut1 and PSD95 puncta along visualized dendrites, magnified in B′) in Sestd1fl/fl; EmxCre+;Thy1-GFP mice is reduced compared with Sestd1fl/fl;Thy1-GFP littermate controls. (C–C″) Inhibitory synapse density (colocalized VGAT and Gephyrin puncta along visualized dendrites, magnified in C′) in Sestd1fl/fl; EmxCre+;Thy1-GFP mice is no different than in Sestd1fl/fl;Thy1-GFP littermate controls. (D,E) Organotypic hippocampal slices from 2 litters of P7 Sestd1fl/fl mice (methods) were biolistically transfected with Cre:GFP at DIV2, and simultaneous whole-cell recordings obtained from Cre-expressing and neighboring untransfected (control) CA1 neurons within the same slice on DIV14-18. Peak amplitudes of AMPAR-EPSCs (left) or NMDAR-EPSCs (middle) from simultaneously recorded experimental and control cell pairs (open circles) and mean ± SEM (filled circles/green dashed line). Dashed lines represent linear regression and 95% confidence interval. Bar graph (right) depicts mean of the EPSC amplitude from the transfected cell as a percentage of the control cell from each pair. In each graph, the solid gray line represents the null hypothesis. (E) Mean ± SEM of the AMPAR-EPSC paired-pulse ratio. Scale bars: (A–C) 10 μm; (B′,C′) 2 μm. The number within each bar = n, 35 neurons/condition in (A′,B″,C″); 8 pairs of simultaneously recorded neurons in (D); 5 pairs in (E). ns, P > 0.05; *P ≤ 0.05; ***P ≤ 0.001.
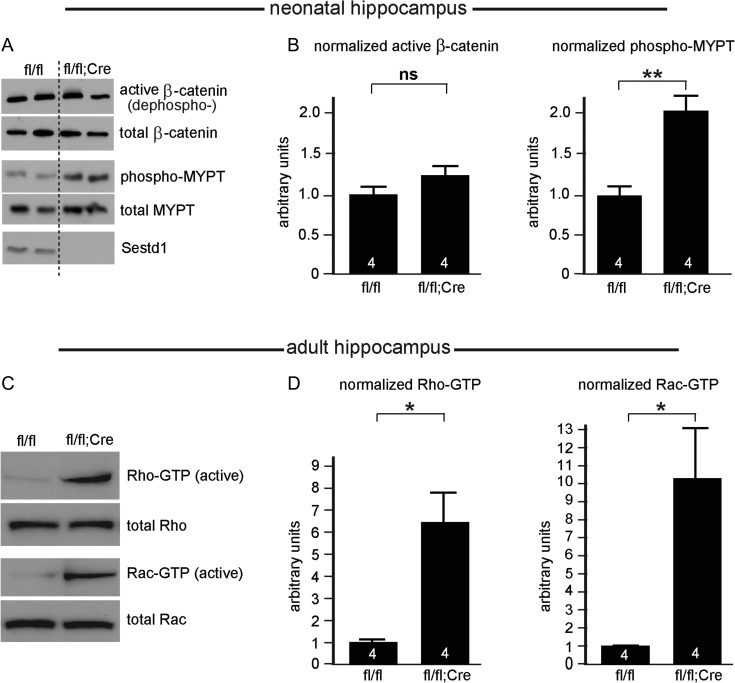
Figure 4.
Loss of Sestd1 does not affect β-catenin but increases activity of Rac1 and RhoA in the neonatal and mature hippocampus. (A) Representative immunoblots of signaling target read-outs conducted using hippocampal lysates obtained from both conditional mutant (Sestd1fl/fl; EmxCre+) and littermate controls (Sestd1fl/fl). (B) Quantification: Levels of β-catenin specifically unphosphorylated at serine 37 and threonine 41 (Wnt-pathway activitated β-catenin, “ABC”) are not significantly increased in the mutants, whereas levels of phosphorylated MYPT, a target of the Rho Kinase downstream of activated Rho GTPase are significantly increased. (C) Representative immunoblots of GTP-bound (active) forms of RhoA (top) and Rac1 (bottom), compared with total RhoA and Rac1, respectively, from adult hippocampal samples. (D) Quantification; active RhoA and Rac1 are both significantly increased in the mutants. The number within each bar = n, 4 animals/condition. ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01. n.b. A significant increase in GTP-bound Rac1 was also observed in neonatal cortical samples from constitutive Sestd1KO mice compared with WT littermate controls (data not shown).
Figure 5.
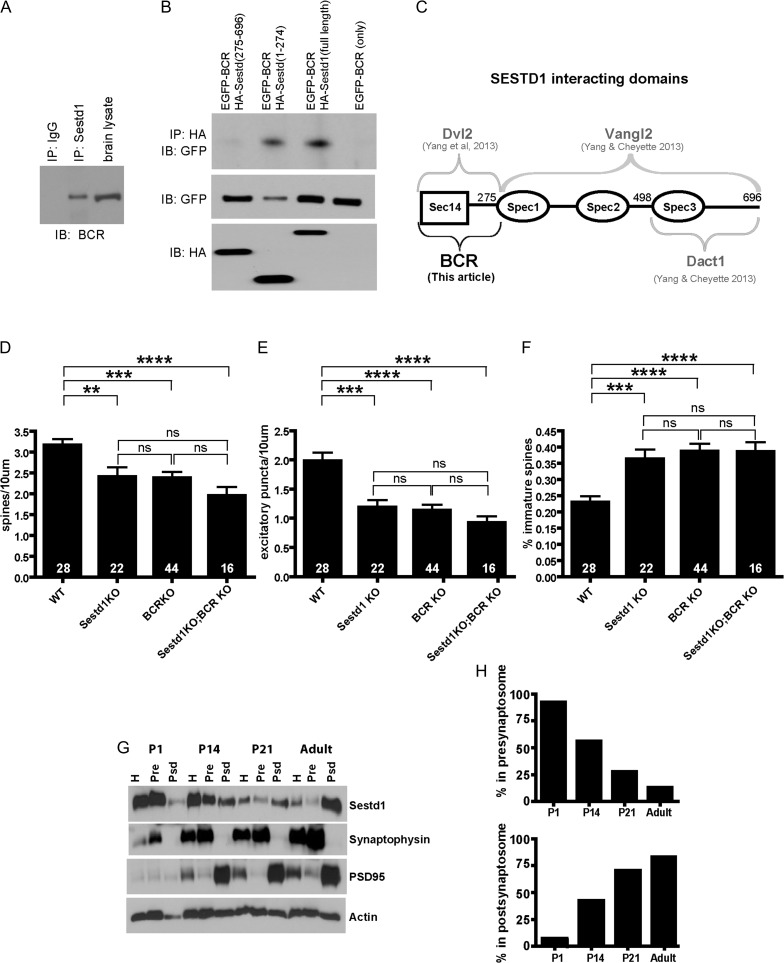
Sestd1 forms physiological complexes with BCR and has a developmentally dynamic presynaptic versus postsynaptic distribution. (A) Adult hippocampal lysate (wild type, C57Bl/6 J) immunoprecipitated with anti-Sestd1 antibody; co-precipitated BCR detected with anti-BCR antibody. (B) HA-tagged full-length Sestd1 and deletion mutants tested for ability to pull down ECFP-tagged BCR when the proteins are recombinantly expressed in HEK293T cells. (C) Schematic representation of Sestd1 domains contributing to interaction with BCR (this study), compared with similar determinations for domains contributing to interactions with Dvl2, Vangl2 and Dact1 (prior studies, Cheyette lab). (D–F) Deletion of either Sestd1 or BCR alone produce similar phenotypic effects with regard to dendritic spine density (D), excitatory synapse density (E), and spine maturity (F), in cultured forebrain neurons. Phenotypic effects of simultaneous Sestd1 and BCR loss are not significantly different from effects resulting from the single mutants. The number within each bar = n, animals/genotype. ns, P > 0.05; **P ≤ 0.01; ***P ≤ 0.001, ****P ≤ 0.0001. (G) Sestd1 synapse localization shifts during postnatal development. Presynaptic and postsynaptic compartment proteins were separated in a brain lysate from a WT (CD-1) mouse using biophysical techniques and the distribution of Sestd1 determined by immunoblotting. Synaptophysin and PSD95 were used as presynaptic and postsynaptic markers respectively for the quality of biophysical separation of the desired synaptosome fractions. (H) Quantification of distribution of Sestd1 into presynaptic and postsynaptic biophysical fractions at different time points.
Culture Slices and Electrophysiology
As previously described (Gray et al. 2011), cultured slices were prepared from hippocampi dissected from 2 complete litters of postnatal day 7 (P7) Sestdfl/fl mice derived from intercross of Sestdfl/fl parents (C57Bl/6 J background). Slices were biolistically transfected with pFUGW-Cre:GFP vector expressing a nuclear-targeted Cre:GFP fusion protein after 2 days in culture. Slices were cultured for an additional 14–18 days. Slices were recorded in a submersion chamber on an upright Olympus microscope, perfused in room temperature normal ACSF saturated with 95% O2/5% CO2. Picrotoxin (0.1 mM) and NBQX (10 μM) were added to the ACSF to block GABAA and AMPA receptors respectively. CA1 pyramidal cells were visualized by infrared differential interference contrast microscopy and transfected neurons identified by epifluorescence microscopy. The intracellular solution contained (in mM) CsMeSO4 135, NaCl 8, HEPES 10, Na-GTP 0.3, Mg-ATP 4, EGTA 0.3, and QX-314 5. Cells were recorded with 3–5 MΩ borosilicate glass pipettes, following stimulation of Schaffer collaterals with bipolar placed in stratum radiatum of the CA1 region. Series resistance was monitored and not compensated, and cells in which series resistance varied by 25% during a recording session were discarded. Synaptic responses were collected with a Multiclamp 700B amplifier (Axon Instruments, Foster City, CA), filtered at 2 kHz, digitized at 10 Hz. All paired recordings involved simultaneous whole-cell recordings from transfected (GFP+) neuron and a neighboring untransfected (GFP−) neuron (n = 8 pairs total for Fig. 3D, 5 pairs total for Fig. 3E). AMPAR-EPSCs were recorded at −70 mV and NMDAR-EPSCs were recorded at +40 mV in the presence of 10 μM NBQX. The stimulus was adjusted to evoke a measurable, monosynaptic EPSC in both cells. Paired-pulse ratios were measured by giving two pulses at a 50 ms interval and taking the ratio of the 2 peaks of the EPSCs from an average of 30–50 sweeps.
Image Analysis
Neurons were visualized on a Nikon Spectral C1si confocal or Nikon Spinning Disk Confocal microscope using a ×40 oil, ×60 oil, or 100× oil objective. NIH Image J software was used to count spine and synapse puncta numbers. Spine maturity (Fig. 5E) was quantified in the Cheyette lab as described previously (Okerlund et al. 2010, 2016; Martin et al. 2016).
Immunoblotting for β-catenin and Mypt
Hippocampal lysates were collected from 8 neonatal littermates (2 males + 2 females per genotype) derived from a cross of Sestd1flox/flox × Sestd1flox/+; Emx1-Cre+ parents as described in the animals section. Tissues were lysed in ice-cold buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton supplemented with proteinase inhibitors) for 30 min as previously described (Kivimae et al. 2011). Supernatant was collected, denatured and resolved by SDS-PAGE. The following antibodies were used: mouse anti-β catenin (BD Biosciences), mouse anti-active β catenin (“ABC”) (Millipore), mouse anti-Mypt1 (BD Biosciences), rabbit anti-phopho-Mypt1 (Thr-696) (Millipore), and rabbit anti-Sestd1 (ProSci).
Immunohistochemistry
P30 brain floating sections were treated with 3% H2O2 for 20 min at room temperature (RT). Sections were then rinsed in PBS, followed by PBT (0.5% Triton in PBS) wash and incubation in blocking solution (5% goat serum and 2% BSA in PBT) for 1 h. Sections were incubated with primary antibodies against excitatory synapse markers: PSD95 (Cell Signaling)/VGLUT1 (Synaptic Systems); inhibitory synapse markers: Gephrin (Synaptic Systems)/VGAT (Synaptic Systems) overnight at 4 °C in blocking solution. After 3 washes in PBT, sections were incubated with secondary antibodies (Alexa Fluor 568 and 647) for 2 h at RT. Sections were then washed and cover-slipped using Mowiol for fluorescence microscopy. Four littermate animals (2 males and 2 females) were used for each condition in Figure 3A–C, with 35 neurons counted for each condition.
Co-Immunoprecipitation
Co-Immunoprecipitations (co-IPs) of endogenous protein complexes (Fig. 5A): This was performed using a brain lysate prepared from a 2-month-old (female) C57Bl/6 mouse. All steps up to the final preparation in SDS sample buffer were conducted using ice-cold buffer solutions in a cold room; incubation steps were all conducted with gentle agitation (i.e., rotation). The dissected brain was dissociated in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton and proteinase inhibitors) for 30 min. After centrifugation at 14 000 × g for 15 min, supernatant was transferred to a fresh tube and precleared by incubation with protein G agarose beads for 30 min, followed by centrifugation at 14 000 × g for 5 min. The supernatant was then incubated with Sestd1 antibody overnight. Proteins associated with Sestd1 were pulled down by incubation with protein G agarose beads for 3 h at 4 °C. After washing with lysis buffer (3 × 10 min), beads were resuspended in SDS sample buffer and boiled for 5 min. The supernatant was collected by brief centrifugation and then subjected to SDS-PAGE analysis. Antibodies: rabbit anti-BCR (Santa Cruz), rabbit anti-Sestd1 (ProSci).
Co-IPs of recombinantly expressed proteins (Fig 5B): These were performed using lysates from HEK293T cells transfected with plasmids to express selected epitope-tagged proteins and protein fragments. Cells were cotransfected with ECFP-BCR (Enhanced Cyan Fluorescent Protein-tagged BCR) and HA-Sestd1 (Human influenza hemagglutinin–tagged Sestd1)-expressing plasmids using Lipofectamine 2000 (Life Technologies). Cells transfected with ECFP-BCR alone were used as a negative control. After 48 h, transfected cells were lysed, precleared, and incubated with anti-HA agarose beads for 3 h at 4 °C with gentle agitation as above. Beads were collected and washed as described previously (Kivimae et al. 2011), and protein complexes separated by SDS-PAGE followed by detection using anti-GFP antibody.
Primary Neuron Culture
Hippocampal neuronal cultures were prepared from neonatal brains taken from littermates as previously described (Beaudoin et al. 2012). Hippocampal neurons were plated at 8 × 104 cells/per 24-well on poly-l-lysine (0.5 mg/mL; Sigma) coated cover slips and maintained in NeuroBasal medium supplemented with glutamine and B-27 (Invitrogen). Three littermate animals (1 male and 2 females) were utilized for each condition in Figure 1, with a total of 32 neurons counted for each condition. Four littermate animals (2 males and 2 females) were used for each condition in Figure 2, with 35 neurons counted for each condition.
Proteomics
Hippocampi from 4 adults (P30; 2 males and 2 females) wild type (C57Bl/6J; JAX) mice were lysed in ice-cold lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 0.1% NP-40 and proteinase inhibitors) for 30 min. The supernatant was precleared by protein G agarose bead for 30 min prior to incubation with Sestd1 antibody overnight in cold room. Proteins associated with Sestd1 were then pulled down by protein G agarose bead for 3 h. Non-specific binders were removed by 3 washes in lysis buffer and 2 additional washes in 50 mM NH4HCO3, pH 7.5. Washed beads were then digested overnight with trypsin. The resulting peptides were desalted on Waters Sep-Pak C18 cartridges. Peptides were measured by nano-LC–MS-MS on a Thermo Scientific Q Exactive. Peptides were separated by reverse phase chromatography in a 180 min gradient (1–45% acetonitrile) at 250 nL/min. The Q Exactive was operated in the data-dependent mode with the following settings: 70 000 resolution, 300–2000 m/z full scan, Top 10, and an 1.8 m/z isolation window. Identification and label free quantification of peptides was done with MaxQuant 1.3.0.5 using a 1% false discovery rate (FDR) against the mouse Swiss-Prot/TrEMB database downloaded from Uniprot on 11 October 2013 (Cox and Mann 2008).
Rac1/RhoA Activity Assays
Performed according to manufacturer’s instructions (Cell Signaling #8815 for Rac; Cell Signaling #8820 for Rho). Four adult (P30) animals (2 males and 2 females) were used for each genotype (Fig. 4C,D). Briefly, hippocampi were lysed in 1× washing/binding/lysis buffer (included with kit) +1 mM PMSF. The tissue to buffer ratio was 1 mg tissue/1 mL lysis buffer. Tubes of lysed cells were spun at 16 000 × g for 15 min at 4 °C, and the supernatant was removed from the cell debris. A 1/10 V of the supernatant was set aside for subsequent determination by immunoblot of total Rac1 or RhoA, respectively. The remaining supernatant was then processed per the instructions provided, with the entire assay performed at 4 °C: Glutathione resin + agarose beads were added to the spin cup, and spun to bind them to the filter. The filter was then washed with the washing buffer. For Rac1: 20 ug of GST-PAK1-PBD was added to the spin cup along with the supernatant from the lysis. For RhoA: 400 ug of the GST-Rhotekin-RBD was added to the spin cup along with the supernatant from the lysis. The reaction was incubated for 1.5 h at 4 °C. After spin down and wash, the resulting eluted fraction added to LDS sample reducing agent with 200 mM DTT. This sample was then boiled at 70 °C for 15 min. This sample was then used for standard SDS-PAGE and immunoblotting. Rac was detected using anti-Rac1 mouse AB (Cell Signaling #8631). Rho was detected using anti-Rho rabbit AB (Cell Signaling #8789).
Statistics
Prism software (GraphPad Software) was used for data analysis. All P-values were calculated by unpaired parametric two-tailed t test. Paired EPSC data were analyzed with a Wilcoxon signed-rank test and linear regressions were obtained using the least squares method. Paired-pulse ratio data were analyzed by a two-tailed paired Student’s t test. Numerical values including P-values are reported in the Results and represented graphically in the figures. For all bar graphs, numbers within the bars represent n for that cohort, error bars represent standard error; ns = P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Synaptosome Fractionation
Two pregnant CD-1 (outbred strain) mice were obtained from Charles River Laboratories. Hippocampi from offspring were collected at different developmental stages (P1, P14, P21, and P60) and crude synaptosomes prepared via serial centrifugation (Tai et al. 2010). Briefly, hippocampi were homogenized in ice-cold sucrose/HEPES buffer (0.32 M sucrose, 10 mM HEPES and protease inhibitor, pH 7.4), and centrifuged at 1000 × g for 10 min. The supernatant was collected for crude synaptosomes by centrifuging at 17 000 × g for 15 min. Presynaptic and postsynaptic fractions were extracted from the crude synaptosomes by Triton (1% Triton X-100, 20 mM Tris–HCl and protease inhibitor, pH 8.0), and separated by centrifugation (40 000 × g for 30 min) as described previously (Garside et al. 2009). Presynaptic and postsynaptic fractions were solubilized in 5% SDS and resolved by SDS-PAGE using the following synaptic antibodies: rabbit anti-synaptophysin (Zymed), mouse anti-PSD95 (NeuroMAB), rabbit anti-Sestd1 (ProSci), rabbit anti-β actin (Cell signaling).
Results
Sestd1 Mutant Neurons Have Simpler Dendrite Arbors and Fewer Spines
Constitutively deleted Sestd1KO mice do not survive beyond the first postnatal day because of urogenital abnormalities (Yang and Cheyette 2013) but have grossly normal brain regionalization and neuronal distributions (data not shown). Given similar findings in the Dact1KO (Okerlund et al. 2010), we asked whether dendrite and spine development is affected in Sestd1KO HCNs. Confirming this hypothesis, Sestd1KO HCNs at 14 days in vitro (DIV) had less complex dendrites (Fig. 1A–C) and fewer spines compared with WT (Fig. 1A′,B′,D; Sestd1+/+, 2.7 ± 0.17 vs. Sestd1−/−, 1.67 ± 0.10; P < 0.001).
Sestd1 Mutant Neurons Have Fewer Excitatory Synapses In Vitro
Having observed reduced spine density, we asked whether Sestd1 is also required for the development of synapses, using fluorescent immunocytochemistry and confocal microscopy to quantify colocalized presynaptic and postsynaptic markers along labeled dendrites. We visualized excitatory synapses using VGlut1 (presynaptic) and PSD95 (postsynaptic) (Fig. 2A), finding a significant reduction in excitatory synapse density in Sestd1KO HCNs compared with WT (Fig. 2A′; puncta per 10 μm in Sestd1+/+, 2.45 ± 0.15 vs. Sestd1−/−, 1.72 ± 0.067; P < 0.001). In contrast, when inhibitory synapses were similarly examined using VGAT (presynaptic) and Gephyrin (postsynaptic) (Fig. 2B), we found no significant reduction (Fig 2B″; puncta per 10μm in Sestd1+/+, 1.77 ± 0.12, vs. Sestd1−/−, 1.81 ± 0.12; P > 0.5).
Sestd1 Mutant Neurons in the Intact Hippocampus Have Fewer Spines and Excitatory Synapses
To confirm these phenotypes in vivo, we eliminated Sestd1 specifically within prenatal forebrain glutamatergic neurons using Cre–loxP technology (Gorski et al. 2002; Yang and Cheyette 2013). We then examined spine and synapse density on primary apical dendrites of individual transgenically labeled pyramidal neurons (Feng et al. 2000) in hippocampal brain slices taken from 1-month-old animals (i.e., Sestd1fl/fl;Thy1-GFP vs. Sestd1fl/fl;EmxCre;Thy1-GFP at P30). We found that Sestd1−/− pyramidal neurons had reduced spine density compared with controls (Fig. 3A,A′; puncta per 10 μm in Sestd1fl/fl;Thy1-GFP, 5.74 ± 0.3, vs. Sestd1fl/fl;EmxCre;Thy1-GFP, 4.39 ± 0.17; P < 0.001). Using the same excitatory and inhibitory preynaptic and postynaptic markers as in culture (cf. Fig. 2A,B), we found a significant reduction in excitatory synapse density on primary apical dendrites of Sestd1−/− pyramidal neurons (Fig. 3B,B′,B″; puncta per 10 μm in Sestd1fl/fl; Thy1-GFP, 1.25 ± 0.15, vs. Sestd1fl/fl;EmxCre; Thy1-GFP, 0.82 ± 0.1; P < 0.05), but no change in inhibitory synapse density (Fig 3C,C′,C″; puncta per 10 μm in Sestd1fl/fl; Thy1-GFP, 1.11 ± 0.05, vs. Sestd1fl/fl;EmxCre; Thy1-GFP, 1.1 ± 0.04; P > 0.5).
Our findings indicate significant reductions in spine and excitatory synapse density in forebrain hippocampal neurons lacking Sestd1. To test whether changes in spine and synapse numbers were accompanied by functional changes in these neurons, we performed paired electrophysiological recordings in cultured hippocampal slices. Hippocampal slices were prepared and maintained in culture from postnatal day 7 (P7) Sestd1fl/fl mice; Sestd1 was deleted from sparsely distributed GFP-labeled individual CA1 hippocampal neurons through biolistic transfection at 2 DIV with a plasmid expressing a Cre-GFP fusion protein, then simultaneous whole-cell recordings were obtained from a GFP-positive (i.e., Sestd1−) and a neighboring GFP-negative (i.e., Sestd1+) CA1 neuron at 14–18 DIV. Deletion of Sestd1 in the postsynaptic CA1 neuron led to an approximate 50% reduction in both AMPA receptor-mediated and NMDA receptor-mediated EPSCs (Fig. 3D; mean amplitude (pA) for AMPAR-EPSCs: control 120.1 ± 30.5, Cre 56.2 ± 15.8, transfected as a percentage of control: 52.0 ± 10.6; NMDAR-EPSCs: control 30.9 ± 6.1, Cre 18.0 ± 4.2, transfected as a percentage of control: 55.4 ± 8.5). Loss of Sestd1 did not affect the paired-pulse ratio, a measure of presynaptic neurotransmitter release probability (Fig. 3E; control, 1.63 ± 0.17; Cre, 1.71 ± 0.24, P > 0.5). These results indicate a cell-autonomous postsynaptic requirement for Sestd1 at excitatory synapses in the mouse hippocampus; and together with our immunohistochemistry support a postsynaptic requirement for Sestd1 in excitatory synapse development.
Selective Loss of Sestd1 in Glutamatergic Hippocampal Neurons Increases Activity of Multiple Rho Family GTPases
We have previously shown that Sestd1 interacts genetically and biochemically with the Dact1, Vangl2, and Dvl2 proteins (Yang and Cheyette 2013; Yang et al. 2013), key components in β-catenin-dependent (Dact1 and Dvl2) (Cheyette et al. 2002; Schwarz-Romond et al. 2007) and β-catenin-independent (Dact1, Dvl2, and Vangl2) (Wang et al. 2006; Suriben et al. 2009; Wen et al. 2010; Shafer et al. 2011) forms of Wnt signaling. Accordingly, we examined whether Wnt signaling pathways were disrupted by loss of Sestd1 in neurodevelopmentally affected brain tissue. We first made neonatal hippocampal lysates from Sestd1fl/fl and Sestd1fl/fl; EmxCre+ mice and used immunoblotting to measure levels of “active” β-catenin that is unphosphorylated on Ser37 or Thr41 (a read-out of the Wnt/β-catenin signaling pathway) and of phospho-Mypt1 (a substrate of Rho Kinase, a read-out of β-catenin-independent Rho GTPase signaling pathways). Selective deletion of Sestd1 from glutamatergic neurons in the hippocampus significantly increased phospho-Mypt1 without measurably affecting active β-catenin (Fig. 4A,B; Active β-catenin/total β-catenin [arbitrary units normalized to control] in Sestd1fl/fl, 1.0 ± 0.14, vs. Sestd1fl/fl;EmxCre, 1.2 ± 0.12; P > 0.5; phospho-Mypt/total Mypt [arbitrary units normalized to control] in Sestd1fl/fl, 1.0 ± 0.11, vs. Sestd1fl/fl;EmxCre, 2.1 ± 0.18; P < 0.01). This suggests that Sestd1, rather than contributing to Wnt/β-catenin signal regulation, instead contributes to regulation of Rho GTPase activity in the postnatal hippocampus. To confirm this, we directly measured active RhoA and Rac1 in adult hippocampal lysates, using a standard pull-down assay to specifically isolate and quantify the GTP-bound forms compared with total levels of these proteins. We found that the ratio of active (GTP-bound):total RhoA and Rac1 were both increased in conditional mutant (Sestd1fl/fl;EmxCre+) hippocampi compared with littermate control (Sestd1fl/fl) hippocampi (Fig. 4C,D; Rho-GTP/total Rho [arbitrary units normalized to control] in Sestd1fl/fl, 1.0 ± 0.12, vs. Sestd1fl/fl;EmxCre, 6.4 ± 0.15; P < 0.05; Rac-GTP/total Rac [arbitrary units normalized to control] in Sestd1fl/fl, 1.0 ± 0.03, vs. Sestd1fl/fl;EmxCre, 10.1 ± 3.0; P < 0.05).
The BCR Rac1-GAP is an Endogenous Partner of Sestd1 in Neurons
To gain insight into the molecular mechanism of Sestd1 function in neurons, we employed a proteomic approach to reveal its endogenous binding partners in the postnatal hippocampus. We used a specific anti-Sestd1 antibody to pull down proteins physiologically associated with Sestd1 in hippocampal lysates; these Sestd1-associated proteins were then identified by MS. This approach identified BCR as a Sestd1-associated protein in the hippocampus. BCR is a serine/threonine kinase and GAP previously demonstrated to antagonize dendrite, spine, and synapse formation in HCNs through inhibition of the RhoA family member Rac1 (Oh et al. 2010; Park et al. 2012; Um et al. 2014). We confirmed that BCR forms physiological complexes with Sestd1 by co-IP of the endogenous proteins from adult hippocampal lysates (Fig. 5A), then mapped the Sestd1 domain that contributes to Sestd1–BCR complex formation via further co-IP experiments using truncated Sestd1 proteins recombinantly expressed along with full-length BCR in immortalized cultured cells. The Sestd1 carboxyl-terminal region, comprised of all 3 of the protein’s spectrin-like repeats (Sestd1 [275–696]) failed to form complexes with BCR, whereas a polypeptide consisting of the amino-terminal 274 amino acids of Sestd1 including the Sec14-like domain (Sestd1 [1–274]), as well as the full-length Sestd1 protein, readily formed complexes with BCR in this assay (Fig. 5B). Together these experiments indicate that BCR is a physiological partner of Sestd1 in the hippocampus, and suggest that the N-terminal region of Sestd1 including the Sec14-like domain mediates this interaction. Along with previously published findings (Fig. 5C), this supports that Sestd1 serves as one component of a multiprotein Rho GTPase regulatory pathway in differentiating forebrain neurons.
To further assess the neurodevelopmental relevance of this finding, we obtained a targeted BCR knockout mouse line (BCRKO), crossed it to the Sestd1KO mouse line, and examined neurodevelopmental phenotypes in cultured forebrain (cortical and hippocampal) pyramidal neurons derived from neonatal mice lacking both Sestd1 and BCR (Sestd1−/−; BCR−/−), compared with those from littermates that were WT at both loci (Sestd1+/+; BCR+/+) and littermates lacking only one or the other locus (i.e., Sestd1−/−; BCR+/+ and Sestd1+/+; BCR−/−) using techniques identical to those we previously used to assess Sestd1KO HCNs (Fig. 2). Using these methods, we found that constitutive loss of either Sestd1 or BCR produced indistinguishable deficits in spine and excitatory synapse density (Fig. 5D,E), as well as an indistinguishable increase in the percentage of spines with immature (filopodial or thin) morphology (Fig. 5F). Moreover, although there was a trend toward slightly increased phenotypic severity in Sestd1/BCR double mutant neurons, these phenotypes were not significantly different from those in single mutant (Sestd1KO only or BCRKO only) littermates (Fig. 5D–F; for spine density: WT 3.16 ± 0.15, Sestd1KO 2.41 ± 0.23, BCRKO 2.38 ± 0.15, Sestd1KO/BCRKO 1.96 ± 0.21; Sestd1KO vs. WT P < 0.01, BCRKO vs. WT P < 0.001, Sestd1KO/BCRKO vs. WT P < 0.0001, Sestd1KO vs. BCRKO P > 0.5, Sestd1KO vs. Sestd1KO/BCRKO P = 0.2, BCRKO vs. Sestd1KO/BCRKO P = 0.1; for excitatory synapse puncta density: WT 1.98 ± 0.14, Sestd1KO 1.19 ± 0.12, BCRKO 1.14 ± 0.09, Sestd1KO/BCRKO 0.93 ± 0.11; Sestd1KO vs. WT P < 0.001, BCRKO vs. WT P < 0.0001, Sestd1KO/BCRKO vs. WT P < 0.0001, Sestd1KO vs. BCRKO P > 0.5, Sestd1KO vs. Sestd1KO/BCRKO P = 0.1, BCRKO vs. Sestd1KO/BCRKO P = 0.2; for spine maturity: WT 0.23 ± 0.02 vs. Sestd1KO 0.36 ± 0.03; vs. BCRKO 0.38 ± 0.02; vs. Sestd1KO/BCRKO 0.39 ± 0.03; Sestd1KO vs. WT P < 0.001, BCRKO vs. WT P < 0.0001, Sestd1KO/BCRKO vs. WT P < 0.0001, Sestd1KO vs. BCRKO P > 0.5, Sestd1KO vs. Sestd1KO/BCRKO P > 0.5, BCRKO vs. Sestd1KO/BCRKO P > 0.5). The phenocopy between the individual Sestd1 and BCR KOs under identical experimental conditions supports that the corresponding gene products have similar activities upstream of spine and excitatory synapse phenotypes; moreover, the epistasis observed in the double mutants compared with each single mutant provides classic genetic support for the 2 gene products operating in a single biochemical pathway upstream of these phenotypes. Considered together with our MS and co-IP results, the data therefore support that Sestd1 and BCR functionally cooperate upstream of dendritic spine and excitatory synapse formation and/or plasticity in forebrain pyramidal neurons, likely via regulation of Rho GTPase activity.
The Subcellular Localization of Sestd1 is Initially Presynaptic, But Shifts to Postsynaptic During Postnatal Hippocampal Development
Finally, we asked whether the subcellular localization of Sestd1 changes as neurons mature by fractionating hippocampal synaptosomes prepared from WT mice at 4 postnatal stages and monitoring relative levels of Sestd1 alongside established presynaptic or postsynaptic markers. Synaptophysin and PSD95 were appropriately enriched in the expected presynaptic and postsynaptic fractions at all stages tested (Fig. 4A). Sestd1 was present in brain homogenates at all stages, but higher levels were present at P1 and P14 compared with P21 and adults (Fig. 5G). Interestingly, the distribution of Sestd1 in these biophysical fractions shifted dramatically over postnatal development: At P1 Sestd1 localized predominantly in the fraction containing synaptophysin, but Sestd1 became progressively more localized to the fraction containing PSD95 as animals aged, with an inflexion point between P14 and P21 (Fig. 5H) (Sestd1 presynaptic vs. postsynaptic ratio: P1 [92.3 vs. 7.7%]; P14 [56.2 vs. 43.8%]; P21 [27.6 vs. 72.7%]; adult [13 vs. 87%]).
Discussion
As in the primitive streak during earlier embryonic stages (Suriben et al. 2009; Yang and Cheyette 2013), loss of Sestd1 closely phenocopies loss of Dact1 in developing hippocampal pyramidal neurons (Okerlund et al. 2010). This includes reductions in dendrite complexity, spines, and excitatory synapses. Data obtained using immunohistochemical labeling of synapses in fixed neurons and brain tissue agree with whole-cell recordings made in a living tissue slice preparation, demonstrating that deletion of Sestd1 from individual neurons in the CA1 region of the hippocampus reduces postsynaptic responses of the affected neuron. The unusual shift in localization of Sestd1 from pre- to postsynaptically associated biophysical fractions as the hippocampus develops postnatally raises fascinating questions about the subcellular requirement for Sestd1 in these compartments as dendrites, spines, and synapses form and mature; especially as our biochemical studies show that phenotypes affecting these structures in Sestd1-deficient neurons correlate with increased RhoA and Rac1 GTPase activity.
As neither Dact1 nor Sestd1 contains a GTPase regulatory domain, we hypothesize that this regulation is determined by separate GAPs or GEFs that form complexes with them. We have shown that a GAP that physiologically interacts with Sestd1 in the postnatal hippocampus is BCR. BCR is a neurodevelopmentally expressed antagonist of Rac1; its deletion leads to increased Rac1 activity in differentiating pyramidal neurons (Oh et al. 2010; Park et al. 2012; Um et al. 2014). This parallels our findings here that deletion of Sestd1 leads to increased Rac1 activity, and together with our genetic and biochemical evidence suggests that Sestd1 and BCR may cooperate in Rac1 regulation upstream of dendritic spine and synapse phenotypes. Along with Rac1, we found that RhoA activity also increased in Sestd1 mutant neurons. This dovetails with a prior report that Sestd1 can partner with a Rho GEF, TRIO8, in these cells (Lee et al. 2015), and with our own prior finding of Sestd1 interactions with another GEF, Arhgef2, based on MS in other tissues (Yang and Cheyette 2013). Taken as a whole, our findings here combined with prior work suggest that Sestd1 can form functional complexes with multiple GAPs and GEFs to modulate RhoA superfamily GTPases. This may be important in processes that involve complex coordination of actin cytoskeletal remodeling, as during the formation, maintenance, and modification of dendritic spines in forebrain pyramidal neurons.
Our neurodevelopmental findings using genetic deletion of Sestd1 and BCR contrast with some prior published studies (Oh et al. 2010; Um et al. 2014; Lee et al. 2015) investigating the same basic molecular and neurodevelopmental questions using different methodologies. In these studies, loss of these gene products led to increased dendrite complexity, spine density, and synapse density. Dendritic spine formation depends on rapid changes in Rho GTPase activity and the consequential intricate dynamics of actin filament assembly and disassembly; slight shifts in the balance and/or timing in different experimental setups can therefore generate apparently opposing biological consequences (Nikolic 2002; Woolfrey and Srivastava 2016; Hedrick and Yasuda 2017). On this basis, the apparent discrepancy between our phenotypic findings and these prior studies is consistent with these proteins operating at the level of Rho GTPase regulation. To summarize, our data and the literature strongly support that Sestd1 partners with several other GTPase regulatory proteins to modulate multiple Rho GTPases upstream of dendritic spine and synapse development within differentiating and mature mammalian forebrain neurons. Ongoing research will elucidate molecular mechanisms underlying this complexity, which is likely to be critical for normal brain function and may also play a role in the pathophysiology of neuropsychiatric conditions, potentially including bipolar disorder (Song et al. 2015, 2017).
Notes
We thank Dr Randall T. Moon of the Howard Hughes Medical Institute and the UW Department of Pharmacology for assistance with MS. The authors also thank members of John Rubenstein’s neighboring laboratory at UCSF for supportive discussions and interactions, and Petros Minasi for laboratory assistance. Confocal microscopy was performed at the UCSF Nikon Imaging Center with advice from Dr Kurt Thorn. The authors declare no competing financial interests. Conflict of Interest: None declared.
Funding
National Institutes of Health (NIH) grants: T32MH089920 (X.Y.Y.), R01HD055300 (X.Y.Y., R.E.S., A.P.R., B.N.R.C.), K08MH100562 (J.A.G.), and U01HL100395 (A.M.R.); it was also supported in part by the UW Proteomics Resource UWPR95794 (A.M.R.) and a NARSAD Independent Investigator Award (B.N.R.C).
References
- Aceti M, Creson TK, Vaissiere T, Rojas C, Huang WC, Wang YX, Petralia RS, Page DT, Miller CA, Rumbaugh G. 2015. Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biol Psychiatry. 77:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello A, Yang X, Vogt D, Stanco A, Rubenstein JL, Cheyette BN. 2013. Dapper antagonist of catenin-1 cooperates with Dishevelled-1 during postsynaptic development in mouse forebrain GABAergic interneurons. PLoS One. 8:e67679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GM 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, Arikkath J. 2012. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc. 7:1741–1754. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. 2002. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell. 2:449–461. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. 2012. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 17:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. 2000. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 28:41–51. [DOI] [PubMed] [Google Scholar]
- Fisher DA, Kivimae S, Hoshino J, Suriben R, Martin PM, Baxter N, Cheyette BNR. 2006. Three dact gene family members are expressed during embryonic development and in the adult brains of mice. Dev Dyn. 235:2620–2630. [DOI] [PubMed] [Google Scholar]
- Garside ML, Turner PR, Austen B, Strehler EE, Beesley PW, Empson RM. 2009. Molecular interactions of the plasma membrane calcium ATPase 2 at pre- and post-synaptic sites in rat cerebellum. Neuroscience. 162:383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. 2002. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 22:6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. 2011. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 71:1085–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick NG, Yasuda R. 2017. Regulation of Rho GTPase proteins during spine structural plasticity for the control of local dendritic plasticity. Curr Opin Neurobiol. 45:193–201. [DOI] [PubMed] [Google Scholar]
- Kivimae S, Yang XY, Cheyette BNR. 2011. All Dact (Dapper/Frodo) scaffold proteins dimerize and exhibit conserved interactions with Vangl, Dvl, and serine/threonine kinases. BMC Biochem. 12(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopaske GT, Lange N, Coyle JT, Benes FM. 2014. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 71:1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Huang CC, Hsu KS. 2015. The phospholipid-binding protein SESTD1 negatively regulates dendritic spine density by interfering with Rac1-Trio8 signaling pathway. Sci Rep. 5:13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PM, Stanley RE, Ross AP, Freitas AE, Moyer CE, Brumback AC, Iafrati J, Stapornwongkul KS, Dominguez S, Kivimae S, et al. . 2016. DIXDC1 contributes to psychiatric susceptibility by regulating dendritic spine and glutamatergic synapse density via GSK3 and Wnt/beta-catenin signaling. Mol Psychiatry. doi: 10.1038/mp.2016.184. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miehe S, Bieberstein A, Arnould I, Ihdene O, Rutten H, Strubing C. 2010. The phospholipid-binding protein SESTD1 is a novel regulator of the transient receptor potential channels TRPC4 and TRPC5. J Biol Chem. 285:12426–12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M. 2002. The role of Rho GTPases and associated kinases in regulating neurite outgrowth. Int J Biochem Cell Biol. 34:731–745. [DOI] [PubMed] [Google Scholar]
- Oh D, Han S, Seo J, Lee JR, Choi J, Groffen J, Kim K, Cho YS, Choi HS, Shin H, et al. . 2010. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J Neurosci. 30:14134–14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerlund ND, Kivimae S, Tong CK, Peng IF, Ullian EM, Cheyette BN. 2010. Dact1 is a postsynaptic protein required for dendrite, spine, and excitatory synapse development in the mouse forebrain. J Neurosci. 30:4362–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerlund ND, Stanley RE, Cheyette BN. 2016. The planar cell polarity transmembrane protein Vangl2 promotes dendrite, spine and glutamatergic synapse formation in the mammalian forebrain. Mol Neuropsychiatry. 2:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AR, Oh D, Lim SH, Choi J, Moon J, Yu DY, Park SG, Heisterkamp N, Kim E, Myung PK, et al. . 2012. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci. 125:4518–4531. [DOI] [PubMed] [Google Scholar]
- Pavlowsky A, Chelly J, Billuart P. 2012. Emerging major synaptic signaling pathways involved in intellectual disability. Mol Psychiatry. 17:682–693. [DOI] [PubMed] [Google Scholar]
- Phillips M, Pozzo-Miller L. 2015. Dendritic spine dysgenesis in autism related disorders. Neurosci Lett. 601:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Romond T, Metcalfe C, Bienz M. 2007. Dynamic recruitment of axin by Dishevelled protein assemblies. J Cell Sci. 120:2402–2412. [DOI] [PubMed] [Google Scholar]
- Sekino Y, Kojima N, Shirao T. 2007. Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem Int. 51:92–104. [DOI] [PubMed] [Google Scholar]
- Shafer B, Onishi K, Lo C, Colakoglu G, Zou Y. 2011. Vangl2 promotes Wnt/planar cell polarity-like signaling by antagonizing Dvl1-mediated feedback inhibition in growth cone guidance. Dev Cell. 20:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bergen SE, Di Florio A, Karlsson R, Charney A, Ruderfer DM, Stahl EA, Chambert KD, Moran JL, Gordon-Smith K, et al. . 2015. Genome-wide association study identifies SESTD1 as a novel risk gene for lithium-responsive bipolar disorder. Mol Psychiatry. 21:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bergen SE, Di Florio A, Karlsson R, Charney A, Ruderfer DM, Stahl EA, Chambert KD, Moran JL, Gordon-Smith K, et al. . 2017. Genome-wide association study identifies SESTD1 as a novel risk gene for lithium-responsive bipolar disorder. Mol Psychiatry. 22:1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence EF, Soderling SH. 2015. Actin out: regulation of the synaptic cytoskeleton. J Biol Chem. 290:28613–28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriben R, Kivimae S, Fisher DA, Moon RT, Cheyette BN. 2009. Posterior malformations in Dact1 mutant mice arise through misregulated Vangl2 at the primitive streak. Nat Genet. 41:977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. 2010. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um K, Niu S, Duman JG, Cheng JX, Tu YK, Schwechter B, Liu F, Hiles L, Narayanan AS, Ash RT, et al. . 2014. Dynamic control of excitatory synapse development by a Rac1 GEF/GAP regulatory complex. Dev Cell. 29:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, van Schaick H, Kaartinen V, Deemer K, Coates T, Landing B, Pattengale P, Dorseuil O, Bokoch GM, Groffen J, et al. . 1995. Increased neutrophil respiratory burst in bcr-null mutants. Cell. 80:719–728. [DOI] [PubMed] [Google Scholar]
- Wang J, Hamblet NS, Mark S, Dickinson ME, Brinkman BC, Segil N, Fraser SE, Chen P, Wallingford JB, Wynshaw-Boris A. 2006. Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development. 133:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Chiang YJ, Gao C, Xue H, Xu J, Ning Y, Hodes RJ, Gao X, Chen YG. 2010. Loss of Dact1 disrupts planar cell polarity signaling by altering dishevelled activity and leads to posterior malformation in mice. J Biol Chem. 285:11023–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfrey KM, Srivastava DP. 2016. Control of dendritic spine morphological and functional plasticity by small GTPases. Neural Plast. 2016:3025948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Cheyette BN. 2013. SEC14 and spectrin domains 1 (Sestd1) and Dapper antagonist of catenin 1 (Dact1) scaffold proteins cooperatively regulate the Van Gogh-like 2 (Vangl2) four-pass transmembrane protein and planar cell polarity (PCP) pathway during embryonic development in mice. J Biol Chem. 288:20111–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Fisher DA, Cheyette BN. 2013. SEC14 and spectrin domains 1 (Sestd1), dishevelled 2 (Dvl2) and dapper antagonist of catenin-1 (Dact1) co-regulate the Wnt/planar cell polarity (PCP) pathway during mammalian development. Commun Integr Biol. 6:e26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. 2012. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 4:a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]