Abstract
Transcription factor (TF) signaling regulates gene transcription and requires a complex network of proteins. This network includes co-activators, co-repressors, multiple TFs, histone-modifying complexes, and the basal transcription machinery. It has been widely appreciated that pioneer factors, such as FoxA1 and GATA1, play an important role in opening closed chromatin regions, thereby allowing binding of a secondary factor. In this review we will focus on a newly proposed model wherein multiple TFs, such as steroid receptors (SRs), can function in a pioneering role. This model, termed dynamic assisted loading, integrates data from widely divergent methodologies, including genome wide ChIP-Seq, digital genomic footprinting, DHS-Seq, live cell protein dynamics, and biochemical studies of ATP-dependent remodeling complexes, to present a real time view of TF chromatin interactions. Under this view, many TFs can act as initiating factors for chromatin landscape programming. Furthermore, enhancer and promoter states are more accurately described as energy-dependent, non-equilibrium steady states.
Keywords: chromatin binding, dynamic assisted loading, estrogen receptor, FoxA1, glucocorticoid receptor, pioneer factors, residence time
Introduction
In eukaryotes, DNA binding proteins are responsible for the regulation of gene expression as well as the development and differentiation of cells. The organization of the eukaryotic genome into chromatin via the association with histones and non-histone proteins in the nucleus is essential for the function of these processes [1, 2]. The modulation of the chromatin structure plays an important role in controlling the transcriptome, specifically, in the activation and repression of steroid receptor (SR) mediated transcription [3].
Pioneer factors, a special class of transcription factors (TFs), have been described as being able to access their DNA target sites in closed chromatin, allowing the binding of other TFs [4]. The first described pioneer factors were FoxA and GATA proteins, observed to occupy liver-specific enhancers prior to hepatic induction [5, 6]. It was later shown that forkhead box A1 (FoxA1) and GATA binding factor 4 (GATA4), members of the FoxA and GATA family, respectively, could target DNA sites in isolated nucleosomes [7, 8], suggesting an ability of these two factors to penetrate closed chromatin. This led to the concept that this special set of TFs have unique “pioneering abilities,” capable of targeting closed chromatin [9]. Although this model suggests long term, static binding events are associated with pioneering activity, recent findings from several groups describe highly dynamic factor/chromatin interactions for many factors, including pioneer proteins [10–14]. We have recently shown that a subset of FoxA1 binding events require the activation of the estrogen or glucocorticoid receptors (ER and GR, respectively), members of the SR family, whereby, ER or GR opens closed chromatin for FoxA1 binding [11], a behavior similar to the properties of a pioneer factor. In agreement, Ballare et al. have demonstrated that the progesterone receptor (PR) can initiate chromatin binding and remodeling of core nucleosomes, and linker histone H1, qualifying it as a bona fide pioneer factor [15]. Moreover, the retinoic acid receptor is able to bind and induce remodeling of highly compacted chromatin in vitro [16].
In addition, we demonstrated through single-molecule tracking (SMT) experiments that FoxA1, like ER and GR, manifests a relatively short DNA residence time (~9 seconds), signifying a highly dynamic TF [11]. This behavior is inconsistent with the putative long duration of open chromatin sites created by a pioneer factor. In light of these findings, we propose that classical pioneer factor models should be reconsidered. TFs can initiate chromatin-remodeling events throughout the genome through the recruitment of ATP-dependent chromatin remodeling factors, thereby providing access to other factors. This event is interchangeable between initiating factors depending on the local chromatin landscape and accessibility of remodeling factors. Here we will discuss the rationale behind this new model, termed “Dynamic Assisted Loading” (DynALoad), and contrast with the classically established role of pioneer factors. We will further review the known involvement of ATP-dependent remodeling factors in modulating the chromatin landscape for TFs, including previously described pioneer factors, and the newly established dynamic nature of most TFs.
Does FoxA1 always function as a pioneer factor for TF recruitment?
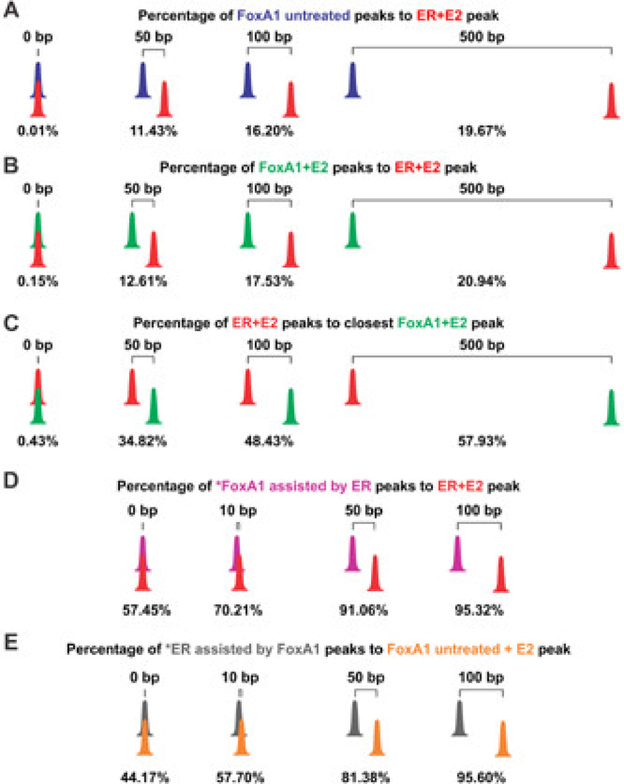
To date, FoxA1 has been well described to play a functional role in breast cancer progression through its known ability to recruit ER to over 50% of its binding sites. Further, 90% of ER sites have been described as inhibited or decreased when FoxA1 is silenced, aiding in the ER proliferative response in breast cancer [17–21]. It was also reported that inhibition of ER did not affect FoxA1 binding events [20, 21], suggesting a factor with a very prominent pioneering function. However, more recent studies suggest that ER or GR can both play a role in regulating FoxA1 binding events. This implies the model is more complex than previously suggested. Specifically, an early finding by Grange et al. [22] showed that GR could modulate FoxA1 binding at a specific glucocorticoid response element (GRE) within the GR-responsive unit of the rat tyrosine aminotransferase gene. Importantly, Grange et al. showed that the two factors competed for binding on pure DNA templates, indicating the two proteins could not co-occupy the GRE. It was next reported that 29% of FoxA1 binding sites stimulated by E2 were dependent on ER binding events in MCF-7 breast cancer cells [23]. A more recent study identified that ER binding sites observed in the unliganded state are frequently occupied by a FoxA1 motif and silencing of ER resulted in a decrease of FoxA1 binding [24]. In support of these findings, we have recently shown that there is a subset of FoxA1 binding events genome-wide that do not display a pioneer role for ER or GR. Rather these two SRs pioneer for FoxA1, putatively through the DynALoad mechanism [11]. A careful examination of ER and FoxA1 binding events in MCF-7 breast cancer cells that we recently published [11] shows that a large proportion of FoxA1 binding events (~80%), regardless of hormone stimulation, are further than 500 base pairs from an ER binding site stimulated by E2 (Fig. 1). We further investigated the location of the reported FoxA1 binding events modulated by ER, and ER binding events modulated by FoxA1 [11]; for this group we see a closer interaction between the two TFs (Fig. 1). Therefore, we suggest that FoxA1’s role is not solely to recruit SRs to chromatin; rather, in some cases the role may be reversed. In the DynALoad model, we propose that the first initiating factor can create an open chromatin state through recruitment of potential chromatin remodeling factors, allowing the binding of a secondary initiating factor. This event is highly dynamic, and creates a transient chromatin conformation [3, 11] with the proposed role of ATP-dependent chromatin remodeling factors playing a major part in these processes (Fig. 2A).
Figure 1.
Classical pioneer factor model of FoxA1 in breast cancer cells is not supported by the distances of estrogen receptor (ER) and FoxA1 binding sites. A and B: Illustration of distances of FoxA1 binding sites in untreated (A) and estradiol (E2) (B) treated cells in relationship to ER binding sites in E2 treated cells. C: Illustration of distances of ER binding sites in E2 treated cells in regards to FoxA1 binding sites in E2 treated cells. These distances indicate more complex action between ER and FoxA1 than originally described. D and E: The distances of FoxA1 assisted by ER (D) and ER assisted by FoxA1 (E) binding sites. The binding sites where FoxA1 binding is assisted by ER are closer together than the sites where ER binding is assisted by FoxA1.
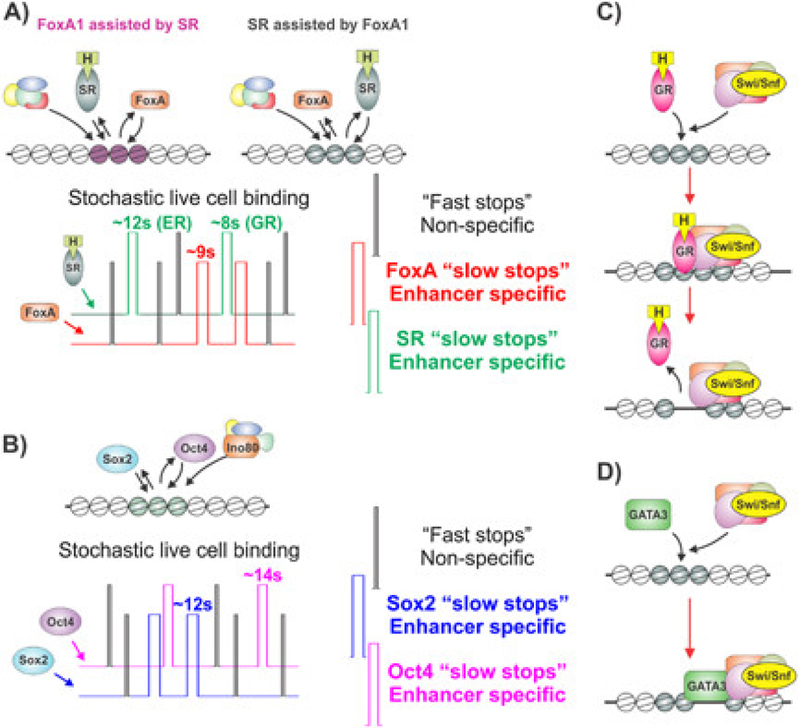
Figure 2.
Regulation of chromatin accessibility by pioneer factors and steroid receptors involve ATP-dependent chromatin remodeling complexes. A: Dynamic assisted loading of steroid receptor (SR) and FoxA1 in breast cancer cells occurs in a symmetric manner (upper panel). Activated SRs are able to open closed chromatin regions, and assist the binding of FoxA1 (left panel). In the classical pioneer factor model, FoxA1 open closed chromatin regions and assist the binding of SRs (right panel). Opening of the closed chromatin regions by SRs and FoxA1 most likely involves ATP-dependent chromatin remodeling complexes. Single-molecule imaging in live cells indicates that the enhancer binding dynamics are comparable between SRs and FoxA1 (lower panel). B: Binding of ES cell master regulators Sox2 and Oct4 occurs in symmetric manner (upper panel). The regulation of chromatin accessibility at Sox2 and Oct4 binding sites are regulated by INO80 remodeling complex. Single-molecule imaging in live cells indicates that the enhancer binding dynamics are comparable between Sox2 and Oct4 (lower panel). C: Glucocorticoid receptor (GR) is capable of binding to closed chromatin sites (upper panel). At these sites GR recruits SWI/SNF chromatin remodeling complexes to modify and open chromatin (middle panel). During the chromatin remodeling reaction, GR is actively displaced from the chromatin by SWI/SNF complex (lower panel). D: Classical pioneer factor, GATA3, has been shown to recruit SWI/SNF complex to the chromatin, which increases chromatin accessibility. Inhibition of interaction between GATA3 and SWI/SNF inhibits the opening of chromatin.
Chromatin remodeling complexes: Classification and function
ATP-dependent chromatin remodeling complexes belong to the SF2 helicase superfamily that share a structural core ATPase subunit [25]. Within the SF2 superfamily is the Snf2 family, which contains four major evolutionarily conserved multi-subunit remodeling complexes; mating type switching/sucrose non-fermenting (SWI/SNF), imitation switch (ISWI), chromodomain helicase (CHD), and inositol (INO80) [26, 27]. Each remodeling complex catalytic subunit, where ATP is hydrolyzed, has a common bipartite DExx and HELICc domain structure [28] but also contains unique domains that are inserted between or flank the two common domains of the ATPase activity. Specifically, in mammals, the SWI/SNF family is largely involved in prompting gene regulation, and contains two possible ATPase core subunits; Brahma (BRM) or Brahma related gene 1 (BRG1). This complex is also associated with a number of additional factors, including BRG1-associated factors (BAFs) [29–31]. The major complexes of the ISWI remodeling family are nucleosome remodeling factor (NURF), chromatin accessibility complex (CHRAC), ATP-utilizing chromatin assembly and remodeling factor (ACF), CECR2-containing remodeling factor (CERF), WSTF-ISWI chromatin remodeling complex (WICH), nucleolar remodeling complex (NoRC), and remodeling and spacing factor (RSF). These complexes are assembled around the ATPase sucrose nonfermenting 2 like (Snf2L) and sucrose nonfermenting 2 homologue (Snf2H), and have been shown to be involved in activation and repression of transcription and assembly of chromatin [26, 27, 32–35]. The CHD remodeling family is divided into three subfamilies denoted by roman numerals [36, 37]. The first subfamily (I) contains CHD1 and CHD2 proteins, which are highly conserved, but have been described to possess distinct functions [38, 39]. The second subfamily (II) includes CHD3 and CHD4, which lack DNA-binding domains: rather, these proteins contain paired N-terminal PHD Zn-finger-like domains [38]. These two proteins form large protein complexes termed nucleosome remodeling deacetylase (NuRD), which contains histone deacetylase and ATP-dependent chromatin remodeling properties. The third subfamily (III) comprises the proteins CHD5, CHD6, CHD7, CHD8, and CHD9. This subfamily is defined by additional functional motifs in the C-terminal regions [38]. The INO80 family is the most complex out of the four remodeling families in terms of the subunit composition across species [26, 40]. It has been shown that INO80 remodeling complexes have several roles in transcription activation; as with ISWI, INO80 remodelers interact with extra-nucleosomal DNA to mobilize nucleosomes. A comprehensive description on the mechanistic action of chromatin remodelers in modifying nucleosomes has been reviewed elsewhere [26, 41, 42].
The remodeling event results in the regulation of chromatin accessibility and the exposure of DNA regulatory elements. Moreover, at least BRG1, CHD4, and Snf2H have been shown to be capable of both opening and closing chromatin [43]. In this sense, these complexes can be considered to be involved in both displacement and repositioning of nucleosomes. This could in turn modulate TF binding in such a way that would lead to the return of the chromatin landscape to a closed state. Similarly to TFs, chromatin remodelers has been characterized as highly dynamic, with only a small fraction of remodelers engaged with chromatin at any point in time [44]. Curiously, the potential role of ATP-dependent remodeling factors in the mechanism of pioneer factor action has been largely ignored [3].
Chromatin remodeling factors aid in the recruitment of pioneer factors during cell development
Chromatin remodeling factors have been shown to play an important role in cell development and embryonic stem (ES) cell self-renewal [26, 41]. A recent study indicated a number of different chromatin remodeling complexes that regulate nucleosome positioning in promoter regions of ES cells [45]. The remodeling of these promoters therefore, influenced the regulation of ES transcriptional programs. Further, it was shown that the binding events of BRG1 correlated with the chromatin binding of the essential ES cell reprogramming factor octamer-binding transcription factor 4 (Oct4) [45]. ES cell reprogramming factors represent a set of TFs capable of transforming adult cells into induced pluripotent stem (iPS) cells [46]. Hence, it is tempting to speculate that other ES cell reprogramming factors such as sex determining region Y-box 2 (Sox2), Kruppel-like factor 4 (Klf4), along with Oct4 [46] could recruit chromatin remodeling factors to chromatin (Fig. 2B), all previously described as pioneer factors. In addition, and correlated to chromatin binding [45], BRG1 and Oct4 co-localize in blastocysts, and knockdown of BRG1 affects ES cell self-renewal and pluripotency [47]. Further, it has also been shown that the expression of BAF complexes can increase the efficiency of ES cell reprogramming [48]. This occurs due to the BAF-facilitated binding of Oct4 to promoters of genes involved in reprogramming. However, it is not clear what directs BAF complexes to these promoters. In addition, it has also been shown that forkhead box D3 (FOXD3), a TF that binds before Oct4, can recruit BRG1 to chromatin, demonstrating an involvement of FOXD3 in the interactions between chromatin remodeling and ES cell reprogramming factors [49]. This suggests that non-reprogramming factors in ES cells can induce chromatin remodeling.
The clearest evidence of reprogramming factors recruiting chromatin-remodeling complexes to chromatin has been shown for the INO80 remodeling complex [50]. The chromatin binding of INO80, which binds mainly at DNaseI hypersensitive (DHS) regions, correlates with the binding of several reprogramming factors, including Oct4, Sox2, Klf4, and Nanog. Interestingly, the chromatin accessibility at Oct4, Sox2, and Nanog binding sites was significantly decreased after INO80 depletion. This suggests that during ES cell self-renewal and reprogramming Oct4, Sox2, and Nanog recruit INO80 to increase chromatin accessibility. These findings implicate a role for chromatin remodeling factors in early cell development, and suggest that the role of remodeling factors in pioneering action has been under appreciated.
Steroid receptor signaling requires chromatin remodeling complexes
BRG1 has been well characterized to regulate the transcriptional response of SRs such as the androgen receptor (AR), PR, ER, and GR (Fig. 2C). Specifically, these receptors interact with BRG1 to initiate its ATP-dependent remodeling function, recruiting this activity to steroid response elements [51–55]. This suggests that the SWI/SNF complex is commonly involved in creating a unique chromatin state during hormone induction. The mouse mammary tumor virus (MMTV) promoter has been used widely as a model to investigate SR mediated chromatin-remodeling events [56–58]. These studies have shown that SWI/SNF dependent chromatin remodeling is required for hormone-regulated transcription at the MMTV promoter [51, 52, 59, 60]. However, it is known that promoters lacking an ordered chromatin structure are able to evade the requirement for remodeling by SWI/SNF in the context of a functional transcriptional response [54, 61]. Specifically, GR can interact with both BRG1 and BRM complexes [52, 62]; however, the interaction between GR and BRG1 is not direct; rather the interaction is reliant on bridges connecting BAF proteins and BRG1 [63–65]. Further investigation of the interaction between GR and BAF proteins has shown that GR can bind to BAF250 through the C-terminal domain [65], and the GR transcriptional function is decreased in cells with a BAF250 C-terminal domain mutation [66]. In addition, in vitro studies identified direct binding of GR and ER to BAF60a and BAF57 as well as PR to BAF60a [63]. We previously demonstrated in reconstituted chromatin assays, utilizing UV laser crosslinking methodology, that GR rapidly binds to the GRE next to the MMTV promoter, but is then actively displaced during the BRG1-induced remodeling event [67]. In direct support of these findings, a recent study using the single-molecule unzipping assay indicated that SWI/SNF is capable of displacing a TF from chromatin during nucleosome remodeling [68]. Interestingly, this is not a global property of the chromatin remodeling complexes, because ISW1a is not capable of displacing the TF. These studies suggest that chromatin remodeling, at least through the SWI/SNF complex, is a dynamic process [67, 68]. Furthermore, utilizing a dominant negative form of either BRG1 or BRM, a marked inhibition of GR mediated transcription was observed. This was associated with a decrease in chromatin remodeling and decondensation of the MMTV promoter, as determined by restriction enzyme accessibility [62]. In support of these findings, in cells that lack BRG1 and BRM, the GR transcriptional response is weak and cannot be restored by the activity of ISWI or NuRD complexes [69]. Together, these findings suggest that the SWI/SNF complex plays a major functional role in the transcriptional responses of SRs creating accessible chromatin states during hormone induction. Interestingly, overexpression of the dominant negative form of BRG1 showed reduction of only 41% of GR regulated genes [62]. This suggests that the collaborative role of other remodeling complexes is likely required for SR transcriptional activation. It is important to note that the ability of SRs to activate transcription is highly cell specific, and this specificity may be due to the abilities of the SRs to recruit individual remodeling complexes.
The collaborative interactions of ATP-dependent remodeling complexes are highly dynamic
In support of the rationale that multiple chromatin remodeling factors are involved in a GR transcriptional response, we have demonstrated, genome-wide, that BRG1, Snf2H, and CHD4 share multiple binding regions [43]. However, failure to detect a direct interaction between the co-localized remodeling complexes suggests that the binding patterns of each chromatin remodeling protein are likely transient and highly dynamic. In agreement, biophysical studies have shown that chromatin remodelers likely find their target sites by a “continuous sampling” mechanism. Here, a combination of high protein concentrations, short residence times in the chromatin bound state, and fast 3D diffusive translocations results in an efficient search and find strategy in the nucleus [70]. These results suggest that, although chromatin remodelers co-localize at the same sites, they do not function simultaneously at any given time.
Investigations of chromatin accessibility by DHS-sequencing (DHS-Seq) revealed 88% of BRG1, 75% of Snf2H, and 85% of CHD4 binding sites correlate to open chromatin. However, there were also a large proportion of binding sites associated with closed chromatin sites [43]. This supports previous findings suggesting that SWI/SNF can function at a closed chromatin structure [71]. Further, utilizing dominant negative variants of each remodeler, a higher level of complexity was revealed. Specifically, upon expression of the dominant negative variants, states were observed where chromatin was (i) unchanged; (ii) deemed inaccessible; or (iii) newly open. Investigation of the affected regions of chromatin accessibility revealed a large number to be occupied by multiple remodelers, and changes in accessibility were either linked to one remodeler, two remodelers functioning together or in opposition, or all three remodelers functioning at that single site [43]. Several reports have demonstrated that TFs can be mobile during chromatin remodeling events [72–74]. In addition, Li et al. [68] reported that the SWI/SNF complex can actively displace a TF, via sliding of a nucleosome, providing evidence of TF regulation in dynamic reposition of nucleosomes. Together, these findings have indicated that multiple remodeling complexes can function collaboratively or individually to open, or close chromatin sites and additionally, they can function in a highly dynamic manner in evicting factors from the chromatin landscape. This suggests a very dynamic model of ATP-dependent chromatin remodeling, whereby multiple remodeling complexes and TFs cycle through a complex series of events.
Interactions between pioneer factors and ATP-dependent remodeling factors
The GATA family are considered pioneer factors because they are able to induce chromatin accessibility [9, 75]. Very recent findings show that GATA factors require other co-factors such as ATP-dependent chromatin remodeling factors to create an accessible landscape for secondary factors to bind (Fig. 2D). Genome-wide studies have determined that GATA binding protein 1 (GATA1) binds to distal regions and there is a global reorganization of chromatin structure that occurs at these potential enhancers during differentiation of hematopoietic stem cells to erythrocytes [76]. In addition, it was shown that BRG1 co-localizes to these binding events and aids in creating a longer hypersensitive region surrounding the GATA1 sites. This nucleosome shifting facilitates the binding of the secondary factor, TAL1, suggesting that GATA1 is initiating the binding of TAL1 through the recruitment of chromatin remodeling factors [76]. In support of this finding, it has also been established that BRG1 can co-localize with GATA binding protein 2 (GATA2), revealing a role for BRG1 in the GATA2 dependent chromatin structural transitions [77]. Very recently, it was shown that GATA binding protein 3 (GATA3) functions as a pioneer factor in the mesenchymal to epithelial transition event in breast cancer cells. It was determined that GATA3 binds to inaccessible chromatin, whereby nucleosome eviction and remodeling of the chromatin architecture occurs. It was further shown that successful reprogramming required stable binding to a nucleosome rich region and then recruitment of co-factors such as BRG1 [78]. Together these studies suggest a prominent role for ATP-dependent remodeling factors in the pioneer factor model, wherein they help to establish chromatin landscape for the factor that is being recruited.
Because the structure of FoxA1 is similar to that of linker histones [79, 80], it is considered that it competes with linker histones to create an accessible chromatin environment. It therefore, has been suggested that FoxA1 can remodel chromatin without the assistance of co-factors [9]. However, this suggests a highly efficient and rather stable binding event. With recent evidence indicating that FoxA1, and other pioneer factors such as Sox2, bind to chromatin in a highly dynamic manner, we propose a role for ATP-dependent chromatin remodeling factors in facilitating the binding events initiated by FoxA1. Clearly, the potential role for remodeling systems in pioneer factor action needs to be studied in much greater depth.
How dynamic are pioneer factors in live cells?
The pioneer factor FoxA1 was described as being able to bind to nucleosomes in a stable manner with slow DNA binding interactions, and this led to the formulation of the classical model for pioneer factor mode of action [7, 81]. The introduction of the fluorescence recovery after photobleaching (FRAP) technique allowed further investigations of the binding nature of FoxA1. These studies suggested a more dynamic interaction with DNA than previously described; however, the binding events were still observed to be more stable than other TFs that had been investigated [82]. In determining a residence time of a TF, it is important to note that there are serious limitations when utilizing the FRAP technique. The time at which the recovery reaches 50% of its final value is termed the recovery half-time, and is often incorrectly interpreted as a residence time [83]. When obtaining a residence time utilizing FRAP, the data must be fitted to the correct model. This model must depict a number of complex events that take place, including binding, diffusion and binding, diffusion and two-binding states, and anomalous diffusion. Therefore, objective determination of the correct model can be quite challenging. Although the residence time of some TFs is comparable to that of the recovery half-time, a number of cases have been described where the two values are largely different [83]. Hence, careful consideration needs to be implemented when assessing TF binding rates via this method. Previous investigations of FoxA1 binding dynamics have only been reported as recovery half-time utilizing the FRAP technique [82].
Alternatively, a single-molecule approach is in principle a more reliable tool to determine residence times, although this approach is also not free from limitations [83, 84]. Utilizing SMT technology, individual bound molecules can be directly observed, enabling a more straightforward approach for the determination of their dynamic binding properties [85]. The recent use of this technique to study TFs binding [10, 12, 14, 85–89] has confirmed the highly transient nature of several TF-chromatin interactions. Interestingly, in all cases, an exponential distribution of bound dwell times is found. Whether the bulk average for the wide range of residence times reflects the dwell time on a single type of chromatin target, or whether the range of residence times reflects binding to different types of chromatin targets remains largely unknown, and likely both options occur in vivo. We recently reported the residence time of FoxA1 through an SMT approach in breast cancer cells. It was determined that FoxA1 chromatin interactions are as transient as other TFs such as ER and GR [11]. Moreover, another well described pioneer factor, Sox2, has been described to present rapid exchange with chromatin through SMT investigations [14, 90]. Together, these recent findings do not support the previously described stable binding events of pioneer factors, such as FoxA1. Therefore, pioneer factors appear to behave in a similar manner to other TFs in terms of their dynamic chromatin interactions.
Conclusions and outlook
Recent studies have begun to reshape our current understanding of TF and chromatin remodeler action, not as a statical system, but as a highly dynamic interaction between multiple SRs, other TFs, and chromatin remodeling complexes. Specifically, the actual dynamic nature of pioneer factors such as FoxA1 has come into question: recent live cell experiments indicate factors with highly transient chromatin interactions. In addition, multiple studies now suggest, on the genome-wide scale, that a number of other TFs, such as ER and GR, can pioneer the binding of FoxA1, hence suggesting a symmetric system with multiple TFs acting as initiating factors, depending on chromatin context. The nature of the chromatin context that ultimately defines whether a TF will behave as an initiator or an assisted factor remains largely unknown. Furthermore, the biological and functional relevance of the newly assisted binding sites remains unexplored. With the recent observation that ATP-dependent chromatin remodeling factors are involved in the pioneering action of GATA factors, major efforts are now needed to establish if this phenomenon is a general requirement for all TFs that act in initiating events. Chromatin structure is an integral component to TF recruitment and action: here we propose that the pioneering mechanism is not restricted to an exclusive set of TFs, but rather can be accomplished by many TFs. We now refer to factors that act in this role as initiating factors, and propose a general mechanism termed “dynamic assisted loading” (DynALoad). This model envisages a highly dynamic system with very quick chromatin interactions, and crucially, involves other co-factors such as ATP-dependent remodeling complexes (Fig. 2). Identifying the potential role of remodelers in initiating factor signaling pathways such as FoxA1 is needed in order to advance our understanding of these complex and essential systems.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), the National Cancer Institute (NCI) and the Center for Cancer Research (CCR). V.P. was supported, in part, by the Sigrid Jusélius Foundation.
Abbreviations:
- ChIP
chromatin immunoprecipitation
- DHS
DNAse I hypersensitive site
- DynALoad
dynamic assisted loading
- ER
estrogen receptor
- FCS
fluorescence correlation spectroscopy
- FoxA1
forkhead box A1
- FRAP
fluorescence recovery after photobleaching
- GR
glucocorticoid receptor
- SMT
single-molecule tracking
- SR
steroid receptor
- TF
transcription factor
Footnotes
The authors have declared no conflict of interest.
The copyright line of this article was updated on October 24, 2016.
References
- 1.Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–71. [DOI] [PubMed] [Google Scholar]
- 2.van Holde KE. 1988. Chromatin. Heidelberg: Springer-Verlag. [Google Scholar]
- 3.Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossard P, Zaret KS. 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125: 4909–17. [DOI] [PubMed] [Google Scholar]
- 6.Gualdi R, Bossard P, Zheng M, Hamada Y, et al. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–82. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo LA, McPherson CE, Bossard P, Stevens K, et al. 1998. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J 17: 244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo LA, Zaret KS. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol Cell 4: 961–9. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo LA, Lin FR, Cuesta I, Friedman D, et al. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–89. [DOI] [PubMed] [Google Scholar]
- 10.Morisaki T, Muller WG, Golob N, Mazza D, et al. 2014. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun 5: 4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swinstead Erin E, Miranda Tina B, Paakinaho V, Baek S, et al. 2016. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugo N, Morimatsu M, Arai Y, Kousoku Y, et al. 2015. Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci Rep 5: 10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Legant WR, Chen BC, Li L, et al. 2014. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. Elife 3: e04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Zhang Z, Li L, Chen BC, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballare C, Castellano G, Gaveglia L, Althammer S, et al. 2012. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell 49: 67–79. [DOI] [PubMed] [Google Scholar]
- 16.Li G, Margueron R, Hu G, Stokes D, et al. 2010. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell 38: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll JS, Liu XS, Brodsky AS, Li W, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43. [DOI] [PubMed] [Google Scholar]
- 18.Laganiere J, Deblois G, Lefebvre C, Bataille AR, et al. 2005. Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA 102: 11651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll JS, Meyer CA, Song J, Li W, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–97. [DOI] [PubMed] [Google Scholar]
- 20.Lupien M, Eeckhoute J, Meyer CA, Wang Q, et al. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, et al. 2010. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaud G, Roux J, Pictet R, Grange T. 1991. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell 67: 977–86. [DOI] [PubMed] [Google Scholar]
- 23.Kong SL, Li G, Loh SL, Sung WK, et al. 2011. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 7: 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caizzi L, Ferrero G, Cutrupi S, Cordero F, et al. 2014. Genome-wide activity of unliganded estrogen receptor-alpha in breast cancer cells. Proc Natl Acad Sci USA 111: 4892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisen JA, Sweder KS, Hanawalt PC. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. [DOI] [PubMed] [Google Scholar]
- 27.Smith CL, Peterson CL. 2005. ATP-dependent chromatin remodeling. Curr Top Dev Biol 65: 115–48. [DOI] [PubMed] [Google Scholar]
- 28.Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20: 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelan ML, Sif S, Narlikar GJ, Kingston RE. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3: 247–53. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Cote J, Xue Y, Zhou S, et al. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15: 5370–82. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Xue Y, Zhou S, Kuo A, et al. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10: 2117–30. [DOI] [PubMed] [Google Scholar]
- 32.Eberharter A, Becker PB. 2004. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci 117: 3707–11. [DOI] [PubMed] [Google Scholar]
- 33.Fyodorov DV, Kadonaga JT. 2002. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418: 897–900. [DOI] [PubMed] [Google Scholar]
- 34.Goldmark JP, Fazzio TG, Estep PW, Church GM, et al. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–33. [DOI] [PubMed] [Google Scholar]
- 35.Langst G, Becker PB. 2001. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–8. [DOI] [PubMed] [Google Scholar]
- 36.Marfella CG, Imbalzano AN. 2007. The Chd family of chromatin remodelers. Mutat Res 618: 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall JA, Georgel PT. 2007. CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85: 463–76. [DOI] [PubMed] [Google Scholar]
- 38.Woodage T, Basrai MA, Baxevanis AD, Hieter P, et al. 1997. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA 94: 11472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stokes DG, Perry RP. 1995. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 15: 2745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao Y, Shen X. 2007. INO80 subfamily of chromatin remodeling complexes. Mutat Res 618: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463: 474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris SA, Baek S, Sung MH, John S, et al. 2014. Overlapping chromatin remodeling systems collaborate genome-wide at dynamic chromatin transitions. Nat Struct Mol Biol 21: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erdel F, Schubert T, Marth C, Langst G, et al. 2010. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc Natl Acad Sci USA 107: 19873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Dieuleveult M, Yen K, Hmitou I, Depaux A, et al. 2016. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature 530: 113–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–76. [DOI] [PubMed] [Google Scholar]
- 47.Kidder BL, Palmer S, Knott JG. 2009. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27: 317–28. [DOI] [PubMed] [Google Scholar]
- 48.Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, et al. 2010. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141: 943–55. [DOI] [PubMed] [Google Scholar]
- 49.Krishnakumar R, Chen AF, Pantovich MG, Danial M, et al. 2016. FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18: 104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Du Y, Ward JM, Shimbo T, et al. 2014. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14: 575–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fryer CJ, Nordeen SK, Archer TK. 1998. Antiprogestins mediate differential effects on glucocorticoid receptor remodeling of chromatin structure. J Biol Chem 273: 1175–83. [DOI] [PubMed] [Google Scholar]
- 52.Fryer CJ, Archer TK. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393: 88–91. [DOI] [PubMed] [Google Scholar]
- 53.Ichinose H, Garnier JM, Chambon P, Losson R. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188: 95–100. [DOI] [PubMed] [Google Scholar]
- 54.Trotter KW, Archer TK. 2004. Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol 24: 3347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vicent GP, Zaurin R, Ballare C, Nacht AS, et al. 2009. Erk signaling and chromatin remodeling in MMTV promoter activation by progestins. Nucl Recept Signal 7: e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perlmann T, Wrange O. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J 7: 3073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pina B, Bruggemeier U, Beato M. 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60: 719–31. [DOI] [PubMed] [Google Scholar]
- 58.Venditti P, Di Croce L, Kauer M, Blank T, et al. 1998. Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res 26: 3657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aoyagi S, Trotter KW, Archer TK. 2005. ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm 70: 281–307. [DOI] [PubMed] [Google Scholar]
- 60.Belandia B, Orford RL, Hurst HC, Parker MG. 2002. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J 21: 4094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee HL, Archer TK. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J 17: 1454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson TA, Elbi C, Parekh BS, Hager GL, et al. 2008. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor regulated promoter. Mol Biol Cell 19: 3308–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsiao PW, Fryer CJ, Trotter KW, Wang W, et al. 2003. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol 23: 6210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeong KW, Lee YH, Stallcup MR. 2009. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J Biol Chem 284: 29298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nie Z, Xue Y, Yang D, Zhou S, et al. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 20: 8879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, et al. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem 277: 41674–85. [DOI] [PubMed] [Google Scholar]
- 67.Nagaich AK, Walker DA, Wolford RG, Hager GL. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. MolCell 14: 163–74. [DOI] [PubMed] [Google Scholar]
- 68.Li M, Hada A, Sen P, Olufemi L, et al. 2015. Dynamic regulation of transcription factors by nucleosome remodeling. Elife 4: e06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muchardt C, Yaniv M. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12: 4279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erdel F, Krug J, Langst G, Rippe K. 2011. Targeting chromatin remodelers: signals and search mechanisms. Biochim Biophys Acta 1809: 497–508. [DOI] [PubMed] [Google Scholar]
- 71.Schnitzler G, Sif S, Kingston RE. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94: 17–27. [DOI] [PubMed] [Google Scholar]
- 72.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, et al. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol 22: 7524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKnight JN, Jenkins KR, Nodelman IM, Escobar T, et al. 2011. Extranucleosomal DNA binding directs nucleosome sliding by Chd1. Mol Cell Biol 31: 4746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagaich AK, Walker DA, Wolford RG, Hager GL. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14: 163–74. [DOI] [PubMed] [Google Scholar]
- 75.Theodorou V, Stark R, Menon S, Carroll JS. 2012. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 23: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu G, Schones DE, Cui K, Ybarra R, et al. 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanalkumar R, Johnson KD, Gao X, Boyer ME, et al. 2014. Mechanism governing a stem cell-generating cis-regulatory element. Proc Natl Acad Sci USA 111: E1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takaku M, Grimm SA, Shimbo T, Perera L, et al. 2016. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol 17:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramakrishnan V, Finch JT, raziano V, Lee PL, et al. 1993. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–23. [DOI] [PubMed] [Google Scholar]
- 80.Clark KL, Halay ED, Lai E, Burley SK. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364: 412–20. [DOI] [PubMed] [Google Scholar]
- 81.Chaya D, Hayamizu T, Bustin M, Zaret KS. 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem 276: 44385–9. [DOI] [PubMed] [Google Scholar]
- 82.Sekiya T, Muthurajan UM, Luger K, Tulin AV, et al. 2009. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev 23: 804–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mueller F, Stasevich TJ, Mazza D, McNally JG. 2013. Quantifying transcription factor kinetics: at work or at play? Crit Rev Biochem Mol Biol 48: 492–514. [DOI] [PubMed] [Google Scholar]
- 84.Stasevich TJ, McNally JG. 2011. Assembly of the transcription machinery: ordered and stable, random and dynamic, or both? Chromosoma 120: 533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazza D, Abernathy A, Golob N, Morisaki T, et al. 2012. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res 40: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Speil J, Baumgart E, Siebrasse JP, Veith R, et al. 2011. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys J 101: 2592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Royen ME, van Cappellen WA, Geverts B, Schmidt T, et al. 2014. Androgen receptor complexes probe DNA for recognition sequences by short random interactions. J Cell Sci 127: 1406–16. [DOI] [PubMed] [Google Scholar]
- 88.Izeddin I, Recamier V, Bosanac L, Cisse II, et al. 2014. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. Elife 3: e02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elf J, Li GW, Xie XS. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316: 1191–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.White MD, Angiolini JF, Alvarez YD, Kaur G, et al. 2016. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165: 75–87. [DOI] [PubMed] [Google Scholar]