Abstract
The von Economo neurons (VENs) are specialized large bipolar projection neurons with restricted distribution in the human brain, and they are far more abundant in humans than in non-human primates. However, VEN functions remain elusive due to the difficulty of isolating VENs and dissecting their connections in the brain. Here, we combined laser-capture-microdissection with RNA sequencing to describe the transcriptomic profile of VENs from human anterior cingulate cortex (ACC). Using pyramidal neurons as reference cells, we identified 344 genes with VEN-associated expression differences, including 215 higher and 129 lower expression genes. Functional enrichment and protein–protein interaction network analyses showed that these genes with VEN-associated expression differences are involved in VEN morphogenesis and functions, such as dendrite branching and axon myelination, and many of them are associated with human social-emotional disorders. With the use of in situ hybridization and immunohistochemistry assays, we validated four novel VEN markers (VAT1L, CHST8, LYPD1, and SULF2). Collectively, we generated a full-spectrum expression profile of VENs from human ACC, greatly enlarging the pool of genes with VEN-associated expression differences that can help researchers to understand the role of VENs in normal and disordered human brains.
Keywords: brain evolution, human cognition, RNA sequencing, transcriptome, von Economo neuron
Introduction
Humans have the most cortical neurons among extant primates, contributing to a superior cognitive ability gained during the origin and evolution of our species (Sousa et al. 2017). The diverse neuronal cell types and their connection patterns provide layers of complexity so that fine-tuned neural circuits can be formed in the human brain. Thus, an understanding of brain cell types is essential to elucidating the properties of neural circuits and how they lead to the generation of behaviors (Ecker et al. 2017).
The von Economo neurons (VENs) are specialized large bipolar projection neurons with a characteristic spindle-shaped soma, thick basal and apical dendrites (Butti et al. 2013). The rediscovery of VENs by Nimchinsky et al. (1995) inspired subsequent efforts of dissecting their functional roles. In the human brain, VENs are primarily distributed in layer 5 of anterior cingulate cortex (ACC) and frontoinsula (FI) (Allman et al. 2011). The average volume of VENs is about 4.6 times larger than that of the layer 5 pyramidal neurons (Allman et al. 2005). VENs are considered an important topic in study of human brain evolution because in primates, they represent a relatively recent evolutionary specialization (appeared in macaque monkeys, apes, and humans) and are far more abundant in humans than in non-human primates (Nimchinsky et al. 1995, 1999; Allman et al. 2010, 2011; Evrard et al. 2012). Later studies found that besides primates, VENs also exist in distantly related mammalian species, including perissodactyla, cetartiodactyl, elephants, and hippopotamus (Butti et al. 2009, 2013, 2014; Hakeem et al. 2009; Raghanti et al. 2015).
VEN functions still remain elusive due to the topographic restriction of VENs and the difficulty of describing their connections in the brain. Since ACC and FI cortices represent key hubs within a large-scale network involved in social-emotional and autonomic functions (Seeley et al. 2007; Guo et al. 2016; Dijkstra et al. 2018), it was speculated that human VENs might have undergone evolutionary changes with specialized functions, for example, social awareness and interoception. VENs may relay output of the processing within ACC and FI to other brain regions and their large size is desirable in relaying a fast intuitive-assessment of complex social situations (Allman et al. 2005), though whether the ACC VENs and the FI VENs are functionally homogeneous are unknown. Consistent with this view, VEN abnormalities have been reported in brain disorders with social-emotional deficits, such as autism spectrum disorders (ASD), schizophrenia (SCZ), the behavioral variant of frontotemporal dementia (bvFTD), and Alzheimer’s disease (AD) (Seeley 2008; Brune et al. 2010; Santos et al. 2011; Kim et al. 2012; Gefen et al. 2018). Given the potentially crucial role of VENs in the brain, dissecting the basic VEN biology is important for understanding normal and disordered human cognition.
Previously, efforts have been made to reveal the molecular profile of VENs, a prerequisite for understanding their functions. However, due to technical and specimen-availability limitations, previous studies were all candidate-based, mainly using in situ hybridization (ISH) and immunohistochemistry (IHC), and only limited candidate genes were tested. Using IHC, Allman et al. (2005) reported positive staining of three neurotransmitter receptor genes (AVPR1A, DRD3, and HTR2B) implicated in the formation of social bonds and reward processing. Stimpson et al. (2011) examined expression of three gut and immune function related proteins (encoded by ATF3, IL4Rα, and NMB, respectively) among different neuron types (pyramidal, VEN, fork, enveloping, and other multipolar), and they found that VENs showed the greatest percentage of immunostaining and a higher proportion of VENs in humans were immunoreactive than in apes. Using mRNA ISH, Cobos and Seeley (2015) characterized the expression patterns of six transcription factors in human ACC and FI, and they found that VENs prominently express FEZF2 and CTIP2 that are involved in regulating the fate and differentiation of subcerebral projection neurons. Recently, by leveraging the Allen Brain Atlas to evaluate mRNA expression of 176 neurotransmitter-related genes, Dijkstra et al. (2018) identified additional three genes (VMAT2, GABRQ, and ADRA1A) as mRNA markers for VENs and fork cells, and they proposed that VENs and fork cells might have a novel, uncharacterized mode of cortical monoaminergic function. These studies provided valuable information for understanding the molecular identity of VENs though they only uncovered “the tip of the iceberg”. A candidate-independent approach is required to fully depict the molecular profile of VENs.
Combining laser-capture-microdissection (LCM) with RNA sequencing, we isolated VENs from human ACC and analyzed their transcriptomic profiles. In comparison to layer 3 and layer 5, pyramidal neurons in ACC, we identified 344 genes showing VEN-associated expression differences, including 215 higher and 129 lower expression genes. Using ISH and IHC assays, we validated four new VEN marker genes with predominant expression in VENs. These data provide a full-spectrum molecular profile of VENs.
Materials and Methods
Postmortem Brain Sample Collection
For LCM RNA sequencing, postmortem brain tissue samples of ten adult males (Han Chinese) were obtained from the Chinese Brain Bank Center (CBBC, http://cbbc.scuec.edu.cn). The ten male donors aged 23–63 years, and the brain samples were collected within 24–72 h after death. The exact PMIs (postmortem intervals) for these samples were not available. We also obtained four additional ACC samples for ISH and IHC analyses, including three adult males and a 1-year-old male (Supplementary Table S1). Female subjects were not included due to sample availability. To avoid the complication of diseasing-causing changes, samples with neurodegenerative changes were excluded according to the pathological evaluation records of CBBC (Supplementary Table S1). The brain samples were flash-frozen by liquid nitrogen and stored in −80 °C freezers before use. According to the protocol of CBBC, written informed consent for brain autopsy and use of the specimen for research was obtained from either the donors or their relatives. Each ACC sample is a tissue block of about 2 cm3, containing area 24b and area 32.
Morphological Identification of Neurons with Nissl Staining
ACC sections of 10-μm thick were prepared using a sliding microtome. The section slides were first placed into microslide box on dry ice, and then stored at −80 °C until use. For Nissl staining, the section slides were placed on a blotting paper for 30 s, and then transferred into the 95% EtOH jar for 30 s. After 30 s in the second jar containing 75% EtOH, and 30 s in the third jar containing 50% EtOH, the slides were placed on a blotting paper. We added 400-μl Nissl stain and 10-μl RNase inhibitor mixture onto the slide, and incubated it at room temperature for 40 s. We then proceeded through the dehydration series (50%, 75%, and 95% EtOH) till the two rounds of 100% EtOH, and left the slides in the final xylene jar for 5 min. After air-drying in the fume hood for 5 min, we immediately proceeded with LCM.
Laser-Capture-Microdissection and RNA Sequencing
The section slides and 0.2-ml thin-wall tubes were loaded onto the LCM apparatus (Leica CTR6500). We adjusted the brightness and focus at 5× magnification in order to distinguish layer 5 and layer 3 of the cortex. Then we set the optimal laser thickness and power parameters at 40× magnification. Once the laser spot has been adjusted, we identified VENs and pyramidal neurons for capture. Two measures were taken during LCM: (1) In order to keep integrity of RNA, capture period should not exceed 1 h; (2) To avoid mis-characterization of cell types, we only capture cells with typical shape. The length of VEN soma should be over 70 μm, and there are no other cells overlapping with it. VENs are mainly located in area 24a and area 24b, and they are more abundant in area 24b than in area 24a (Vogt et al. 1995). For each tissue specimen, we captured 50 cells for each cell type, including layer 5 VENs from area 24b, layer 5 pyramidal neurons from area 24b, layer 5 pyramidal neurons from area 32 and layer 3 pyramidal neurons from area 24b. Each 50-cell capture resulted in approximately 30 pg RNA. We prepared three slides of each tissue sample for cell capture. We added 10 μl freshly prepared cell lysis buffer into the tube containing the 50 captured cells. After a fast centrifuge, the tube was incubated at 70 °C for 2 min, and then transferred on ice immediately. We took 4.5 ul cell lysis for synthesis and amplification of cDNA using previously published protocol with minor modifications (Tang et al. 2010). A total of 40 samples were prepared (4 cell types from each of the 10 brain specimens).
For each sample, 1.5 μg cDNA was used for library construction and next generation sequencing. Library preparation was performed following Illumina’s manufacture instructions. Libraries were sequenced on the Illumina Hiseq 2000 platform (BGI, Shenzhen, China). We generated ~5 G data for each 50 cells capture of four cell types from the 10 male individuals. Low quality reads were removed, and ployA and adapter sequences were cut before mapping. The raw RNAseq data (FASTQ) was deposited in the Genome Sequence Archive (GSA) database (Project ID: PRJCA000919).
Reads Mapping and Expression Data Adjustment Using AC-PCA
The filtered reads were mapped to the human reference genome (GRCh38) using Bowtie (version 2.3.1) and Tophtat (version 2.1.1) (Trapnell et al. 2009; Kim et al. 2013) with default parameters. We filtered the data using a criterion of >50% mapping rate, resulting in valid data from 39 samples. One dropout of low mapping rate (33%) (sample ID: 201558X) was removed from further analysis. Cufflinks was employed to assemble the transcripts and to output gtf files, which were then merged with cuffmerge (Trapnell et al. 2010). The FPKM data matrix was acquired using cuffdiff (Trapnell et al. 2010).
The FPKM data matrix obtained above was filtered using the criterion of at least three samples with FPKM > 1, which led to the identification of 14 745 expressed genes. To remove potential confounding factors either biologically or technically originated, we utilized the AC-PCA method for data adjustment so that the shared variation among samples of the same cell type can be captured (Lin et al. 2016).
Identification of Differentially Expressed Genes
The AC-PCA adjusted expression matrix was taken as input to perform differential expression analysis with limma (Smyth 2004). A linear model was fitted for the adjusted expression matrix. To make pair-wise comparison among the cell types (VENs, the layer 5 pyramidal neurons and the layer 3 pyramidal neurons), the contrast matrix was created to expend the linear model fit and then to compute empirical Bayes statistics. The matrix of pair-wise differentially expressed genes (DEGs) were extracted. Two criteria for DEG identification were applied: (1) the pair-wise fold change is larger than 2 (|log2FC| > 1); (2) the adjusted P value is smaller than 0.05 after multiple test correction using the Benjamini and Hochberg method. To see the VEN-associated expression difference, we overlapped the two DEG matrix (VEN vs. layer 5 pyramidal neurons and VEN versus layer 3 pyramidal neurons). After removing 10 genes showing opposite direction of expression changes, we obtained a total of 344 VEN-associated DEGs (216 higher and 128 lower expression genes). Similarly, the pyramidal-neuron-associated DEGs were also identified. GraphPad Prism5 was recruited to conduct scatter plot analysis for differentially expressed genes FPKM matrix. MGI (Mouse Genome Informatics) (Smith et al. 2018) was used to search mouse knockout phenotypes for the VEN-associated DEGs.
RNA In situ Hybridization
For ISH using the fresh-frozen ACC samples, we used the commercial kit (RNAscope® 2.5 HD Detection Reagents-BROWN, #322 310). For each gene, ~20 target-specific double Z probes were designed. We employed the ISH protocol provided by the manufacture (Advanced Cell Diagnos tics, Inc., California, USA). Tissue sections (15 μm thick) were obtained using cryostat at −20 °C, and stored at −80 °C before use. Briefly, the cryostat sections were air-dried at room temperature for 20 min and fixed by 4% pre-chilled (at 4 °C) paraformaldehyde (PFA) in phosphate-buffered saline solution (0.1 M, pH 7.4) for 60 min. The sections were dehydrated through 50%, 70% and 100% EtOH (5 min for each concentration and twice for 100% EtOH) and air-dried for 5 min. The sections were digested using RNAscope Protease IV for 30 min at room temperature and washed twice with 1× PBS. Hybridization was performed using the HybEZ Humidity Control Tray at 40 °C for 2 h. To amplify the signals, the sections were subject to further hybridization to a cascade of signal amplification molecules, culminating in binding of horseradish peroxidase (HRP)-labeled probes. A chromogenic substrate (DAB) was used to detect the target RNA. After dehydration using EtOH (2 min for 70% and 95% respectively) and xylene (5 min), the sections were mounted. Each single RNA transcript appears as a distinct dot of chromogen precipitate visible using a common bright field microscope at 5–40× magnification.
To quantify the ISH results, using the ImageJ tool (version_1.8.0; https://imagej.nih.gov/ij/index.html), we calculated and compared the signal intensities among VENs, L5Ps and L3Ps. Three adult male brain samples were analyzed (Supplementary Table S1). Global calibration and background subtraction were performed before assessing the signal intensity (gray value). For each cell type of a sample, we analyzed more than 50 cells from at least five randomly selected microscope fields (under 5× magnification).
IHC and IF Analyses
The ACC frozen blocks were cut into 25 μm serial sections at −20 °C with cryostat. The sections were separated in five sets of six serial sections. The first four sets were used for IHC, and the last set was Nissl-stained for counting the proportion of cells with positive immunoreactivity. Sections were air-dried over night to ensure complete desiccation.
For IHC, sections were soaked in 0.01 M PBS at pH 7.2 for 5 min and fixed in 4% paraformaldehyde at room temperature for 30 min, and washed 3 × 5 min in 0.01 M PBS. Sections were soaked for 20 min in 0.01 M PBS containing 0.6% H2O2 and 80% methanol at room temperature, washed 3 × 5 min in 0.01 M PBS, and then incubated for 20 min at room temperature (RT) in 0.01 M PBS containing 0.5% TritonX-100, and incubated for 30 min in 3% BSA (CAS # 9048-46-8, Amresco). The primary antibodies are mouse monoclonal or rabbit polyclonal antibodies against VAT1L (1:25; orb1026, Biorbyt), LYPD1 (1:25; ab157516, Abcam), SULF2 (1:25; GTX38989, GeneTex), and CHST8 (1:25; HPA016004, Atlas Antibodies). The validation data (western blotting) of these antibodies can be found at the manufacturers’ webpages (https://www.abcam.com; https://www.genetex.com; https://atlasantibodies.com). Sections were incubated with primary antibodies in PBT-Az for 60 h at RT. After 3 × 5 min washing, sections were incubated with biotinylated secondary antibody in PBT-Az (Biotinylated Goat Anti-Polyvalent 1:50, ab64264, Abcam) for 2 h at RT, washed 3 × 5 min in 0.01 M PBS and then incubated with Streptavidin Peroxidase (1:100, ab64264, Abcam) in PBT-Az for 2 h at RT. After 3 × 5 min washing, immunostaining was visualized with DAB kit (DAB-1031, MXB Biotechnologies). Finally, sections were counterstained with hematoxylin (BF06064M, Beijing Biodragon Immunotechnologies), dehydrated and coverslipped. Negative controls for testing the specificity of the secondary antibodies were included by omitting the primary antibody, and no immunoreactivity was detected in the negative controls. IHC images were acquired using Leica CTR6500 at 10–40× magnification. To calculate the ratio of cells with positive immuoreactivity, we analyzed three ACC samples, including two adult males and a 1-year-old male (Supplementary Table S1). For each sample, we calculated the percentage of cells with positive immuoreactivity from three sections that contained at least 50 VENs.
For immunofluorescence (IF), six 16-μm thick frozen ACC sections were fixed in 4% paraformaldehyde at RT for 30 min, washed 3 × 5 min in 0.01 M PBS, and then incubated for 20 min in 0.01 M PBS containing 0.3% TritonX-100 at RT. Sections were incubated for 30 min in 3% BSA. Three sections were incubated with primary antibodies (VAT1L: 1:50, ab68926; LYPD1:1:50, ab157516, Abcam; MAP2:1:1000, AB5543, Millipore), and the other sections were incubated in primary antibodies (SULF2:1:50, GTX38989, GeneTex; CHST8:1:50, HPA016004, Atlas Antibodies; MAP2:1:1000, AB5543, Millipore) in PBT-Az for 36 h at RT. Sections were washed 3 × 5 min in 0.01 M PBS and incubated for 2 h in secondary antibodies in PBT-Az. The secondary antibodies are: Alexa Fluor 647 Donkey anti-Rabbit IgG (1:200, Catalog # A-31 573, Thermo Fisher Scientific), Alexa Fluor 568 Donkey anti-Mouse IgG (1:200, Catalog # A-10 037, Thermo Fisher Scientific), and Alexa Fluor 488 Goat anti-Chicken IgY (1:500, Catalog # A-11 039, Thermo Fisher Scientific). Sections were washed 3 × 5 min in 0.01 M PBS and incubated in DAPI diluent (2 μg/ml, D8417-1MG, Sigma-Aldrich) for 5 min. Sections were rinsed and coverslipped. The IF images were acquired using Nikon Confocal laser scanning microscopy (A1MP+).
Supervised Clustering, Enriched Gene Ontology and Network Analyses
We first extracted the FPKM values of those DEGs. To find VEN-higher DEGs with large expression difference, we chose those genes with logFC > 1.5 and then performed hierarchical clustering using pheatmap (https://cran.r-project.org/web/packages/pheatmap). Enriched gene ontology (GO) terms (q-value < 0.05) for VEN-associated DEGs were calculated using Consensus PathDB (Herwig et al. 2016).
For network analysis, we used String (Szklarczyk et al. 2015) to construct protein-protein interaction network so that the relationship among VEN-specific DEGs can be evaluated.
Results
Isolation of VENs from ACC and RNA Sequencing
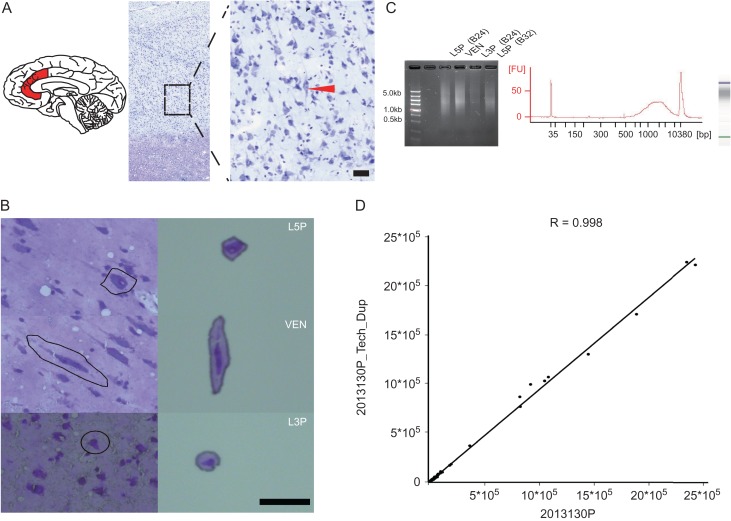
The morphology of VENs and pyramidal neurons were identified by Nissl staining (Fig. 1A). The sampled ACC tissue blocks contain both Brodmann’s area 24b and Brodmann’s area 32, and we collected VENs from layer 5 of area 24b by LCM (Fig. 1B). We also collected three reference cell types, including the neighboring pyramidal neurons from layer 5 of area 24b, the pyramidal neurons from layer 5 of area 32, and the pyramidal neurons from layer 3 of area 24b (abbreviated as L3Ps). A total of 10 adult males were subject to LCM capture (Supplementary Table S1). Consequently, 40 LCM captures (4 cell types from 10 individuals) were generated. After amplification (see Methods for details), we obtained more than 1.5 μg cDNA for each LCM capture, and the cDNA fragments ranging from 500 bp to 5 000 bp were collected by gel electrophoresis (Fig. 1C). For RNA-sequencing (BGI, Shenzhen, China), we first performed a technical replication, and the result turned out to be highly consistent (R2 = 0.989) (Fig. 1D). For each LCM capture, we acquired >5 G RNAseq data with >50 million reads (Supplementary Table S2).
Figure 1.
Laser-capture-microdissection (LCM) and RNA sequencing of VENs from human ACC. (A) Image of Nissl stain showing layer 5 VENs. The black bar indicates 100 μm; (B) Isolation of VENs, L5Ps, and L3Ps using LCM under microscope; (C) RNA extraction and quantification; (D) Technical replication of RNA sequencing using L5P cells (sample ID: 2013130). The correlation coefficient between the two technical replicates is high (R = 0.998).
Comparison of Transcriptional Profiles Between VENs and Pyramidal Neurons
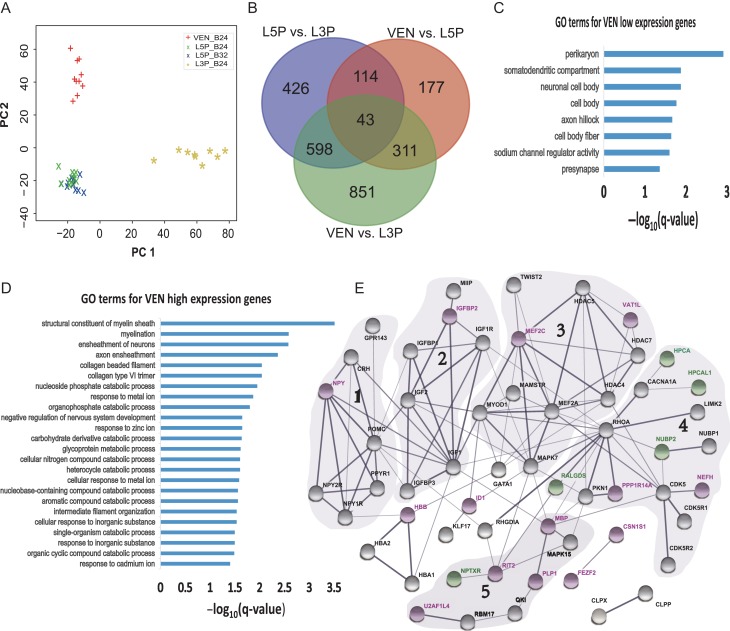
We filtered the data using a criterion of >50% mapping rate and one layer 3 pyramidal neuron sample with low mapping rate was removed from further analysis (sample ID: 201 558X, mapping rate = 33.0%) (Supplementary Table S2). We used a relatively conserved cutoff to determine genes expressed in VENs and pyramidal neurons, i.e., at least three samples with FPKM > 1. This led to the identification of 14 745 expressed genes. According to the AC-PCA plot (see Methods for details), VENs were clearly separated from the pyramidal neurons from layer 3 and layer 5 (Fig. 2A). The layer 5 pyramidal neurons from area 24b and area 32 were tightly clustered (Fig. 2A), suggesting that they have highly similar transcriptomic profiles and therefore merged as one cell type (abbreviated as L5Ps) for further analysis. Of note, VGLUT1 is highly expressed in VENs and pyramidal neurons (average FPKM > 500; Supplementary Figure 1A), confirming the glutamatergic nature of VENs, as suggested in the previous report (Evrard et al. 2012).
Figure 2.
Transcriptome comparison among VENs, L5Ps, and L3Ps, and functional enrichment and protein-protein interaction (PPI) analyses of VEN-associated differentially expressed genes (VA-DEGs). (A) The AC-PCA map showing clustering of VENs and pyramidal neurons (L5Ps and L3Ps); (B) Counts of cell-type-specific genes with expression changes; (C) Functional enrichment of the 129 lower expression VA-DEGs; (D) Functional enrichment of the 215 higher expression VA-DEGs; (E) The PPI network with five clusters of genes associated with VEN morphology and functions. The five PPI clusters were classified based on the local PPI patterns and functional relatedness of the genes. The purple circles are the high expression VA-DEGs, and the green circles are the low expression VA-DEGs. The gray circles are genes showing direct or indirect interactions with VA-DEGs.
We next performed pair-wise comparisons among VENs, L5Ps, and L3Ps, and a total of 3 629 differentially expressed genes (DEGs) were identified, including 645 DEGs between VENs and L5Ps, 1 803 DEGs between VENs and L3Ps, and 1 181 DEGs between L3Ps and L5Ps (Fig. 2B). Clearly, VENs and L5Ps are closely related in view of the number of DEGs between them, consistent with the speculation that VENs are likely derived from layer 5 pyramidal neurons (Allman et al. 2011).
Identification of VEN-Associated Expression Differences
Using both L5Ps and L3Ps as the reference cell types, we found 354 VEN-associated DEGs (VA-DEGs) either higher expressed or lower expressed in VENs. Notably, 344 out of the 354 VA-DEGs showed the same direction of expression changes when compared VENs with L5Ps and L3Ps respectively, and only 10 genes showed an opposite direction, suggesting that a great majority of the identified VA-DEGs have characteristic expression in VENs (Supplementary Table S3). Among the 344 VA-DEGs with consistent expression patterns in VENs, there were 215 higher expression and 129 lower expression genes. For the 215 higher expression VA-DEGs, the fold changes were highly correlated between VEN-L5P and VEN-L3P comparisons (R2 = 0.41, P < 0.0001), contrasting the much lower correlation for the 128 lower expression VA-DEGs (R2 = 0.04, P = 0.021) (Supplementary Figure S2), implying that these higher expression VA-DEGs are likely the key genes defining the molecular profile of VENs as they showed relatively consistent between-cell-type expression divergence. With the same approach, we also identified 125 L5P-associated and 640 L3P-associated DEGs, respectively (Supplementary Tables S4 and Supplementary Table S5).
Previous studies using candidate-based approaches have reported 11 genes showing either VEN-positive or VEN-enriched expression (Allman et al. 2005; Stimpson et al. 2011; Cobos and Seeley 2015; Dijkstra et al. 2018). We therefore checked the status of these genes in our RNAseq data. Notably, FEZF2 and CTIP2 showed VEN-predominant expression, significantly higher than the expression in L5Ps and L3Ps, consistent with the previous report (Cobos and Seeley 2015) (Supplementary Figure S1B-C). Similarly, among the three recently reported VEN marker genes (VMAT2, GABRQ, and ADRA1A) (Dijkstra et al. 2018), GABRQ showed a significant VEN-higher expression. However, the expression of VMAT2 and ADRA1A were low (average FPKM < 0.5) and no difference was seen between VENs and L5Ps/L3Ps (Supplementary Figure S1D-F). Interestingly, we detected VEN-enriched expression of two other genes (VAT1 and ADRA1D) from the same gene family containing VMAT2 and ADRA1A (Supplementary Figure S1G-H). For the other reported genes, the expression of ATF3, IL4Rα and NMB (Stimpson et al. 2011) were low, and VENs showed relatively higher expression than L5Ps/L3Ps for IL4Rα and NMB though statistically not significant (Supplementary Figure S1I-K), but the expression of AVPR1A, DRD3, and HTR2B (Allman et al. 2005) were quite low (average FPKM < 0.2), and no between-cell-type difference was detected (Supplementary Figure 1L-N). Collectively, the RNAseq data confirmed the VEN-enriched expression of five previously reported genes, among which FEZF2 and GABRQ were included in the 215 higher expression VA-DEGs, suggesting that the transcriptomic data generated is informative to reveal the molecular profile of VEN and to identify new VEN marker genes.
Gene Ontology and Protein–Protein Interaction Network Analyses
To understand functional enrichment of the 344 VA-DEGs, we conducted GO analysis. For the 129 lower expression VA-DEGs, there were eight significantly enriched GO terms (adjusted P < 0.05), mostly related with cell body and dendrite morphology such as perikaryon, somatodendritic compartment and presynapse (Fig. 2C and Supplementary Table S6). For the 215 higher expression VA-DEGs, there were 24 significantly enriched GO terms (adjusted P < 0.05), with the top four terms associated with myelination and the other terms involved in varied neural processes such as intermediate filament organization, response to metal ions and catabolic process (Fig. 2D and Supplementary Table S7). In general, these enriched functional categories reflected the known morphological characters of VENs. For example, there were 11 lower expression VA-DEGs that were enriched for somatodendritic compartment (Supplementary Table S6), which might contribute to the large spindle-shaped perikarya and thick basal dendrites of VENs. The enriched GO terms of myelination contained nine higher expression VA-DEGs (Supplementary Table S7), and the presumably enhanced myelination for VENs indicated their need of long-range projection and impulse propagation (Craig 2009; Allman et al. 2010; Evrard et al. 2012). We also analyzed GO enrichment for the 90 L5P-associated and 284 L3P-associated higher expression genes, which showed different functional categories. The enriched GO terms are mostly associated with lipid biosynthesis for L5Ps and with synapse for L3Ps (Supplementary Figure S3).
We next performed protein-protein interaction (PPI) network analysis using String (Szklarczyk et al. 2015). We identified a large PPI network involving 53 proteins, as well as two separate paired PPIs (Fig. 2E). The large PPI network containing 12 higher and five lower expression VA-DEGs were divided into five clusters based on the network structure and gene functions (Fig. 2E). Cluster-1, 2 and 3 included four VEN higher expression genes (NPY, IGFBP2, MEF2C, and VAT1L), which were closely related with social-emotional-autonomic functions such as anxiety, stress, arousal and circadian rhythm (Kuo et al. 2007; Haberland et al. 2009; Fernandez and Torres-Aleman 2012; Zhao et al. 2015; Singh et al. 2017; Longo et al. 2018). In Cluster-4, there were four VEN lower expression genes (HPCA, HPCAL1, NUBP2, and RALGDS) that interact with the hub genes RHOA and CDK5. RHOA can cause reduction of dendritic complexity while CDK5 modulates actin cytoskeletal dynamics, which may regulate VEN somatodendritic morphogenesis (Lee et al. 2000; Fu et al. 2007). Cluster-5 contained two higher expression VA-DEGs (MBP and PLP1) that were involved in myelination (Popko et al. 1987; Inoue 2005; Wang et al. 2008; Lutz et al. 2014; Raasakka et al. 2017). Additionally, the previously identified VEN marker gene FEZF2 (Cobos and Seeley 2015) had a separate PPI with CSN1S1, a newly identified higher expression VA-DEG in this study (Fig. 2E). Taken together, the PPI network provided useful information in dissecting the roles of these VA-DEGs in shaping up VEN morphology and functions.
ISH, IHC and IF Analyses of Novel VEN Markers
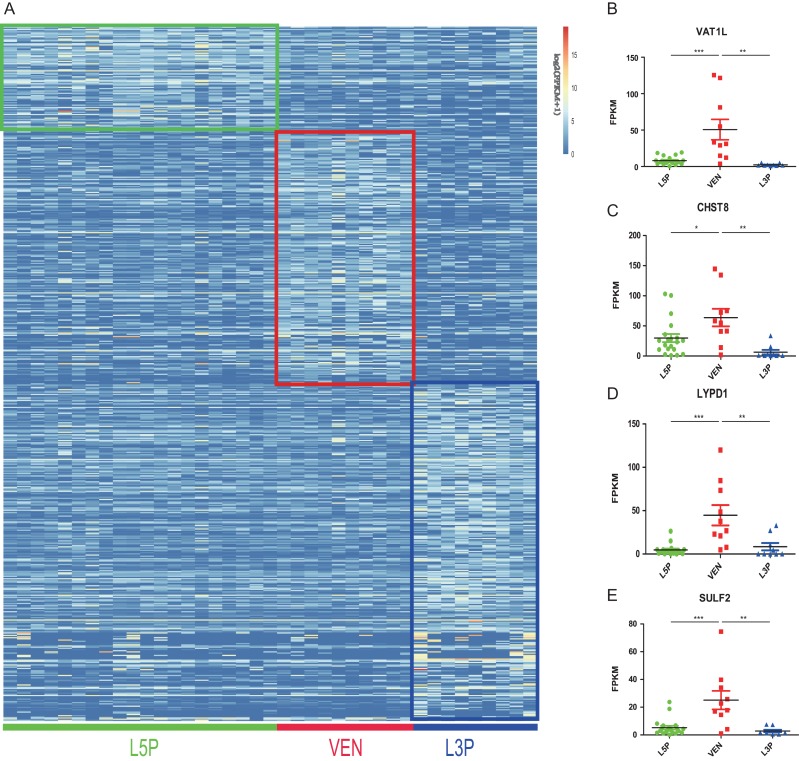
The cell-type-associated higher expression genes may serve as cell markers. There are 215 higher expression genes in VENs, contrasting 90 in L5Ps and 284 in L3Ps (Supplementary Tables S4 and S5). The expression heatmap of the cell-type-specific higher expression genes is shown in Fig. 3A. We listed the top 20 higher expression VA-DEGs in Table 1, including 16 protein-coding genes, two RNA genes and two pseudogenes. These genes showed the largest VEN-associated higher expression (average log2FC > 2), and therefore could serve as candidates of VEN markers. Among the 16 protein-coding genes, besides the previously reported FEZF2 (Cobos and Seeley 2015), the other 15 genes are new candidates. We therefore checked the known functions of these genes (DisGeNET, http://www.disgenet.org/web/DisGeNET/menu/home), and we found that nine genes were associated with human mental disorders, such as ASD, SCZ, AD, and depression, consistent with the reported VEN abnormalities in social-emotional diseases. More importantly, in the mouse gene-knockout models (Mouse Genome Informatics, http://www.informatics.jax.org), 10 genes showed abnormal behavioral or neural defects (Table 1). Based on the fold changes and their known functions, we selected four candidate VEN marker genes for further validation analyses, including VAT1L (Vesicle amine transport 1 like), CHST8 (Carbohydrate sulfotransferase 8), LYPD1 (LY6/PLAUR domain containing 1), and SULF2 (Sulfatase 2) (highlighted in Table 1). The VEN-predominant expression of these four genes are shown in Figure 3B–E.
Figure 3.
Identification of VEN-higher expression genes. (A) The expression heatmap of genes with cell-type-associated expression changes in VENs (215 genes), L5Ps (90 genes), and L3Ps (284 genes); (B–E) The four novel VEN marker genes showing VEN-predominant expression compared with L5Ps and L3Ps. The statistical significance levels are labeled as *(P < 0.01), **(P < 0.01) and ***(P < 0.001) (two-tailed t test).
Table 1.
The top 20 VEN-associated higher expression genes
| Gene symbol | Gene name | LogFC | Gene function | Mouse gene-knockout phenotype | Human neurological disorder/phenotype |
|---|---|---|---|---|---|
| RNA5SP352 | RNA, 5S Ribosomal Pseudogene 352 | 4.48 | Pseudogene | — | — |
| Y_RNA | Y_RNA 809_201 | 3.64 | Resistance to UV irradiation, DNA replication | — | — |
| VAT1L | Vesicle Amine Transport 1 Like | 3.28 | Oxidoreductase activity, chromatin hub | Abnormal behavior | — |
| FEZF2 | FEZ Family Zinc Finger 2 | 3.03 | Regulate the differentiation, fate and axon targeting of layer five neurons | Abnormal axon guidance and cerebral cortex morphology | ASD, Microcephaly |
| CHST8 | Carbohydrate Sulfotransferase 8 | 2.91 | Sulfotransferase to mediate sulfation of chondroitin | Abnormal barbering behavior; pup cannibalization | Tobacco use disorder |
| ACA64 | Small Nucleolar RNA ACA64 | 2.77 | Guide chemical modifications of other RNAs | — | — |
| LYPD1 | LY6/PLAUR Domain Containing 1 | 2.73 | Control anxiety | Increased fear; anxiety-related response | Tobacco use disorder |
| SULF2 | Sulfatase 2 | 2.48 | Remove sulfate groups from heparan sulfate; axon guidance signaling | Neurite outgrowth; embryonic lethality | — |
| ALKBH3 | AlkB Homolog 3, Alpha-Ketoglutaratedependent Dioxygenase | 2.47 | DNA alkylation damage repair; demethylate m1A in tRNA and mRNA | Abnormal DNA repair; mortality/aging | — |
| FABP6 | Fatty Acid Binding Protein 6 | 2.42 | Binding to bile acids | Abnormal bile salt homeostasis | — |
| PLP1 | Proteolipid Protein 1 | 2.41 | Myelin protein | Abnormal axon morphology; demyelination; neurodegeneration; seizures | PMD, SCZ, Intellectural disability, Cocaine abuse, Explosive speech |
| METRNL | Meteorin Like, Glial Cell Differentiation Regulator | 2.28 | Induces white adipose browning;neurite outgrowth | Decreased lumbar vertebrae number and neutrophil cell number | — |
| MBP | Myelin Basic Protein | 2.19 | Myelin sheath;neurite outgrowth and neuronal survival | Abnormal myelination; abnormal spatial learning; ataxia | SCZ, Demyelinating diseases, Multiple sclerosis |
| LINC00982 | Long Intergenic Non-Protein-Coding RNA 982 | 2.17 | Cell proliferation and cell cycle progression | — | — |
| RNU6-1240P | RNA, U6 Small Nuclear 1240, Pseudogene | 2.15 | Pseudogene | — | — |
| RGMA | Repulsive Guidance Molecule Family Member A | 2.15 | Neural tube closure and neuroepithelial morphology; guide axons | Exencephaly; prenatal lethality | Sciatic neuropathy, Eplepsy, Trauma, Depression |
| PRR5 | Proline Rich 5 | 2.15 | Cell growth and tumorigenesis | — | Tobacco use disorder |
| NPY | Neuropeptide Y | 2.09 | Anxiolytic-like effects;food intake | Decreased body weight; increased anxiety-related response | Depression, SCZ, AD, Alcohol abuse, Anxiety |
| HAPLN4 | Hyaluronan And Proteoglycan Link Protein 4 | 2.09 | Stabilize extracellular matrix | Decrease brevican localization in PNNs | SCZ |
| SOHLH1 | Spermatogenesis And Oogenesis Specific Basic Helix-Loop-Helix 1 | 2.08 | Regulate oogenesis and spermatogonial differentiation | Abnormal spermiogenesis | — |
Note: the selected four candidate VEN markers are highlighted. The fold change is displayed as LogFC, that is, the average log2(fold change) of VENs/L5Ps and VENs/L3Ps.
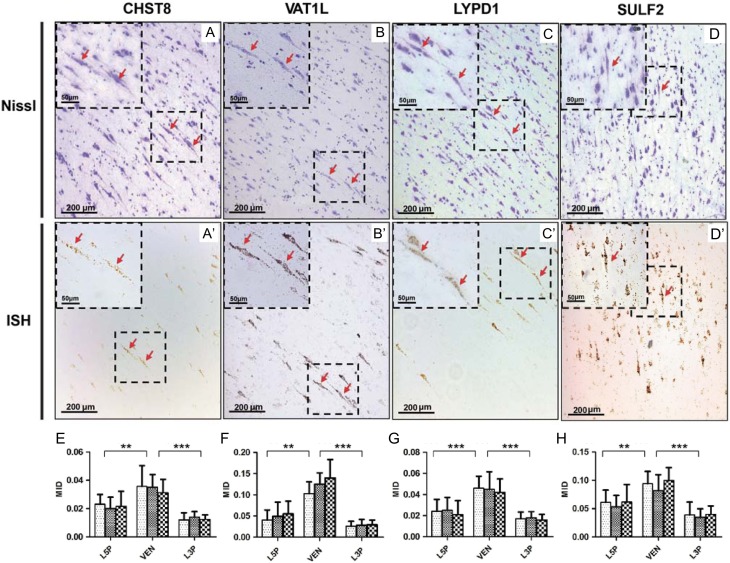
To validate the expression pattern of the four selected candidates, we first performed mRNA ISH by testing three additional adult male samples (Supplementary Table S1). As expected, all four genes showed positive ISH signals in VENs (Fig. 4), among which three genes (CHST8, VAT1L, and LYPD1) were predominantly expressed in VENs and much less in neighboring L5Ps (Figs 4A–C, 4A′-C′). The expression of SULF2 was also high in VENs, and less strong in neighboring L5Ps (Fig. 4D, D′). Using the ImageJ tool (version_1.8.0; https://imagej.nih.gov/ij/index.html), we quantified the ISH signal intensity of each cell type. Consistently, we detected 1.45–4.81 times higher expression for the four genes in VENs compared to L5Ps and L3Ps (Fig. 4E–H).
Figure 4.
In situ hybridization (ISH) analysis of four genes as the novel candidates of VEN markers. (A–D) Nissl stain images of layer 5 neurons. VENs with typical spindle shape are indicated with red arrows; (A′–D′) ISH images of the corresponding microscopic fields with Nissl stain, showing predominant mRNA expression of the four genes in VENs; (E–H) Comparison of quantified signal intensities of the four genes between VENs and L5Ps/L3Ps. Three adult male samples were analyzed (Supplementary Table S1). The statistical significance levels are labeled as **(P < 0.01) and ***(P < 0.001) (two-tailed t test).
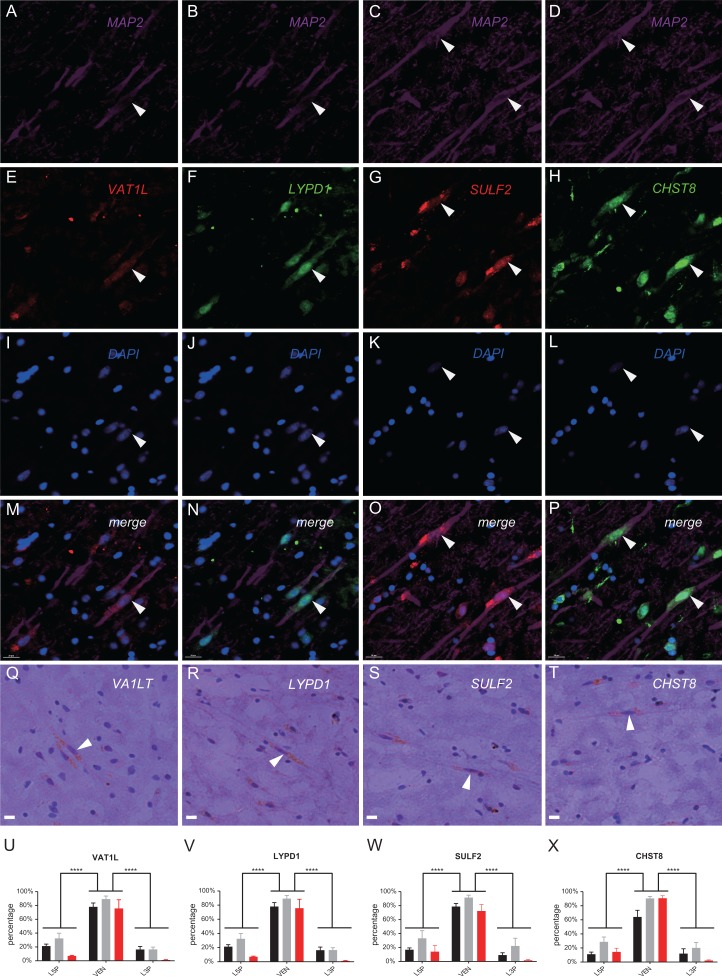
Using IHC and IF, we tested the expression of these four genes at protein level in two adult male samples (Supplementary Table S1). The IF images showed that all four genes were predominantly expressed in VENs with three of them (LYPD1, SULF2, and CHST8) showing strong nuclear signals (Fig. 5A–P). Consistently, the IHC images showed the same expression pattern of VEN-enriched expression (Fig. 5Q–T) and the immunoreactivity signals were mainly located within the cell body, and not seen in the nucleus. In contrast, the signals in neighboring pyramidal neurons were either weak or absent (Fig. 5M–T). We also performed IHC in a 1-year-old male, and the pattern was the same though the overall signal intensity was relatively weak (Supplementary Figure S4). Quantification analysis using the IHC data showed consistent patterns. VENs had about 60–95% positive stain for the four genes, 3.8–7.3 times higher than those in L5Ps and L3Ps (Fig. 5U–X). Taken together, the results of ISH and IHC assays further confirmed the high expression VA-DEGs identified in the RNAseq data, and the top genes may serve as new VEN markers and help understanding VEN functions.
Figure 5.
Immunohistochemistry (IHC) and immunofluorescence (IF) analyses of the four candidate VEN markers. (A–P) IF-stain images of the four candidate genes (VAT1L, LYPD1, SULF2, and CHST8). The MAP2 gene is a neuron-specific maker and was used to show cell morphology (A–D), and DAPI was used to show cell nucleus (I–L). The immunoreactivity signals of the four genes are shown in panels E–H, and the merged signals are shown in panels M–P; (Q–T) IHC-stain images of the four candidate genes. Typical VENs with positive IHC-stain are indicated by arrows; (U–X) Comparison of ratios of positive-stain cells between VENs and L5Ps/L3Ps. Three male samples were analyzed (Supplementary Table S1). The black and gray bars are the two adult males, and the red bar is the 1-year-old male. The statistical significance level is labeled as ****(P < 0.0001) (two-tailed t test).
Discussion
VENs are present in humans, apes, and macaques, as well as several other mammalian lineages, representing parallel evolutionary innovations in the central nervous system. In primates, since humans have far more abundant VENs than apes and macaques, it is likely that during the origin of our own species, VENs might have undergone human-specific functional specialization, contributing to our higher cognitive abilities (Allman et al. 2011). To probe into the functions of human VENs, using LCM RNA-seq, we generated by far the most complete molecular profile of human VENs from ACC. In total, we identified 344 genes with VEN-associated expression differences, and 215 of them showed VEN-increased expression, contrasting the previously reported 11 genes with VEN-positive or VEN-enriched expression (Allman et al. 2005; Stimpson et al. 2011; Cobos and Seeley 2015; Dijkstra et al. 2018). Using ISH, IHC, and IF, we further validated four novel VEN markers, including VAT1L, CHST8, LYPD1, and SULF2. Hence, these identified VEN-associated expression difference genes and their expression patterns are highly informative to defining the molecular identity of VENs and dissecting functions of these evolutionarily specialized projection neurons.
The GO enrichment analyses indicated that among the 129 VEN-lower expression genes, many of them are involved in modulating VEN morphology (Fig. 2C). This is likely processed through lower expression genes associated with perikarya and dendrites. In particular, four lower expression genes (HPCA, HPCAL1, RALGDS, and NUBP2) had direct interactions with RHOA, an important regulator of dendritic morphogenesis. It was shown that in Drosophila, neurons lacking RhoA overextended their dendrites while activated RhoA expression caused a reduction of dendritic complexity (Lee et al. 2000). The single large basal dendrite of VEN might have resulted from a transformation of the genetic programs during evolution for pyramidal neuron development to modify the basal dendrite in order to concentrate its growth in the primary component and suppress the secondary and tertiary branching (Allman et al. 2011). Consistently, HPCA was reported as a possible partner of MLK2/3 which interacts with small GTPase Rac and Cdc42 to regulate cellular cytoskeleton dynamics (Tibbles and Woodgett 1999), and HPCAL1 was implicated in clathrin-dependent membrane trafficking processes (Braunewell and Klein-Szanto 2009). It is possible that these VEN lower expression genes may interact with RHOA in a way to incease its expression, resulting in a reduction of dendritic complexity. As expected, VENs showed a relatively higher RHOA expression compared to L5Ps and L3Ps though statistically not significant (P > 0.05, two-tailed t test; the average FPKMs are 83.8 ± 107.3 for VENs, 51.2 ± 76.2 for L5Ps, and 36.8 ± 29.9 for L3Ps). Taken together, the identified VEN lower expression genes are possibly involved in suppressing VEN dendrite branching and conduction of nerve signals.
By contrast, the VEN-higher expression genes likely define VEN functions as many of them are related with social-emotional-autonomic functions. For example, NPY (Neuropeptide Y) is among the top-20 VEN-higher expression genes (Table 1). By interacting with its receptors (NPY1R, NPY2R, and PPYR1; Fig. 2E), NPY can influence food intake, anxiety, stress, pain perception, and circadian rhythm, consistent with the known functions of ACC, the brain area with rich VEN distribution (Kuo et al. 2007; Loktev and Jackson 2013; Sohn et al. 2013; Singh et al. 2017; Longo et al. 2018). In addition, NPY can regulate sleep by modulating noradrenergic signaling. Interestingly, the noradrenaline receptor ADRA1D is also VEN-higher expressed (Supplementary Figure S1H), implying that it may work together with NPY to regulate the related functions. Furthermore, the increased expression of IGFBP2 (Insulin-like growth factor-binding protein 2) in the VENs is intriguing because this gene was identified in the IGF signaling which modulates hippocampal neurogenesis, synapse formation, neuritogenesis, axon guidance, brain reward system, arousal, spatial memory, and associative activity (Hodge et al. 2004; Aberg et al. 2006; Babri et al. 2007; Ciucci et al. 2007; Chiu et al. 2008).
Strikingly, an inter-chromosomal interaction was reported between the IGF2/H19 locus and the VAT1L locus, and the recruitment of IGF2/H19 and VAT1L loci to lamina was regulated to promote circadian oscillations in transcription (Zhao et al. 2015). Importantly, VAT1L was listed as the top one VEN-higher expression gene (Table 1), and it can react with chromatin remodeling transcription factors (such as MEF2A, MYOD1, and MEF2C) and histone deacetylases (HDAC4, HDAC5, and HDAC7) which are recruited to chromatin hubs to regulate chromatin 3D dynamics and transcription (Fig. 2E) (Haberland et al. 2009; Pliner et al. 2017).
We also detected a few VEN-higher expression genes that may contribute to VEN morphology. For example, MEF2C can promote excitatory synapse elimination in hippocampal pyramidal neurons (Harrington et al. 2016), and MEF2C loss of function led to an increase in dendritic spine density (Barbosa et al. 2008). Hence, MEF2C high expression is expected to reduce the density of dendritic spines of VENs. Another example is CPLX1, which regulates spontaneous neurotransmitter release and synaptic growth (Kümmel et al. 2012).
VENs are projection neurons and they bear large, rapidly conducting axons (Allman et al. 2002). Accordingly, we saw many myelination-related genes (such as MBP and PLP1) showing VEN-increased expression (Fig. 2D and Supplementary Table S7). It is known that a myelin sheath is to increase the speed at which impulses propagate along the myelinated fiber through preventing the electric current from leaving the axon. VENs’ axonal connections extend through the underlying white mater to other parts of the brain (Nimchinsky et al. 1995), and they may have diverse cortical and subcerebral targets (Cobos and Seeley 2015) that require long-distance communications. Therefore, an enhanced myelination is necessary for VENs to perform this function (Hartline 2008). Consistently, we detected VEN-increased expression of NEFH (neurofilament protein, heavy polypeptide) (Supplementary Table S3), previously found to be expressed in the VENs of ACC using ISH (Dijkstra et al. 2018). NEFH is strongly and positively related to axon caliber, a key determinant of conduction velocity in nerve cells (Walker et al. 2001). Hence, the VEN-increased expression of NEFH is consistent with the proposal that VENs have large and rapidly conducting axons supporting quick social decision-making (Allman et al. 2005).
There is a possibility that the VEN-increased expression of myelination-related genes (such as MBP and PLP1) might be caused by oligodendrocyte cell body contamination in the microdissections since oligodendrocytes produce myelin, rather than neurons. To evaluate this situation, we checked the expression status of ten known oligodendrocyte-high-expression genes, including five oligodendrocyte marker genes (OLIG2, MOBP, MAG, MOG, and SOX10) and five myelination-related genes (ERMN, MYT1, MYRF, CNTN2, and PDGFRA) (Li et al. 2018). Overall, these genes had relatively low expression in all tested neuron types (average FPKM <20), suggesting that the suspected contamination was unlikely (Supplementary Figure S5). Also, besides of oligodendrocytes, neurons also express MBP and PLP1 (Sarret et al. 2010; Lutz et al. 2014)
Additionally, the VEN-higher expression genes may also contribute to VEN specification during development. In our RNAseq data, both FEZF2 and CTIP2 showed VEN-higher expression (Table 1 and Supplementary Figure S1), confirming the previous findings (Cobos and Seeley 2015). It was reported that FEZF2 is necessary and sufficient for the differentiation of layer 5 neurons that project subcortically to the striatum, superior colliculus, basal pons, and spinal cord, and the other known VEN marker gene CTIP2 is a major downstream effector of FEZF2 (Chen et al. 2008; Rouaux and Arlotta 2013; Cobos and Seeley 2015).
Among the top 20 VEN-higher expression genes, about half of them have known association with human social-emotional disorders, such as ASD, SCZ, AD, and depression, and this observation was further reinforced by the phenotypic outcomes of gene-knockout mice showing abnormal behavioral or neural defects (Table 1). This information is useful in understanding VENs’ role in human social-emotional disorders.
Technically, LCM allows a clear separation of individual VENs under microscope so that the entire transcriptome (instead of limited number of candidate genes) of VENs can be analyzed. Compared to the currently fast-advancing single-cell-sequencing (SCS) techniques (Woodworth et al. 2017), LCM-RNAseq is a cost-effective method in studying molecular profiles of restrictively distributed neuronal types in the brain. For example, even in human ACC and FI, VENs only account for about 1–2% of the total neurons, and if using SCS, an enormously large number of single neurons from ACC need to be analyzed in order to capture enough VENs. Also, LCM uses frozen tissues while SCS usually require fresh tissues that are rather difficult to obtain for humans. Although recent advances in SCS allowed dissecting transcriptome of cell nucleus using frozen tissues, the genes analyzed were still limited (around 1 000–2 000 genes per nucleus) (Lake et al. 2017). By contrast, with LCM-RNAseq, we could quantify the expression of a great proportion of genes in the genome. Among the four cell types from 10 brain samples, we quantified expression of 14 745 genes using a relatively conserved cutoff. Finally, the topographic positions of the LCM-isolated cells are known while this information cannot be directly obtained for SCS.
It should be noted that the LCM-RNAseq method also has limitations. The microdissected cells are in fact cell parts, not the entire cells, and there were PCR-based cDNA amplifications before RNA-seq. Consequently, these factors may increase expression variation so that a few previously identified VEN markers (e.g., VMAT2) (Dijkstra et al. 2018) using ISH and IHC showed low expression levels in our RNAseq data. To cope with this drawback, a large number of biological replicates combined with multiple techniques would be ideal.
Supplementary Material
Notes
We thank Yan Guo for her technical assistance in this study. Conflict of Interest: None declared.
Authors’ Contributions
L.Y. and B.S. designed the study; L.Y., Y.Y., and J.Y. performed experiments; Y.S. and J.D. provided brain samples; L.Y., Y.Y., and J.Y. performed data analysis; L.Y., Y.Y., J.Y., and B.S. wrote the paper.
Funding
This study was supported by grants from the Strategic Priority Research Program (XDB13010000) and the National Natural Science Foundation of China (31730088 and 31621062).
Conflict of interest
The authors declare no competing financial interests.
References
- Aberg ND, Brywe KG, Isgaard J. 2006. Aspects of growth hormone and insulin-like growth factor-I related to neuroprotection, regeneration, and functional plasticity in the adult brain. ScientificWorldJournal. 6:53–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman JM, Hakeem AY, Waston K. 2002. Two phylogenetic specializations in the human brain. Neuroscientist. 8:335–346. [DOI] [PubMed] [Google Scholar]
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, Park S, Goubert V, Hof PR. 2010. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct Funct. 214:495–517. [DOI] [PubMed] [Google Scholar]
- Allman JM, Tetreault NA, Hakeem AY, Park S. 2011. The von Economo neurons in apes and humans. Am J Hum Biol. 23:5–21. [DOI] [PubMed] [Google Scholar]
- Allman JM, Watson KK, Tetreault NA, Hakeem AY. 2005. Intuition and autism: a possible role for Von Economo neurons. Trends Cogn Sci. 9:367–373. [DOI] [PubMed] [Google Scholar]
- Babri S, Badie HG, Khamenei S, Seyedlar MO. 2007. Intrahippocampal insulin improves memory in a passive-avoidance task in male wistar rats. Brain Cogn. 64:86–91. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Kim M-S, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel Duby R, et al. 2008. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. PNAS. 105:9391–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunewell KH, Klein-Szanto AJ. 2009. Visinin-like proteins (VSNLs): interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2+sensor proteins. Cell Tissue Res. 335:301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune M, Schobel A, Karau R, Benali A, Faustmann PM, Juckel G, Petrasch-Parwez E. 2010. Von Economo neuron density in the anterior cingulate cortex is reduced in early onset schizophrenia. Acta Neuropathol. 119:771–778. [DOI] [PubMed] [Google Scholar]
- Butti C, Fordyce RE, Raghanti MA, Gu X, Bonar CJ, Wicinski B, Wong EW, Roman J, Brake A, Eaves E, et al. 2014. The cerebral cortex of the pygmy hippopotamus, Hexaprotodon liberiensis (Cetartiodactyla, Hippopotamidae). Anat Rec. 297:670–700. [DOI] [PubMed] [Google Scholar]
- Butti C, Santos M, Uppal N, Hof PR. 2013. Von Economo neurons: clinical and evolutionary perspectives. Cortex. 49:312–326. [DOI] [PubMed] [Google Scholar]
- Butti C, Sherwood CC, Hakeem AY, Allman JM, Hof PR. 2009. Total number and volume of Von Economo neurons in the cerebral cortex of cetaceans. J Comp Neurol. 515:243–259. [DOI] [PubMed] [Google Scholar]
- Chen B, Wang S, Hattox A, Rayburn H, Nelson S, McConnell S. 2008. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 105:11382–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SL, Chen CM, Cline HT. 2008. Insulin receptor signaling regulates synapse number, dendritic plasticity, and circuit function in vivo. Neuron. 58:708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci F, Putignano E, Baroncelli L, Landi S, Berardi N, Maffei L. 2007. Insulin-like growth factor 1 (IGF-1) mediates the effects of enriched environment (EE) on visual cortical development. PLoS One. 2:e475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Seeley WW. 2015. Human von Economo neurons express transcription factors associated with Layer V subcerebral projection neurons. Cereb Cortex. 25:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AD. 2009. How do you feel—now? The anterior insula and human awareness. Nat Rev Neurosci. 10:59–70. [DOI] [PubMed] [Google Scholar]
- Dijkstra AA, Lin LC, Nana AL, Gaus SE, Seeley WW. 2018. Von economo neurons and fork cells: a neurochemical signature linked to monoaminergic function. Cereb Cortex. 28:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H. 2017. The BRAIN initiative cell census consortium: lessons learned toward generating a comprehensive brain cell Atlas. Neuron. 96:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard HC, Forro T, Logothetis NK. 2012. Von Economo neurons in the anterior insula of the macaque monkey. Neuron. 74:482–489. [DOI] [PubMed] [Google Scholar]
- Fernandez AM, Torres-Aleman I. 2012. The many faces of insulin-like peptide signalling in the brain. Nat Rev Neurosci. 13:225–239. [DOI] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, et al. 2007. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 10:67–76. [DOI] [PubMed] [Google Scholar]
- Gefen T, Papastefan ST, Rezvanian A, Bigio EH, Weintraub S, Rogalski E, Mesulam MM, Geula C. 2018. Von Economo neurons of the anterior cingulate across the lifespan and in Alzheimer’s disease. Cortex. 99:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CC, Sturm VE, Zhou J, Gennatas ED, Trujillo AJ, Hua AY, Crawford R, Stables L, Kramer JH, Rankin K, et al. 2016. Dominant hemisphere lateralization of cortical parasympathetic control as revealed by frontotemporal dementia. Proc Natl Acad Sci USA. 113(17):E2430–E2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. 2009. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeem AY, Sherwood CC, Bonar CJ, Butti C, Hof PR, Allman JM. 2009. Von Economo neurons in the elephant brain. Anat Rec (Hoboken). 292:242–248. [DOI] [PubMed] [Google Scholar]
- Harrington AJ, Raissi A, Rajkovich K, Berto S, Kumar J, Molinaro G, Raduazzo J, Guo Y, Loerwald K, Konopka G, et al. 2016. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. Elife. 5:e20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline D. 2008. What is myelin? Neuron Glia Biol. 4:153–163. [DOI] [PubMed] [Google Scholar]
- Herwig R, Hardt C, Lienhard M, Kamburov A. 2016. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 11:1889–1907. [DOI] [PubMed] [Google Scholar]
- Hodge RD, D’Ercole AJ, O’Kusky JR. 2004. Insulin-like growth factor-I accelerates the cell cycle by decreasing G1 phase length and increases cell cycle reentry in the embryonic cerebral cortex. J Neurosci. 24:10201–10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. 2005. PLP1-related inherited dysmyelinating disorders: Pelizaeus-Merzbacher disease and spastic paraplegia type 2. Neurogenetics. 6:1–16. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Sidhu M, Gaus SE, Huang EJ, Hof PR, Miller BL, DeArmond SJ, Seeley WW. 2012. Selective frontoinsular von Economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb Cortex. 22(2):251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümmel D, Krishnakumar SS, Radoff DT, Li F, Giraudo CG, Pincet F, Rothman JE, Reinisch KM. 2012. Complexin cross-links pre-fusion SNAREs into a zig-zag array: a structure-based model for complexin clamping. Nat Struct Mol Biol. 18:927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo LE, Kitlinska JB, Tilan JU, Li L, Baker SB, Johnson MD, Lee EW, Burnett MS, Fricke ST, Kvetnansky R, et al. 2007. Neuropeptide Y acts directly in the periphery on fat tissue and mediates stress-induced obesity and metabolic syndrome. Nat Med. 13:803–811. [DOI] [PubMed] [Google Scholar]
- Lake B, Cheng S, Sos B, Fan J, Yung Y, Kaeser G, Duong T, Gao D, Chun J, Kharchenko P. 2017. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol. 36:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke S, Lee A, Luo L. 2000. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 25:307–316. [DOI] [PubMed] [Google Scholar]
- Li L, Tian E, Chen X, Chao J, Klein J, Qu Q, Sun G, Sun G, Huang Y, Warden CD, et al. 2018. GFAP mutations in astrocytes impair oligodendrocyte progenitor proliferation and myelination in an hiPSC model of Alexander disease. Cell Stem Cell. 23:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Yang C, Zhu Y, Duchi J, Fu Y, Wang Y, Jiang B, Zamanighomi M, Xu X, Li M, et al. 2016. Simultaneous dimension reduction and adjustment for confounding variation. Proc Natl Acad Sci USA. 113:14662–14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loktev AV, Jackson PK. 2013. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5:1316–1329. [DOI] [PubMed] [Google Scholar]
- Longo A, Fadda M, Brasso C, Mele P, Palanza P, Nanavaty I, Bertocchi I, Oberto A, Eva C. 2018. Conditional inactivation of Npy1r gene in mice induces behavioural inflexibility and orbitofrontal cortex hyperactivity that are reversed by escitalopram. Neuropharmacology. 133:12–22. [DOI] [PubMed] [Google Scholar]
- Lutz D, Loers G, Kleene R, Oezen I, Kataria H, Katagihallimath N, Braren I, Harauz G, Schachner M. 2014. Myelin basic protein cleaves cell adhesion molecule L1 and promotes neuritogenesis and cell survival. J Biol Chem. 289:13503–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky E, Gilissen E, Allman J, Perl D, Erwin J, Hof P. 1999. A neuronal morphologic type unique to humans and great apes. Proc Natl Acad Sci USA. 96:5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky E, Vogt B, Morrison J, Hof P. 1995. Spindle neurons of the human anterior cingulate cortex. J Comp Neurol. 355:27–37. [DOI] [PubMed] [Google Scholar]
- Pliner H, Packer J, McFaline-Figueroa J, Cusanovich D, Daza R, Srivatsan S, Qiu X, Jackson D, Minkina A, Adey A, et al. 2017. Chromatin accessibility dynamics of myogenesis at single cell resolution. bioRxiv.
- Popko B, Puckett C, Lai ESH, Readhead C, Takahashi N, Hunt SW, Sidman RL, Hood L. 1987. Myelin deficient mice expression of myelin basic protein and generation of mice with varying levels of myelin. Cell. 48:713–721. [DOI] [PubMed] [Google Scholar]
- Raasakka A, Ruskamo S, Kowal J, Barker R, Baumann A, Martel A, Tuusa J, Myllykoski M, Burck J, Ulrich AS, et al. 2017. Membrane association landscape of myelin basic protein portrays formation of the myelin major dense line. Sci Rep. 7:4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Spurlock LB, Treichler FR, Weigel SE, Stimmelmayr R, Butti C, Thewissen JG, Hof PR. 2015. An analysis of von Economo neurons in the cerebral cortex of cetaceans, artiodactyls, and perissodactyls. Brain Struct Funct. 220:2303–2314. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. 2013. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat Cell Biol. 15:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Uppal N, Butti C, Wicinski B, Schmeidler J, Giannakopoulos P, Heinsen H, Schmitz C, Hof PR. 2011. Von Economo neurons in autism: a stereologic study of the frontoinsular cortex in children. Brain Res. 1380:206–217. [DOI] [PubMed] [Google Scholar]
- Sarret C, Combes P, Micheau P, Gelot A, Boespflug-Tanguy O, Vaurs-Barriere C. 2010. Novel neuronal proteolopid protein isforms encoded by the human myelin proteolipid protein 1 gene. Neuroscience. 166:522–538. [DOI] [PubMed] [Google Scholar]
- Seeley WW. 2008. Selective functional, regional, and neuronal vulnerability in frontotemporal dementia. Curr Opin Neurol. 21:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. 2007. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 27(9):2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C, Rihel J, Prober DA. 2017. Neuropeptide Y regulates sleep by modulating noradrenergic signaling. Curr Biol. 27:3796–3811 e3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Blake J, Kadin J, Richardson J, Bult C. 2018. Mouse Genome Database(MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 46:D836–D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- Sohn JW, Elmquist JK, Williams KW. 2013. Neuronal circuits that regulate feeding behavior and metabolism. Trends Neurosci. 36:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Santpere G, Gulden FO, Sestan N. 2017. Evolution of the human nervous system function, structure, and development. Cell. 170:226–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson CD, Tetreault NA, Allman JM, Jacobs B, Butti C, Hof PR, Sherwood CC. 2011. Biochemical specificity of von Economo neurons in hominoids. Am J Hum Biol. 23:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. 2015. STRING v10: protein–protein interaction networks integrated over the tree of life. Nucleic Acids Res. 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. 2010. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 5:516–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbles LA, Woodgett JR. 1999. The stress-activated protein kinase pathways. Cell Mol Life Sci. 55:1230–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt BA, Nimchinsky EA, Vogt LJ, Hof PR. 1995. Human cingulate cortex: surface features, flat maps, and cytoarchitecture. J Comp Neurol. 359:490–506. [DOI] [PubMed] [Google Scholar]
- Walker KL, Yoo HK, Undamatla J, Szaro BG. 2001. Loss of neurofilaments alters axonal growth dynamics. J Neurosci. 21:9655–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Dimova N, Sperle K, Huang Z, Lock L, McCulloch MC, Edgar JM, Hobson GM, Cambi F. 2008. Deletion of a splicing enhancer disrupts PLP1/DM20 ratio and myelin stability. Exp Neurol. 214:322–330. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Girskis KM, Walsh CA. 2017. Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat Rev Genet. 18:230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millan-Arino L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS, et al. 2015. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell. 59:984–997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.