Abstract
Since a fecal occult blood test for colorectal cancer (CRC) does not offer sufficient diagnostic power for CRC, novel non-invasive biomarkers are hopeful for CRC screening. We conducted the current study to discover non-invasive urinary biomarkers for diagnosing CRC. Among urine samples from 258 patients (CRC, n = 148; healthy controls, n = 110), a cohort of 176 patients composed of 88 patients with GC and 88 healthy controls was selected after age- and sex-matching using propensity score. This cohort was then randomly divided into 2 groups: 53 pairs (106 patients) in the training cohort, and 35 pairs (70 patients) in the validation cohort. No significant differences were found for baseline characteristics between the CRC and healthy control groups in both training and validation cohorts. On multivariate analysis in the training cohort, urinary levels of cysteine-rich protein 61 (uCyr61) and trefoil factor 3 (uTFF3) were identified as independent significant diagnostic markers for CRC. Moreover, uCyr61 alone and the combination of uCyr61 and uTFF3 allowed significant differentiation between healthy controls and CRC groups in the training set (uCyr61: area under the curve (AUC) = 0.745 [95% CI, 0.653–0.838]; uCyr61 + uTFF3: AUC = 0.753 [95% CI, 0.659–0.847]). In the validation cohort, uCyr61 and uTFF3 were significantly higher in the CRC group than in the healthy control group, and they also allowed significant differentiation between healthy control and CRC groups (uCyr61: AUC = 0.696 [95% CI, 0.571–0.822]; uTFF3: AUC = 0.639 [95% CI, 0.508–0.770]; uCyr61 + uTFF3: AUC = 0.720 [95% CI, 0.599–0.841]), as in the training cohort. A panel combining uCyr61 and uTFF3 offers a promising non-invasive biomarker for diagnosing CRC.
Introduction
Colorectal cancer (CRC) is the third most common malignancy and the fourth leading cause of cancer-related deaths worldwide.1 Despite recent developments in medical treatment and new agents, the prognosis of CRC after diagnosis has reached a ceiling. Early diagnosis remains the most critical issue and colonoscopy is currently the gold standard for diagnosing CRC. However, colonoscopy is unsuitable for mass-screening due to the high invasiveness, risk, and financial and time costs. Elucidation of non-invasive diagnostic biomarkers for CRC is thus needed to improve prognosis.
The fecal occult blood test (FOBT) has been used in screening for CRC in medical check-ups, because use of the FOBT reduced mortality from CRC in previous epidemiological studies.[2], [3], [4] However, the FOBT does not offer sufficient diagnostic power for CRC due to its low sensitivity (30–87%).[5], [6], [7], [8] Moreover, the FOBT is cumbersome for both patients and investigators in handling stool samples, and the quality of sample collection by patients would affect the results.
Serum tumor markers such as CEA and CA19–9 have sometimes been used in clinical practice, but use of these biomarkers has not been recommended because of their very low sensitivity. Although CEA is the most commonly used tumor marker for CRC, the sensitivity of CEA is less than 50% for any-stage CRC.9 Moreover, early-stage CRC does not usually show high levels of CEA. Many studies have thus sought serum or plasma biomarkers for CRC, predominantly focusing on protein, miRNA, circulating DNA and exosomes. However, all these biomarkers are still under investigation.
Urine is a completely non-invasive sample that can be collected without pain or risk, making the sample very suitable for mass-screening of healthy individuals. However, no urinary biomarkers have been currently applied to the clinical setting of malignancies. We have previously reported on the utility of urinary biomarkers to predict the presence10 or operability11 of gastric cancer. We therefore investigated the usefulness of urinary biomarkers for detecting CRC.
Materials and Methods
Patients and Study Design
All samples were collected from September 2012 to April 2017 at 3 participating Japanese institutions (Nagoya City University Hospital, Japanese Red Cross Nagoya Daini Hospital, and Okazaki Public Health Center). Patients who met the following inclusion criteria were enrolled in this study: age between 20 and 90 years; histologically confirmed adenocarcinoma using endoscopic biopsy for the CRC group; no treatment before study enrollment for the CRC group; and no neoplasms of any type for the healthy control (HC) group. Patients with a history of neoplasms of any type and/or with multiple neoplasms were excluded from enrollment in this study. The HC cohort comprised individuals who were asymptomatic and had no evidence of neoplasms at their annual checkup.
To ensure the accuracy and comprehensiveness of reporting in this case–control biomarker study, the present study complied with both the REMARK guidelines12 and the STROBE statement.13 The study protocol was approved by the ethics committee at each participating institution and was conducted according to the ethical guidelines of the 1975 Declaration of Helsinki (6th revision, 2008). All patients provided written, informed consent before study entry. This study was registered with the University Hospital Medical Information Network Clinical Trials Registry (UMIN000021350).
Samples and Definition
All urine samples were collected before any treatment for CRC, immediately frozen and stored at −80 °C until assayed, as previously reported.[10], [11] Clinical stage was determined by the final pathological diagnosis after resection, according to the 7th edition of the Union for International Cancer Control tumor-node-metastasis classification.14
Enzyme-Linked Immunosorbent Assays (ELISAs)
We measured the urinary protein concentration of each of the proteins of interest using mono-specific ELISAs, in accordance with the instructions from the manufacturers. To measure each protein concentration, we used a Quantikine ELISA kit (R&D Systems, Minneapolis, MN) for cysteine-rich protein 61 (Cyr61) and trefoil factor 3 (TFF3), a SimpleStep ELISA Kit (Abcam, Cambridge, UK) for insulin-like growth factor-binding protein 3 (IGFBP3) and alpha 1 antitrypsin (SERPINA1), a human EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2) ELISA kit (LSBio, Seattle, WA) for EFEMP2, a human angiopoietin-like protein 2 (ANGPTL2) ELISA kit (Aviva Systems Biology, San Diego, CA) for ANGPLT2 and a Parameter Creatinine Assay (R&D Systems) for creatinine. All urinary protein levels were normalized to urinary creatinine.
Statistical Analysis
The aim of this study was to identify urinary proteins that can diagnose the presence of CRC. Representative variables were described with mean or median value and analyzed using the t-test or Mann–Whitney U test, as appropriate. Other data were analyzed using the chi-squared test or Fisher's exact probability test, as appropriate. The nonparametric Spearman's rank correlation coefficient (r) was used as a measure of correlation.
We estimated the propensity score (PS) with a logistic regression model, including 2 factors (age and sex). We randomly matched between 2 groups one by one, using the nearest-neighbor method within a caliper of width of 0.02 of the standard deviation of the logit of the PS.
Receiver operating characteristic (ROC) curve analysis was used to calculate the area under the curve (AUC) for each biomarker, and the representative value was shown as the AUC value with 95% confidence interval (CI). Logistic regression modeling was used to estimate the odds ratio (OR) with 95% CI and construct a composite score, which in turn was used to calculate the AUC for the combination biomarker. A two-tailed P value of less than 0.05 was considered statistically significant.
Results
Patients
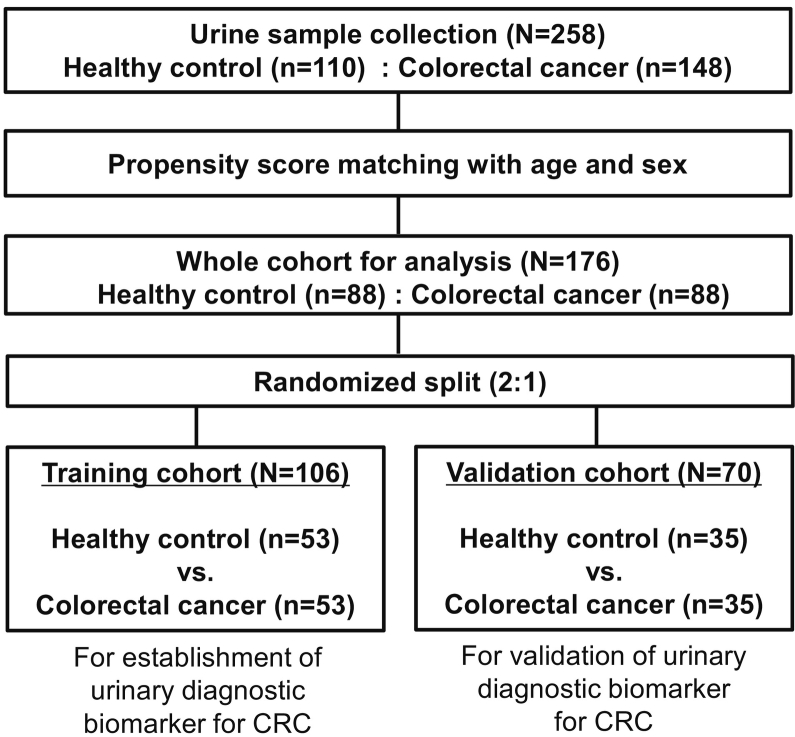
In total, 258 patients were enrolled from September 2012 to April 2017 at three Japanese institutions, consisting of 110 patients with HC and 148 patients with CRC. After one-by-one PS matching, 176 patients (88 pairs of patients from both groups) were selected as a whole cohort in the present study. A whole cohort was randomly divided into 2 groups: 53 pairs (106 patients) in the training cohort; and 35 pairs (70 patients) in the validation cohort. Urinary biomarkers were identified and a biomarker panel was established in the training cohort, then established biomarkers were tested in the independent validation cohort (Figure 1).
Figure 1.
Consort diagram. CRC, colorectal cancer.
As shown in Table 1, baseline characteristics were well balanced between both HC and GC groups. No significant differences in age, sex or serum creatinine were found between groups, in either training and validation cohorts. The training and validation cohorts showed even distributions of every stage.
Table 1.
Characteristics of the Study Cohort
| Training Cohort (n = 106) |
Validation Cohort (n = 70) |
||||||
|---|---|---|---|---|---|---|---|
| HC (n = 53) |
CRC (n = 53) |
P | HC (n = 35) |
CRC (n = 35) |
P | ||
| Age (years) | Median (range) | 65 (43–82) | 66 (46–78) | 0.962 #1 | 67 (39–78) | 67 (38–77) | 0.954 #1 |
| Sex | Male | 28 | 30 | 0.696 #2 | 23 | 22 | 0.803 #2 |
| Female | 25 | 23 | 12 | 13 | |||
| Serum Cr (mg/dl) | Mean ± SD | 0.75 ± 0.16 | 0.77 ± 0.38 | 0.661 #3 | 0.76 ± 0.16 | 0.80 ± 0.22 | 0.392 #3 |
| Stage, n | 0 | 6 (11.3%) | 1 (2.9%) | ||||
| I | 14 (26.4%) | 6 (17.1%) | |||||
| II | 7 (13.2%) | 8 (22.9%) | |||||
| III | 11 (20.8%) | 8 (22.9%) | |||||
| IV | 15 (28.3%) | 12 (34.3%) | |||||
CRC, colorectal cancer; HC, healthy control; n, number; Cr, creatinine.
Clinical stage is according to the seventh edition of the UICC-TNM classification.
Mann–Whitney U test.
χ2 test.
t test.
Establishment of Urinary Biomarker
To discover urinary diagnostic biomarkers for CRC, we thoroughly searched the previous literature on serum protein biomarkers for the diagnosis of CRC using PubMed, and identified 6 small protein biomarker candidates with a high AUC and high accuracy: Cyr 61, TFF3, IGFBP3, ANGPTL2, SERPINA1 and EFEMP2.[15], [16], [17], [18], [19], [20] Based on the results from the literature search, we analyzed urinary protein concentrations of these 6 proteins in the training cohort, using quantitative mono-specific ELISAs.
Urinary levels of Cyr61 (uCyr61) and IGFBP3 (uIGFBP3) were significantly higher in the CRC group than in the HC group, on univariate analysis. On the other hand, multivariate analysis identified uCyr61 and urinary levels of TFF3 (uTFF3) as independent significant proteins for the diagnosis of CRC (uCyr61, OR: 1.146 [95% CI, 1.048–1.252], P = .003; uTFF3, OR: 1.055 [95% CI, 1.004–1.109], P = .033), but uIGFBP3 was not significant. As a result, uCyr61 and uTFF3 were considered as diagnostic biomarkers for CRC (Table 2).
Table 2.
Urinary Protein Level in the Training Cohort
| Univariate Analysis |
Multivariate Analysis |
|||||
|---|---|---|---|---|---|---|
| HC (Median (IQR)) |
CRC (Median (IQR)) |
P | Odds Ratio (95% CI) |
P | ||
| Cyr61 | (pg/g.Cr) | 0.8 (0–2.4) | 3.6 (1.0–13.0) | 0.000 | 1.146 (1.048–1.252) | 0.003 |
| TFF3 | (ng/g.Cr) | 10.7 (7.4–14.9) | 10.9 (6.7–19.0) | 0.177 | 1.055 (1.004–1.109) | 0.033 |
| IGFBP3 | (pg/g.Cr) | 28.3 (0.5–55.6) | 93.3 (31.8–211.3) | 0.000 | ||
| ANGPLT2 | (pg/g.Cr) | 46.3 (33.1–73.4) | 35.6 (18.1–64.0) | 0.099 | ||
| SERPINA1 | (ng/g.Cr) | 34.6 (2.4–86.0) | 51.8 (4.8–157.4) | 0.542 | ||
| EFEMP2 | (ng/g.Cr) | 2.6 (1.9–3.2) | 2.3 (1.3–4.3) | 0.726 | ||
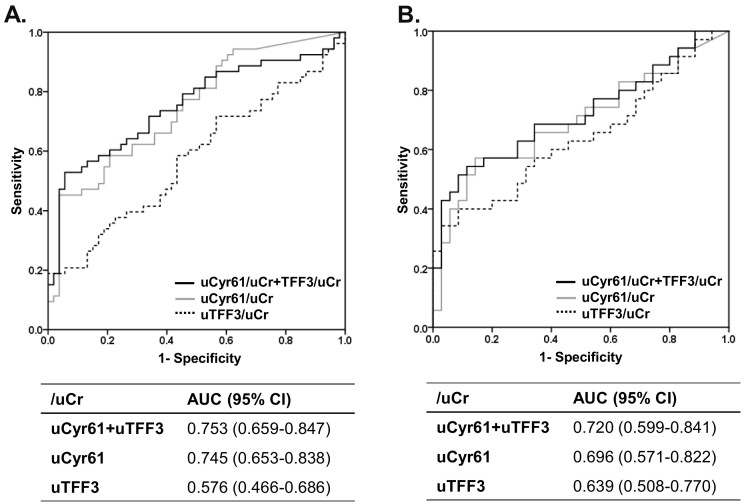
Moreover, uCyr61 allowed significant differentiation between HC and CRC groups in ROC analyses for the training set (AUC: 0.745 [95% CI, 0.653–0.838]). When uTFF3 was combined with uCyr61, this combination biomarker distinguished between HC and CRC (uCyr61 + uTFF3: AUC = 0.753 [95% CI, 0.659–0.847]) (Figure 2A). When cut-off values were determined as ≥5 pg/g.Cr for uCyr61 or ≥15 ng/g.Cr for uTFF3, CRC could be detected with 75.5% sensitivity, 69.8% specificity, and 72.6% accuracy (Table 3).
Figure 2.
Receiver operating characteristic curves.
ROC curves were obtained from normalized urinary cysteine-rich protein 61 (uCyr61/uCr), trefoil factor 3 (uTFF3/uCr) and the combination (uCyr61 + uTFF3/uCr).
A. Training cohort.
B. Validation cohort.
Table 3.
Diagnostic Power of Urinary Biomarker Panel
| uCyr61 ≥ 5 or TFF3 ≥ 15 | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| Training cohort (n = 106) | 75.5% (40/53) | 69.8% (37/53) | 72.6% (77/106) |
| Validation cohort (n = 70) | 71.4% (25/35) | 74.3% (26/35) | 72.9% (51/70) |
Validation of Urinary Biomarker
As shown in Table 4, uCyr61 and uTFF3 were significantly higher in the CRC group than in the HC group, in the validation cohort (HC vs. CRC: median uCyr61 (pg/g.Cr), 1.5 vs. 6.5, P = .005; median uTFF3 (ng/g.Cr), 9.7 vs. 14.0, P = .045). ROC analysis of uCyr61 and uTFF3 for the validation cohort also showed significant differentiation between HC and CRC groups (uCyr61: AUC = 0.696 [95% CI, 0.571–0.822])(uTFF3: AUC = 0.639 [0.508–0.770]). A biomarker panel combining uCyr61 and uTFF3 revealed a good AUC of 0.720 [95% CI, 0.599–0.841], as well as the training cohort (Figure 2B). This combination urinary biomarker panel also showed 71.4% sensitivity, 74.3% specificity and 72.9% accuracy in the validation cohort, similar to the results from the training cohort (Table 3).
Table 4.
Urinary Protein Level in the Validation Cohort
| Univariate Analysis |
||||
|---|---|---|---|---|
| HC (Median (IQR)) |
CRC (Median (IQR)) |
P | ||
| Cyr61 | (pg/g.Cr) | 1.5 (0.5–4.0) | 6.5 (1.2–19.0) | 0.005 |
| TFF3 | (ng/g.Cr) | 9.7 (5.4–15.3) | 14.0 (6.7–29.7) | 0.045 |
| IGFBP3 | (pg/g.Cr) | 115.1 (81.2–160.3) | 144.5 (96.9–301.5) | 0.073 |
Correlation to Disease Stage
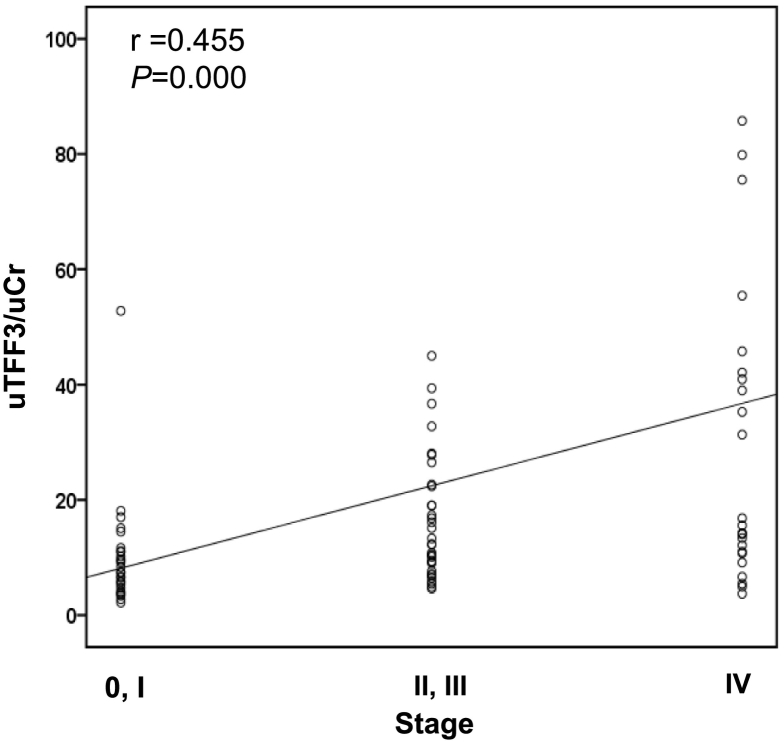
Next, we analyzed correlations between urinary biomarkers and disease stage. As shown in Figure 3, uTFF3 showed a significant positive correlation with disease stage (r = 0.454, P = .000), but uCyr61 did not. Median concentrations of uTFF3 (ng/g.Cr) were 7.4 [IQR, 5.0–11.0] in stage 0/I, 12.8 [IQR, 9.0–22.6] in stage II/III and 16.8 [IQR, 11.6–50.6] in stage IV.
Figure 3.
Correlation between urinary trefoil factor 3 and disease stage of colorectal cancer.
Data were analyzed using the Spearman rank correlation.
Discussion
This study offered the first evidence that the combination of uCyr61 and uTFF3 provides a potential non-invasive biomarker of CRC. Urine sampling is a straightforward and non-invasive procedure with low risks and costs, representing an attractive option for biomarker detection. We have previously shown the utility of urinary biomarkers for the early detection10 and prediction of curability11 against gastric cancer. Urinary biomarker is thus an attractive, non-invasive tool for the detection of malignancies.
Cyr61, also called CCN1, is matricellular protein and a member of the CCN protein family along with the 5 fellow members: CCN2 (connective tissue growth factor, CTGF), CCN3 (nephroblastoma overexpressed, NOV), CCN4 (Wnt-inducible secreted protein-1, WISP-1), CCN5 (WISP-2) and CCN6 (WISP-3).[21], [22] Cyr61 has many diverse functions related to cell proliferation, apoptosis, adhesion, angiogenesis and tumorigenesis through interaction with distinct integrins and heparan sulfate proteoglycans depending on the cells and environment.[21], [22] In terms of tumorigenesis, Cyr61 has been reported to promote tumor growth in many malignancies including breast cancer,23 pancreatic cancer,24 gastric cancer,25 prostate cancer26 and ovarian cancer,27 but acts as a tumor suppressor in some cancers such as non-small-cell lung cancer,28 hepatocellular carcinoma29 and endometrial cancer.30 A few studies related to Cyr61 and CRC showed Cyr61 was more expressed in tumor tissues than in normal tissues.[31], [32] As described above, some biomarker studies have examined Cyr61 using tissues, but other cancer biomarker studies have analyzed circulating Cyr61, identifying serum Cyr61 as a prognostic biomarker for prostate cancer.33 Among these, only one diagnostic biomarker study for CRC utilized serum Cyr61.15 Moreover, no studies have previously investigated urinary levels of Cyr61. The present study is the first to demonstrate the presence of Cyr61 in the urine of cancer patients. In the present study, Cyr61 levels were significantly higher in the urine of CRC patients than in that of healthy controls. Among the potential biomarkers examined in our study, uCyr61 demonstrated the highest diagnostic power, with consistent significance through two independent (training and validation) cohorts. Thus, uCyr61 appears promisingly placed for a key role in the urinary diagnosis of CRC. On the other hand, uCyr61 was not associated with disease stage in our study, supporting a previous report that found no correlation between Cyr61 mRNA expression and clinical stage of CRC.31
TFF3 is a member of the trefoil factor family, along with TFF1 and TFF2. All three are small secreted molecules mainly expressed in gastrointestinal epithelial cells and play roles in mucosal repair and mucus polymerization.34 In this family, TFF1 and TFF3 are reportedly expressed in tissues of the normal colon and rectum,35 but the major site of TFF1 and TFF2 expression is generally the stomach.34 Although TFF3 is intermediately expressed in some sites including the airways, salivary glands and small intestine, TFF3 expression is more specific for the large intestine among the TFF family.35 Based on this knowledge, some studies have identified TFF3 as a biomarker for CRC. In fact, recent studies have reported the utility of serum TFF3 as a diagnostic biomarker with a high AUC (0.889–0.930).[16], [36], [37] In addition, serum levels of TFF3 correlated with clinical stage and poor prognosis.[16], [37] Although uTFF3 has previously been reported as a biomarker for kidney disease,38 no studies have examined uTFF3 as a cancer biomarker. In the current study, uTFF3 alone was not a particularly powerful biomarker for CRC, but contributed to increasing the diagnostic power by compensating for another strong biomarker, uCyr61. Interestingly, uTFF3 correlated with clinical stage, consistent with the results of previous serum biomarker studies.[16], [37] Since Cyr61 and TFF3 have different functions, these 2 factors appear to offer a good combination.
A key limitation in the current study was that the sample size was not very large. However, since our study was a case–control study with a random-matching method using PSs, analysis of the 176 age- and sex-matched 176 should have provided satisfactory quality. Moreover, the consistent significance between the 2 independent cohorts suggests that our urinary biomarker panel might be worthy of consideration. In addition, as early-stage CRC comprised around 30% of cases in the current study, this biomarker panel should enable early discovery of CRC.
Conclusions
This completely non-invasive urinary biomarker panel combining uCyr61 with uTFF3 provides a promising method for identifying the presence of CRC.
Acknowledgments
Acknowledgments
We wish to thank Yukimi Hashidume-Itoh for handling urine samples and Takako Onodera for data management of enrolled patients in this study (Department of Gastroenterology and Metabolism, Nagoya City University Graduate School of Medical Sciences). We are grateful to our clinical colleagues who assisted in sample collection. We are also grateful to the Kobayashi Foundation for Cancer Research (to T. Shimura) for supporting this study.
Footnotes
Funding: This work was supported by the Kobayashi Foundation for Cancer Research (to T. Shimura).
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Saito H, Soma Y, Nakajima M, Koeda J, Kawaguchi H, Kakizaki R, Chiba R, Aisawa T, Munakata A. A case-control study evaluating occult blood screening for colorectal cancer with hemoccult test and an immunochemical hemagglutination test. Oncol Rep. 2000;7:815–819. doi: 10.3892/or.7.4.815. [DOI] [PubMed] [Google Scholar]
- 3.Saito H, Soma Y, Koeda J, Wada T, Kawaguchi H, Sobue T, Aisawa T, Yoshida Y. Reduction in risk of mortality from colorectal cancer by fecal occult blood screening with immunochemical hemagglutination test. A case-control study. Int J Cancer. 1995;61:465–469. doi: 10.1002/ijc.2910610406. [DOI] [PubMed] [Google Scholar]
- 4.Hiwatashi N, Morimoto T, Fukao A, Sato H, Sugahara N, Hisamichi S, Toyota T. An evaluation of mass screening using fecal occult blood test for colorectal cancer in Japan: a case-control study. Jpn J Cancer Res. 1993;84:1110–1112. doi: 10.1111/j.1349-7006.1993.tb02809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison JE, Tekawa IS, Ransom LJ, Adrain AL. A comparison of fecal occult-blood tests for colorectal-cancer screening. N Engl J Med. 1996;334:155–159. doi: 10.1056/NEJM199601183340304. [DOI] [PubMed] [Google Scholar]
- 6.Rozen P, Knaani J, Papo N. Evaluation and comparison of an immunochemical and a guaiac faecal occult blood screening test for colorectal neoplasia. Eur J Cancer Prev. 1995;4:475–481. doi: 10.1097/00008469-199512000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Robinson MH, Kronborg O, Williams CB, Bostock K, Rooney PS, Hunt LM, Hardcastle JD. Faecal occult blood testing and colonoscopy in the surveillance of subjects at high risk of colorectal neoplasia. Br J Surg. 1995;82:318–320. doi: 10.1002/bjs.1800820310. [DOI] [PubMed] [Google Scholar]
- 8.Walter SD, Frommer DJ, Cook RJ. The estimation of sensitivity and specificity in colorectal cancer screening methods. Cancer Detect Prev. 1991;15:465–469. [PubMed] [Google Scholar]
- 9.Sun LC, Chu KS, Cheng SC, Lu CY, Kuo CH, Hsieh JS, Shih YL, Chang SJ, Wang JY. Preoperative serum carcinoembryonic antigen, albumin and age are supplementary to UICC staging systems in predicting survival for colorectal cancer patients undergoing surgical treatment. BMC Cancer. 2009;9:288. doi: 10.1186/1471-2407-9-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimura T, Dagher A, Sachdev M, Ebi M, Yamada T, Yamada T, Joh T, Moses MA. Urinary ADAM12 and MMP-9/NGAL complex detect the presence of gastric cancer. Cancer Prev Res (Phila) 2015;8:240–248. doi: 10.1158/1940-6207.CAPR-14-0229. [DOI] [PubMed] [Google Scholar]
- 11.Shimura T, Ebi M, Yamada T, Yamada T, Katano T, Nojiri Y, Iwasaki H, Nomura S, Hayashi N, Mori Y. Urinary kallikrein 10 predicts the incurability of gastric cancer. Oncotarget. 2017;8:29247–29257. doi: 10.18632/oncotarget.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 13.Vandenbroucke JP, von Elm E, Altman DG, Gotzsche PC, Mulrow CD, Pocock SJ, Poole C, Schlesselman JJ, Egger M. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Epidemiology. 2007;18:805–835. doi: 10.1097/EDE.0b013e3181577511. [DOI] [PubMed] [Google Scholar]
- 14.Sobin LH, Gospodarowicz MK, Wittekind C. 7th ed. John Wiley & Sons, Inc; Hoboken, NJ: 2009. TNM classification of malignant tumours. [Google Scholar]
- 15.Song YF, Xu ZB, Zhu XJ, Tao X, Liu JL, Gao FL, Wu CL, Song B, Lin Q. Serum Cyr61 as a potential biomarker for diagnosis of colorectal cancer. Clin Transl Oncol. 2017;19:519–524. doi: 10.1007/s12094-016-1560-7. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Wang K, Su C, Fang J. Serum Trefoil factor 3 as a protein biomarker for the diagnosis of colorectal cancer. Technol Cancer Res Treat. 2017;16:440–445. doi: 10.1177/1533034616674323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankaj J, Kumari JR, Kim W, Lee SA. Insulin-like Growth Factor-1, IGF-binding Protein-3, C-peptide and Colorectal Cancer: a Case-control Study. Asian Pac J Cancer Prev. 2015;16:3735–3740. doi: 10.7314/apjcp.2015.16.9.3735. [DOI] [PubMed] [Google Scholar]
- 18.Toiyama Y, Tanaka K, Kitajima T, Shimura T, Kawamura M, Kawamoto A, Okugawa Y, Saigusa S, Hiro J, Inoue Y. Elevated serum angiopoietin-like protein 2 correlates with the metastatic properties of colorectal cancer: a serum biomarker for early diagnosis and recurrence. Clin Cancer Res. 2014;20:6175–6186. doi: 10.1158/1078-0432.CCR-14-0007. [DOI] [PubMed] [Google Scholar]
- 19.Peltier J, Roperch JP, Audebert S, Borg JP, Camoin L. Quantitative proteomic analysis exploring progression of colorectal cancer: Modulation of the serpin family. J Proteomics. 2016;148:139–148. doi: 10.1016/j.jprot.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Yao L, Lao W, Zhang Y, Tang X, Hu X, He C, Hu X, Xu LX. Identification of EFEMP2 as a serum biomarker for the early detection of colorectal cancer with lectin affinity capture assisted secretome analysis of cultured fresh tissues. J Proteome Res. 2012;11:3281–3294. doi: 10.1021/pr300020p. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Du XY. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007;100:1337–1345. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- 22.Lau LF. CCN1/CYR61: the very model of a modern matricellular protein. Cell Mol Life Sci. 2011;68:3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris LG, Pannell LK, Singh S, Samant RS, Shevde LA. Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene. 2012;31:3370–3380. doi: 10.1038/onc.2011.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maity G, Mehta S, Haque I, Dhar K, Sarkar S, Banerjee SK, Banerjee S. Pancreatic tumor cell secreted CCN1/Cyr61 promotes endothelial cell migration and aberrant neovascularization. Sci Rep. 2014;4:4995. doi: 10.1038/srep04995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babic AM, Kireeva ML, Kolesnikova TV, Lau LF. CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci U S A. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzen CA, Chen CC, Todorovic V, Juric V, Monzon RI, Lau LF. Matrix protein CCN1 is critical for prostate carcinoma cell proliferation and TRAIL-induced apoptosis. Mol Cancer Res. 2009;7:1045–1055. doi: 10.1158/1541-7786.MCR-09-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, Scott D, Yi WS, Popoviciu ML, Said JW. Ovarian carcinomas: CCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res. 2005;11:7243–7254. doi: 10.1158/1078-0432.CCR-05-0231. [DOI] [PubMed] [Google Scholar]
- 28.Tong X, O'Kelly J, Xie D, Mori A, Lemp N, McKenna R, Miller CW, Koeffler HP. Cyr61 suppresses the growth of non-small-cell lung cancer cells via the beta-catenin-c-myc-p53 pathway. Oncogene. 2004;23:4847–4855. doi: 10.1038/sj.onc.1207628. [DOI] [PubMed] [Google Scholar]
- 29.Feng P, Wang B, Ren EC. Cyr61/CCN1 is a tumor suppressor in human hepatocellular carcinoma and involved in DNA damage response. Int J Biochem Cell Biol. 2008;40:98–109. doi: 10.1016/j.biocel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 30.Chien W, Kumagai T, Miller CW, Desmond JC, Frank JM, Said JW, Koeffler HP. Cyr61 suppresses growth of human endometrial cancer cells. J Biol Chem. 2004;279:53087–53096. doi: 10.1074/jbc.M410254200. [DOI] [PubMed] [Google Scholar]
- 31.Ladwa R, Pringle H, Kumar R, West K. Expression of CTGF and Cyr61 in colorectal cancer. J Clin Pathol. 2011;64:58–64. doi: 10.1136/jcp.2010.082768. [DOI] [PubMed] [Google Scholar]
- 32.Jeong D, Heo S, Sung Ahn T, Lee S, Park S, Kim H, Park D, Byung Bae S, Lee SS, Soo Lee M. Cyr61 expression is associated with prognosis in patients with colorectal cancer. BMC Cancer. 2014;14:164. doi: 10.1186/1471-2407-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terada N, Kulkarni P, Getzenberg RH. Cyr61 is a potential prognostic marker for prostate cancer. Asian J Androl. 2012;14:405–408. doi: 10.1038/aja.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong WM, Poulsom R, Wright NA. Trefoil peptides. Gut. 1999;44:890–895. doi: 10.1136/gut.44.6.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madsen J, Nielsen O, Tornoe I, Thim L, Holmskov U. Tissue localization of human trefoil factors 1, 2, and 3. J Histochem Cytochem. 2007;55:505–513. doi: 10.1369/jhc.6A7100.2007. [DOI] [PubMed] [Google Scholar]
- 36.Xie H, Guo JH, An WM, Tian ST, Yu HP, Yang XL, Wang HM, Guo Z. Diagnostic value evaluation of trefoil factors family 3 for the early detection of colorectal cancer. World J Gastroenterol. 2017;23:2159–2167. doi: 10.3748/wjg.v23.i12.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vocka M, Langer D, Petrtyl J, Vockova P, Hanus T, Kalousova M, Zima T, Petruzelka L. Trefoil factor family (TFF) proteins as potential serum biomarkers in patients with metastatic colorectal cancer. Neoplasma. 2015;62:470–477. doi: 10.4149/neo_2015_056. [DOI] [PubMed] [Google Scholar]
- 38.O'Seaghdha CM, Hwang SJ, Larson MG, Meigs JB, Vasan RS, Fox CS. Analysis of a urinary biomarker panel for incident kidney disease and clinical outcomes. J Am Soc Nephrol. 2013;24:1880–1888. doi: 10.1681/ASN.2013010019. [DOI] [PMC free article] [PubMed] [Google Scholar]