Abstract
Improved therapy of cancer has significantly increased the lifespan of patients. However, cancer survivors face an increased risk of cardiovascular complications due to adverse effects of cancer therapies. The chemotherapy drug doxorubicin is well known to induce myofibril damage and cardiac atrophy. Our aim was to test potential counteracting effects of the pro-hypertrophic miR-212/132 family in doxorubicin-induced cardiotoxicity. In vitro, overexpression of the pro-hypertrophic miR-212/132 cluster in primary rodent and human iPSC-derived cardiomyocytes inhibited doxorubicin-induced toxicity. Next, a disease model of doxorubicin-induced cardiotoxicity was established in male C57BL/6N mice. Mice were administered either adeno-associated virus (AAV)9-control or AAV9-miR-212/132 to achieve myocardial overexpression of the miR-212/132 cluster. AAV9-mediated overexpression limited cardiac atrophy by increasing left ventricular mass and wall thickness, decreased doxorubicin-mediated apoptosis, and prevented myofibril damage. Based on a transcriptomic profiling we identified fat storage-inducing transmembrane protein 2 (Fitm2) as a novel target and downstream effector molecule responsible, at least in part, for the observed miR-212/132 anti-cardiotoxic effects. Overexpression of Fitm2 partially reversed the effects of miR-212/132. Overexpression of the miR-212/132 family reduces development of doxorubicin-induced cardiotoxicity and thus could be a therapeutic entry point to limit doxorubicin-mediated adverse cardiac effects.
Keywords: doxorubicin, cardiotoxicity, microRNA, atrophy, apoptosis, AAV
Doxorubicin is known to cause dose-dependent cardiotoxicity in cancer patients. Here, Thum and colleagues show that overexpression of a pro-hypertrophic microRNA cluster, miR-212/132, can reduce the doxorubicin-induced cardiotoxic effects by reversing cardiac atrophy and apoptosis.
Introduction
Cancer is one of the leading causes of death globally and was responsible for nearly 9 million deaths in 2016.1 The number of cancer survivors is increasing due to early detection, awareness, and improved therapeutic interventions.2 In the United States alone, 15.5 million cancer survivors were reported until 2015, which is projected to increase to 20.3 million in the next decade.2 Post-cancer therapy, cancer survivors are prone to develop heart failure due to the cardiotoxic effects of chemotherapy drugs like doxorubicin.3
Rodent models of doxorubicin-induced heart failure show reduction in heart weight, cardiomyocyte apoptosis, myocardial damage, and atrophy as hallmarks of cardiotoxicity.4 Similar to animal models, patients with anthracycline-induced cardiomyopathy also showed dose-related decline in left ventricular mass.5 Prevention strategies to reduce doxorubicin-induced cardiotoxicity involve the usage of liposomal doxorubicin or an iron chelator dexrazoxane, but their use is limited due to the risk of secondary malignancies.6 Furthermore, conventional heart failure drugs are unable to completely reverse the anthracycline-induced cardiotoxicity,7 as cardiac atrophy rather than pathological hypertrophy is mechanistically involved. In a study on a heterogeneous cohort of 2,625 patients receiving anthracycline-containing therapy, 9% of the patients developed cardiotoxicity and heart failure.7 Of these patients, only 11% showed complete recovery from cardiotoxicity with conventional heart failure therapy.7 This shows an urgent need for the development of new therapies focused on preventing anthracycline-induced cardiotoxicity.
Recent studies have shown Topoisomerase 2β, Quaking, and PI3Kγ proteins as key mediators of doxorubicin-induced cardiotoxicity.4, 6, 8 Apart from the protein-coding genes, whether non-coding RNAs like microRNAs might also play a protective role in doxorubicin-induced cardiotoxicity remains largely unknown. microRNAs are small, non-coding RNA molecules that exert their regulatory function either by translational repression or by mRNA degradation of their targets.9 In contrast to doxorubicin, which induces cardiomyocyte atrophy, the miR-212/132 cluster was found to induce cardiomyocyte hypertrophy.4, 10, 11 Whether overexpression of the pro-hypertrophic miR-212/132 cluster could prevent doxorubicin cardiotoxicity remains to be tested.
Results
In Vitro Overexpression of miR-212/132 Inhibits Doxorubicin-Induced Cardiotoxicity
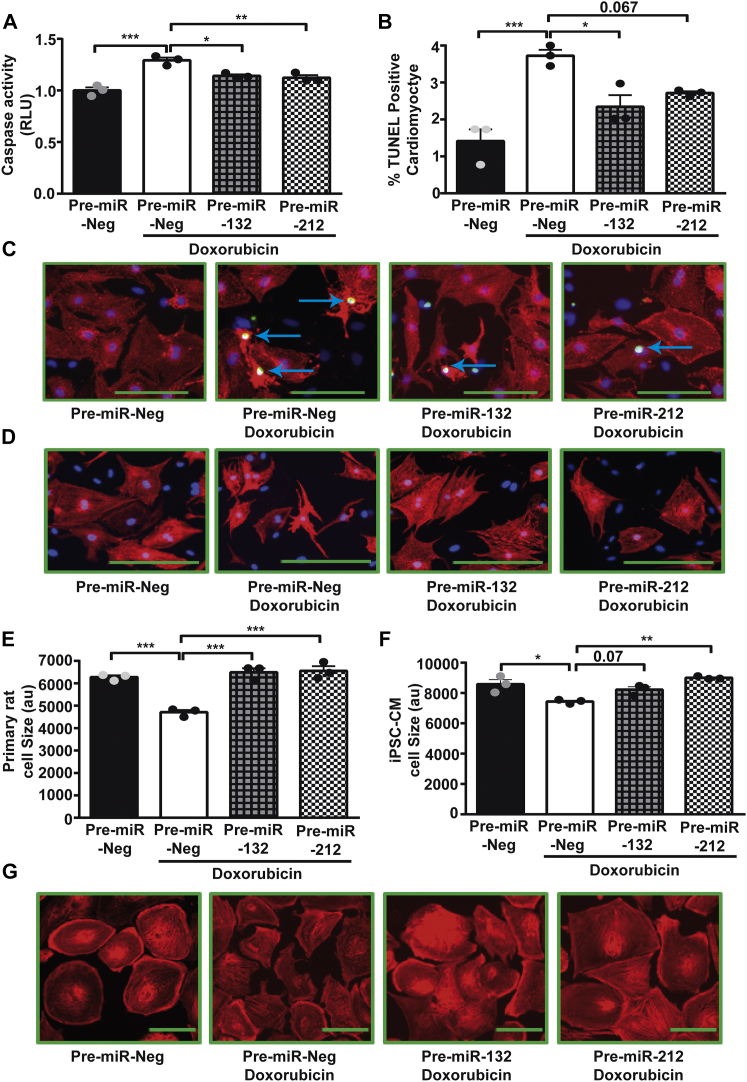
Doxorubicin induces atrophy in cardiomyocytes, while miR-212/132 was shown to exert a pro-hypertrophic phenotype.4, 10, 11 Thus, we hypothesized that overexpression of the pro-hypertrophic miR-212/132 in cardiomyocytes would constrain the detrimental effects in the setting of doxorubicin-mediated weakening of the heart because of muscular atrophy. To test a potential protective effect, we overexpressed miR-132 and miR-212 in primary neonatal rat cardiomyocytes (Figures S1A and S1B) and measured apoptosis and atrophy after treatment with doxorubicin. As expected, doxorubicin treatment of primary cardiomyocytes led to a significant increase in caspase 3/7 activity in control transfected cells (Figure 1A). This increased apoptotic response to doxorubicin was significantly inhibited by the overexpression of miR-132 and miR-212 (Figure 1A). Likewise, overexpression of miR-132 and miR-212 decreased apoptosis induced by doxorubicin, as documented by a lower percentage of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cardiomyocytes (Figures 1B and 1C). Primary cardiomyocytes treated with doxorubicin undergo atrophy, as indicated by a significant decline in cardiomyocyte size (Figures 1D and 1E). miR-132 and miR-212 overexpression significantly inhibited this doxorubicin-induced atrophy in rat cardiomyocytes (Figures 1D and 1E). Previously, we have shown that the miR-212/132 cluster inhibits cardiomyocyte autophagy,10 and dysregulation of autophagy has been reported in doxorubicin-induced cardiotoxicity.12 Therefore, we checked the levels of p62, an autophagic substrate, and found it to be downregulated in cardiomyocytes treated with doxorubicin, indicating activation of autophagy (Figures S1C and S1D). Overexpression of miR-212/132 inhibited autophagy and partially reduced loss of p62 in response to doxorubicin (Figures S1C and S1D). Doxorubicin is known to exert its cardiotoxic effects via oxidative damage.13 Therefore, we checked whether the miR-212/132 cluster has any effect on oxidative damage induced by doxorubicin. We measured the levels of MnSOD, which is upregulated by oxidative stress and functions as a detoxifying agent to remove free radicals.14 Doxorubicin increased levels of MnSOD in neonatal rat cardiomyocytes, and overexpression of miR-212 inhibited the upregulation of MnSOD in response to doxorubicin, while miR-132 had only minor effects (Figures S1E and S1F). To check for translational potential of the anti-atrophic impact of the miR-212/132 cluster, we performed additional experiments in human induced-pluripotent-stem-cell (iPSC)-derived cardiomyocytes. Similar to rat cardiomyocytes, overexpression of miR-132 and miR-212 (Figures S1G and S1H) also inhibited the atrophic effects of doxorubicin in human iPSC-derived cardiomyocytes (Figures 1F and 1G). These results indicate a protective effect of miR-212/132 on doxorubicin-induced cardiotoxicity.
Figure 1.
miR-212/132 Inhibits Doxorubicin-Induced Apoptosis and Atrophy in Cardiomyocytes
(A) Caspase 3/7 activity in primary neonatal rat cardiomyocytes after pre-miR-Neg, pre-miR-132, and pre-miR-212 transfection followed by 0.1 μM doxorubicin treatment for 48 hr. Percentage of TUNEL-positive cardiomyocytes (B and C) and cardiomyocyte cell size (D and E) in pre-miR-Neg-, pre-miR-132-, and pre-miR-212-transfected primary neonatal rat cardiomyocytes, with or without doxorubicin, are indicated. (F and G) Cell size measurements of human iPSC-derived cardiomyocytes transfected with pre-miR-Neg, pre-miR-132, and pre-miR-212, in the presence or absence of doxorubicin, are indicated. *p < 0.05; **p < 0.01; ***p < 0.001. RLU, relative luminescence unit; iPSC-CM, iPSC-derived cardiomyocyte.
AAV9-miR-212/132 Reduces Doxorubicin-Associated Cardiotoxicity
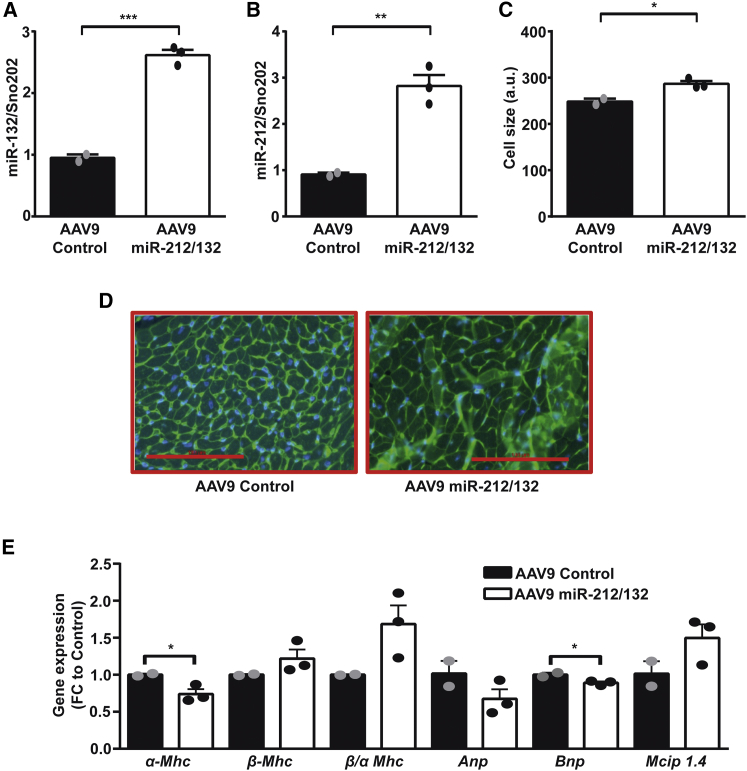
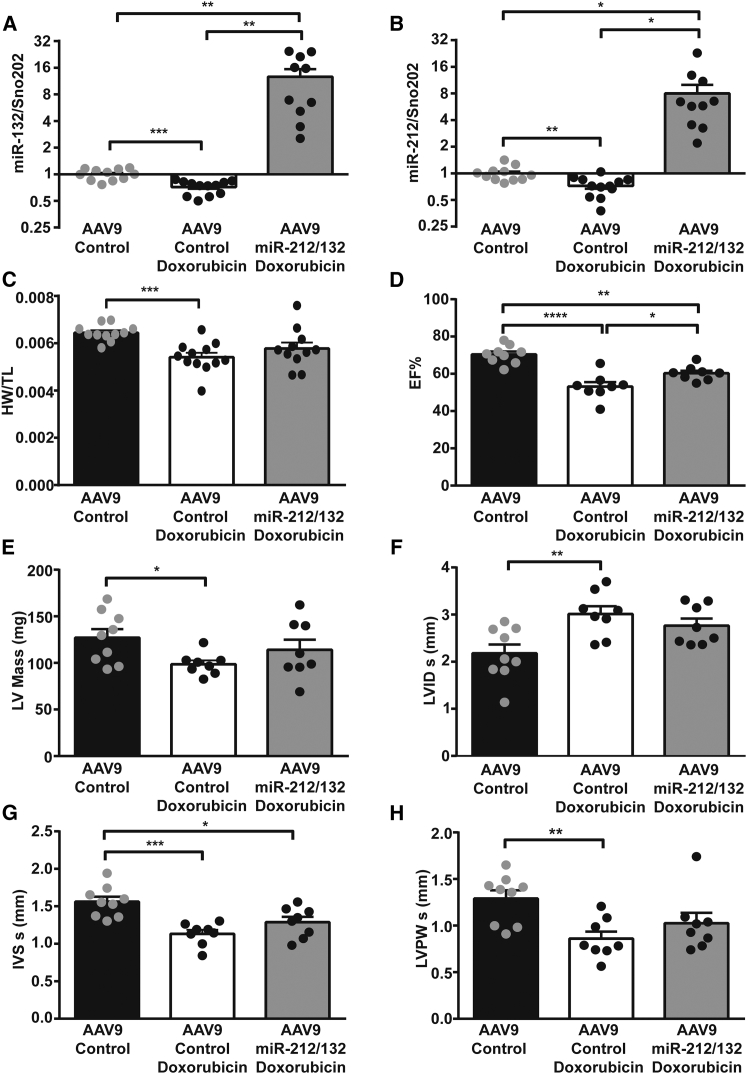
Based on our in vitro results, we speculated that in vivo overexpression of the miR-212/132 cluster in the heart would be able to prevent cardiotoxic effects of doxorubicin, due to its ability to increase the cardiac muscle mass. To achieve cardiac overexpression in vivo in murine heart, we used the adeno-associated virus (AAV)9 delivery system.15 AAV9-miR-212/132 viral particles were injected in healthy adult mice at a dose of 2 × 1012, and expression levels were evaluated 2 weeks later. AAV9-miR-212/132 increased the expression of miR-132 and miR-212 in murine myocardium compared to AAV9-control animals (Figures 2A and 2B). Overexpression of the miR-212/132 cluster induced cardiac hypertrophy in mice, as previously shown by our group (Figures 2C and 2D). Increased levels of miR-212/132 also decreased the expression of α-Mhc (myosin heavy chain) and showed an increased ratio of β/α Mhc with cardiac remodeling (Figure 2E). Anp and Bnp were only mildly regulated, while an increase in the NFAT-pathway downstream effector molecule Mcip1.4 was seen (Figure 2E). To test our hypothesis that in vivo overexpression of the miR-212/132 cluster would be protective against doxorubicin-mediated cardiotoxic effects, we overexpressed the miR-212/132 cluster in a previously established mouse model of doxorubicin-induced cardiotoxicity.4 Doxorubicin treatment in mice led to a significant decline in cardiac expression of miR-132 and miR-212, pointing to a potential direct involvement of the miR-212/132 cluster in doxorubicin-induced cardiotoxicity (Figures 3A and 3B). In contrast, no significant difference was observed in the expression of miR-212/132 in response to doxorubicin in primary rat (Figures S1I and S1J) and human iPSC-derived cardiomyocytes (Figures S1K and S1L), indicating an in vivo scenario needed for the overall regulation. AAV9-miR-212/132 treatment led to a significant increase in the myocardial levels of miR-132 and miR-212 (Figures 3A and 3B). Body weight declined in both groups of animals receiving doxorubicin, with no evident difference induced by the AAV9-miR-212/132 therapy (Figure S2A). The ratios of heart weight to tibia length, however, significantly declined with doxorubicin exposure in the AAV9-control group, while AAV9-miR-212/132 treatment attenuated the significant decline (Figure 3C). Echocardiographic analysis of cardiac function showed a significant decline in ejection fraction (EF) with doxorubicin (Figure 3D). Animals that received AAV9-miR-212/132 showed an improved ejection fraction, compared to AAV9-control, suggesting prevention from doxorubicin-induced cardiotoxicity based on muscle mass increase (Figure 3D). Indeed, left ventricular mass was significantly decreased with doxorubicin, but this was partially prevented in the AAV9-miR-212/132 group (Figure 3E). Similarly, left ventricular volume was significantly increased with doxorubicin, while mice with AAV9-miR-212/132 treatment were protected from doxorubicin-mediated toxicity (Figure 3F). Wall thickness of myocardium as another parameter for atrophy was significantly decreased with doxorubicin (Figures 3G and 3H), whereas AAV-miR-212/132 treatment moderately increased muscle thickness (Figures 3G and 3H). During diastolic measurements, no differences in the AAV-miR-212/132 group, compared to control, were observed (Figures S2B–S2D).
Figure 2.
Overexpression of miR-212/132 in Healthy Adult Mice
miR-132 (A) and miR-212 (B) expression in hearts of healthy mice treated with AAV9-miR-212/132 compared to AAV9-control. (C and D) Cardiomyocyte cell size measured by wheat germ agglutinin (WGA) staining of AAV9-miR-212/132 and AAV9-control animals. (E) Expression of cardiac remodeling genes in AAV9-miR-212/132 and AAV9-control hearts. AAV9-control, n = 2; AAV9-miR-212/132, n = 3. *p < 0.05; **p < 0.01; ***p < 0.001. FC, fold change.
Figure 3.
AAV9-Mediated miR-212/132 Overexpression Improves Cardiac Function in Doxorubicin-Induced Cardiotoxicity Model
miR-132 (A) and miR-212 (B) expression levels in AAV9-control hearts (n = 11), AAV9-control hearts with doxorubicin (n = 12), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 10). (C) Heart weight to tibia length ratio in AAV9-control healthy hearts (n = 11), AAV9-control hearts with doxorubicin (n = 12), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 10). Echocardiographic analyses of AAV9-control hearts (n = 9), AAV9-control hearts with doxorubicin (n = 8), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 8) showing ejection fraction (D), left ventricular mass (E), left ventricular internal diameter systole (F), interventricular septum systole (G), and left ventricular posterior wall thickness systole (H). *p < 0.05; **p < 0.01; ***p < 0.001.
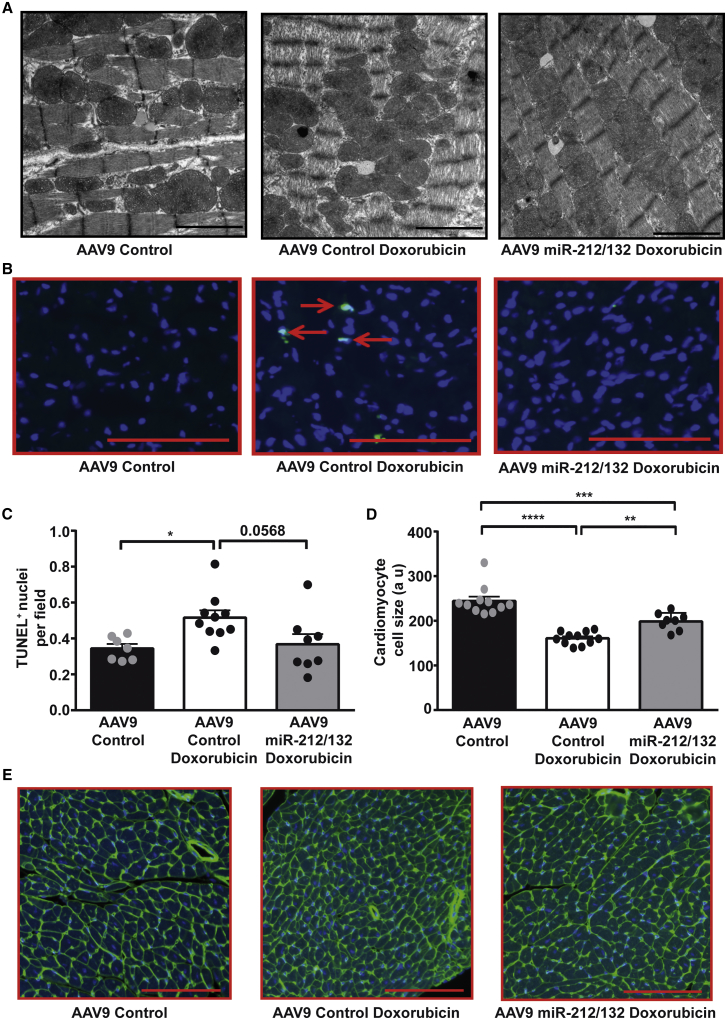
Direct myofibril damage can be visualized by electron microscopy and is a hallmark of doxorubicin cardiotoxicity.4 To assess the myofibril damage in our study, we analyzed the heart sections by transmission electron microscopy. Doxorubicin-induced myofibril damage was evident in electron-microscopic images of control-treated mouse, depicted by less dense and damaged myofibrils, compared to control without doxorubicin treatment (Figure 4A). Heart sections from AAV9-miR-212/132-treated animals showed preserved myofibrils, indicating rescue from doxorubicin-induced myofibril damage (Figure 4A). Doxorubicin treatment also induced cellular apoptosis, as detected by TUNEL staining (Figures 4B and 4C). Effects on cardiac apoptosis in mice treated with AAV9-miR-212/132 only showed a trend for less apoptosis (Figures 4B and 4C). Doxorubicin treatment also led to a significant decrease in cardiomyocyte size, confirming atrophy visualized by echocardiography measurements (Figures 4D and 4E). However, treatment with AAV9-miR-212/132 prevented doxorubicin-mediated atrophy and led to significantly enlarged cardiomyocytes (Figures 4D and 4E). The autophagic marker protein p62 showed a trend for downregulation in hearts exposed to doxorubicin, while higher p62 levels were seen in the AAV-miR-212/132 group (Figures S3A and S3B). Similarly, the oxidative stress marker MnSOD exhibited some increase in AAV-control hearts that received doxorubicin, compared to vehicle control, but no changes were seen with AAV9-miR-212/132 (Figure S3C). Furthermore expression of remodeling genes Anp, Bnp, β-Mhc, and α-Mhc did not exhibit any significant difference between the AAV-control group and the AAV9-miR-212/132 group treated with doxorubicin (Figures S3D–S3G). Based on these results, we can conclude that overexpression of the miR-212/132 cluster abrogates several, but not all, of the cardiotoxic effects of doxorubicin.
Figure 4.
AAV9-miR-212/132 Prevents Doxorubicin-Induced Cardiac Apoptosis and Atrophy
(A) Representative transmission electron microscope images of AAV9-control hearts, AAV9-control hearts with doxorubicin, and AAV9-miR-212/132 hearts with doxorubicin showing myofibril structures. (B and C) Heart sections showing TUNEL-positive nuclei in AAV9-control healthy hearts (n = 7), AAV9-control hearts with doxorubicin (n = 10), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 8). (D and E) Wheat germ agglutinin staining showing cardiomyocyte cell size in AAV9-control healthy hearts (n = 11), AAV9-control hearts with doxorubicin (n = 11), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 8). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Downstream Effectors of miR-212/132
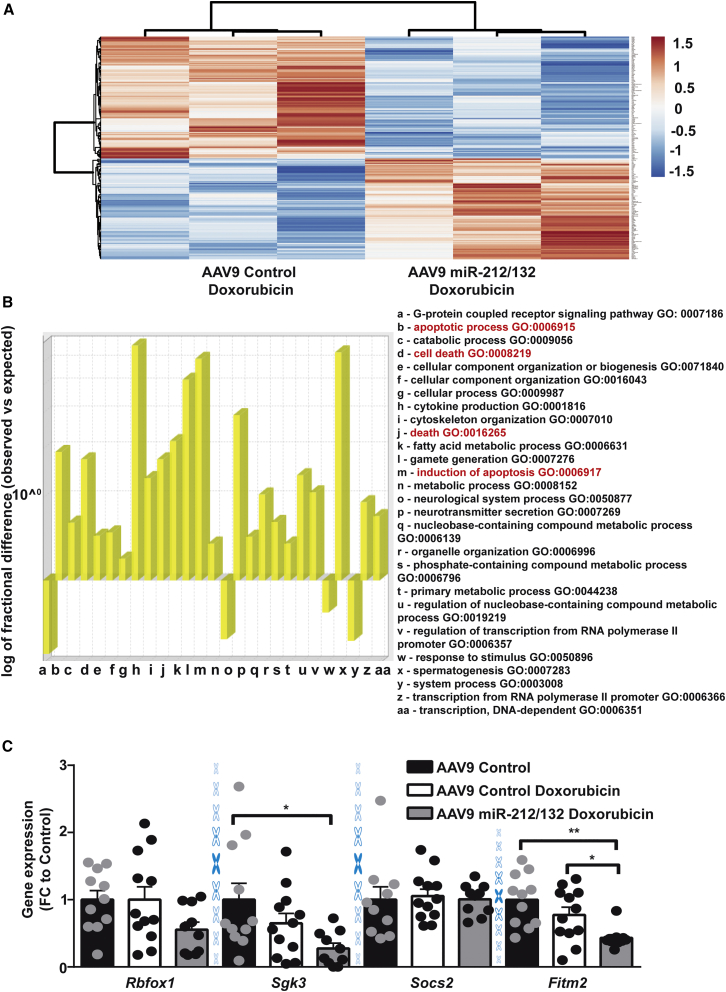
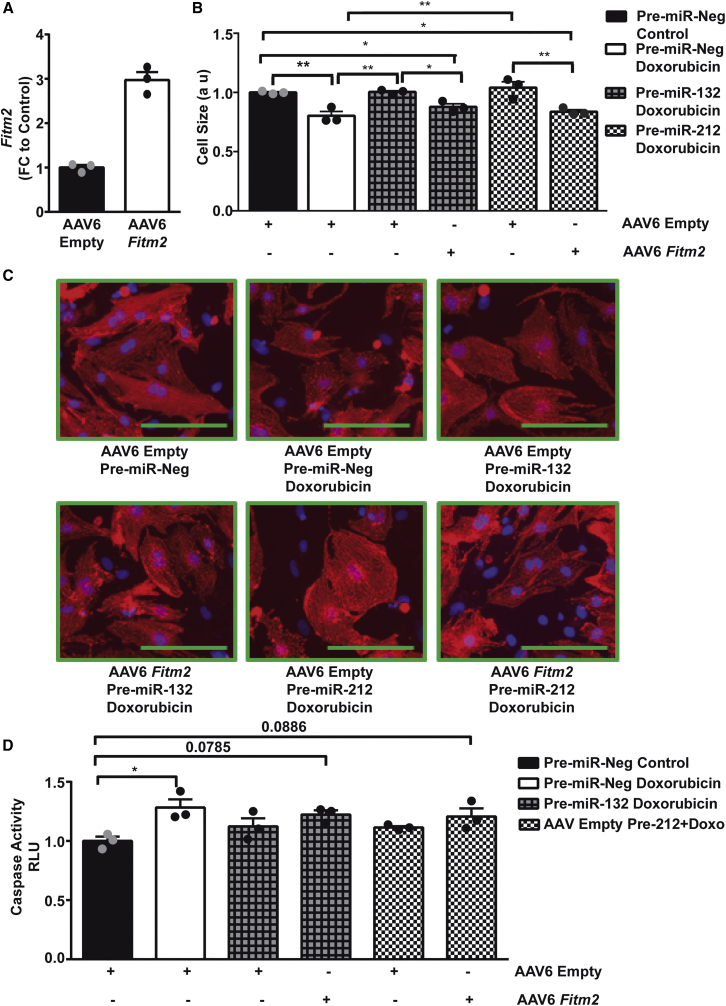
In order to identify potential downstream mediators of the miR-212/132 cluster, we compared the cardiac transcriptome of AAV9-control and AAV9-miR-212/132-treated mice. We identified 134 downregulated and 111 upregulated genes in the AAV9-miR-212/132-treated group compared to AAV9-controls (Figure 5A; Table S2). Gene Ontology analysis of the differentially regulated genes resulted in the enrichment of cell-death-related processes (Figure 5B, highlighted in red) among others. Among the downregulated genes, Fitm2, Rbfox1, Sgk3, Socs2, and L3mbt13 were found to have miR-212/132 binding sites in their 3′ UTR, as predicted by TargetScan. Real-time PCR-based quantification confirmed a significant downregulation of Fitm2 after overexpression of miR-212/132, compared to AAV9-control, both in the presence and the absence of doxorubicin (Figure 5C). Rbfox1 and Sgk3 also showed trends for downregulation with AAV9-miR-212/132, while Socs2 remained unaffected (Figure 5C). We were unable to amplify L3mbt13 with our primer, which might be due to its low expression level seen in the array. In vitro overexpression of miR-212/132 in rat primary cardiomyocytes also resulted in lowered Fitm2 levels (Figure S4A), while in human iPSC-derived cardiomyocytes, a decline in FITM2 was only seen with miR-132 overexpression (Figure S4B). Fitm2 (fat storage-inducing transmembrane protein 2) is ubiquitously expressed, including in the heart, with the highest expression in adipose tissue.16 Fitm2 has been found to be localized in the endoplasmic reticulum and to be involved in lipid droplet accumulation.16 Small interfering RNA (siRNA)-mediated inhibition of Fitm2 in primary rat cardiomyocytes (Figure S4C) slightly reduced doxorubicin-induced cardiac apoptosis and promoted cardiac hypertrophy (Figures S4D and S4E). The limited response seen via siRNA inhibition of Fitm2 might be due to the already existing decline in Fitm2 expression with doxorubicin in rat cardiomyocytes. To confirm Fitm2 as a downstream mediator of miR-212/132, we overexpressed Fitm2 in neonatal rat primary cardiomyocytes (Figure 6A). AAV6-mediated Fitm2 overexpression was able to significantly inhibit the anti-atrophic phenotype of miR-132 and miR-212 (Figures 6B and 6C), proving its direct involvement. Anti-apoptotic effects of the miR-212/132 cluster in response to doxorubicin were also partially prevented with AAV6-Fitm2 (Figure 6D). These results demonstrate that Fitm2 is highly involved in miR-212/132-dependent regulation of doxorubicin-induced cardiotoxicity, and its inhibition might be one of the mechanisms responsible for the observed anti-apoptotic and anti-atrophic effects of the miR-212/132 cluster, as seen in the case of the doxorubicin-stress model applied. Furthermore, FOXO3, previously identified as a direct target of miR-212/132, was also reduced in AAV9-miR-212/132 hearts and could also be responsible for miR-212/132 effects (Figure S4F).
Figure 5.
Downstream Targets of miR-212/132
(A) Heatmap showing differentially regulated genes in the myocardium of AAV9-miR-212/132 with doxorubicin compared to AAV9-control with doxorubicin (n = 3). (B) Gene Ontology (GO) analysis of differentially regulated genes shows overrepresented biological process. (C) Expression levels of Rbfox1, Sgk3, Socs2, and Fitm2 in AAV9-control healthy hearts (n = 11), AAV9-control hearts with doxorubicin (n = 12), and AAV9-miR-212/132 hearts exposed to doxorubicin (n = 10). *p < 0.05; **p < 0.01. FC, fold change.
Figure 6.
Fitm2 Inhibits the Anti-apoptotic and Anti-atrophic Function of miR-212/132
(A) Expression level of Fitm2 in primary neonatal rat cardiomyocytes transduced with AAV6-control and AAV6-Fitm2 (n = 3 technical replicates). (B and C) Cell size of neonatal rat cardiomyocytes transduced with either AAV6-Empty or AAV6-Fitm2 and transfected with pre-miR-Neg, pre-miR-132, and pre-miR-212 in the presence or absence of doxorubicin. (D) Caspase 3/7 activity in neonatal cardiomyocytes after overexpression of miR-132/212 together with AAV6-control or AAV6-Fitm2 transduction in the presence or absence of doxorubicin. *p < 0.05; **p < 0.01. FC, fold change.
Discussion
Here, we demonstrate that miR-212/132 reduces doxorubicin-induced cardiotoxicity and might be a potential therapeutic target. Doxorubicin is known to exert dose-dependent cardiotoxicity in cancer patients.17 Rodents exposed to doxorubicin exhibit loss in body weight and heart weight; increased cardiac apoptosis and atrophy; and, importantly, damaged myofibrils seen under an electron microscope.4 We show that overexpression of the pro-hypertrophic miR-212/132 family improves cardiac function, reduces doxorubicin-induced cardiac apoptosis and atrophy, and maintains cardiomyocyte myofibril structure. Similar to in vitro and in vivo rodent models, overexpression of the miR-212/132 family in human iPSC-derived cardiomyocytes also inhibited doxorubicin-induced atrophy, thus depicting translational potential to humans.
We identified Fitm2 as a novel target of the miR-212/132 cluster. Fitm2 is involved in lipid droplet formation, and its overexpression in skeletal muscle cells leads to a decline in cellular ATP levels and protects mice from high-fat-diet-induced weight gain.18 However, its role in cardiomyocyte biology is unknown. Here, we show that overexpression of Fitm2 partially opposes the anti-apoptotic and anti-atrophic effects of miR-212/132. microRNAs are known to target multiple targets to exert their effects.19 Apart from the Fitm2, we also identified the downregulation of other targets—Rbfox1, Sgk3, and Foxo3—with miR-212/132 overexpression. Rbfox1 deletion had been shown to aggravate stress-induced cardiomyocyte hypertrophy in line with miR-212/132 effects.20 Thus, these additional targets might also be responsible for the miR-212/132 anti-apoptotic and anti-atrophic effects observed in the doxorubicin-cardiotoxicity model.
In this study, we achieved overexpression of miR-212/132 by using the long-acting AAV9 system. As AAVs have been found to show persistent expression for the long term, this might also be harmful in the case of miR-212/132 in the long run.21, 22 Previously, we have shown that transgenic mice with miR-212/132 overexpression developed a heart-failure phenotype due to massive cardiac hypertrophy.10 Thus, AAV9-mediated overexpression of miR-212/132 in the long term could likely show pathological remodeling and induce heart failure. Therefore, alternative strategies to induce short-term overexpression (e.g., during the time of doxorubicin treatment during cancer) are needed to utilize the beneficial effects of miR-212/132 in relieving doxorubicin-induced cardiotoxicity. microRNA mimics are used for short-term overexpression studies in vitro, but their in vivo application is limited due to decreased stability and lower organ-specific uptake. A recent study from Lesizza et al. has shown successful cardiac overexpression of microRNAs by using mimics in combination with lipids when directly delivered to the heart via intra-cardiac injection.23 microRNA mimics were able to maintain increased expression levels for nearly 12 days in the heart.23 In another study, microRNA mimics were coupled with microbubbles, which enable imaging as well as ultrasound-guided burst delivery, and an antibody against VCAM-1 to specifically target endothelial cells.24 Thus, in the near future with an improved technique of cell-specific microRNA mimic delivery, cardiac-specific short-term overexpression of miR-212/132 could be used to prevent doxorubicin-induced cardiotoxicity. Another limitation of the AAV9 viral system that we used is that, despite higher cardiotropism, it also transduces other organs, and we cannot completely rule out effects mediated by transduction of other cell types or tissues.25
Doxorubicin mainly leads to cardiac malfunctioning in a chronic long-term setting, especially in children.17 Our present study only focuses on the immediate cardiotoxic effects of doxorubicin after therapy and does not focus on long-term toxicity, where the mechanism and the therapeutic effect of miR-212/132 overexpression might be completely different.
Materials and Methods
Neonatal Rat Cardiomyocyte and Human iPSC-Derived Cardiomyocytes
Neonatal rat cardiomyocytes were isolated as mentioned previously.26 Briefly, hearts from 1- to 3-day-old neonatal rats were explanted. Explanted hearts were cleaned and cut into small pieces and digested with trypsin (BD Diffco Trypsin) and DNase I (Sigma-Aldrich). Digested cells were centrifuged and pre-plated for 90 min to remove fibroblast population. Unattached cells were collected and seeded in minimum essential medium Eagle (MEM) with 5% fetal bovine serum (FBS), vitamin B-12, L-glutamine, and penicillin-streptomycin. Cells were washed the next day with PBS to remove debris and further processed for experiments. Pre-miR-negative (pre-miR-Neg) control, pre-miR-132, and pre-miR-212 (Ambion) were transfected at a concentration of 100 nM with Lipofectamine 2000 (Invitrogen). Human control iPSC-derived cardiomyocytes were differentiated at the lab of Dr. Katrin Streckfuss-Bömeke, as previously described.27, 28 Doxorubicin (Sigma-Aldrich) was applied at a dose of 100 nM for 48 to 72 hr. All the in vitro experiments were performed as three biological replicates, with technical replicates each time, except for TUNEL and cell size, unless otherwise mentioned.
Caspase Assay
Caspase assay was performed as per the manufacturer’s instructions with the Caspase-Glo 3/7 kit (Promega). Briefly, neonatal rat cardiomyocytes were treated with 100 nM doxorubicin in normal medium with no serum for 48 hr. After 48 hr, cardiomyocytes and caspase assay reagent provided with the kit were left at room temperature for 30 min. Then, an equal amount of caspase assay reagent was added and further incubated for 30 min at room temperature. Luminescence was measured using the HT Synergy (BioTek Instruments) plate reader.
TUNEL
TUNEL was performed per the manufacturer’s instructions, using an in situ cell death detection kit (Roche) as previously described.4 Neonatal rat cardiomyocytes or heart sections were fixed with 4% paraformaldehyde for 20 min at room temperature and then permeabilized with ice-cold 0.1% Triton X-100 in PBS for 2 min at room temperature. Cells or sections were incubated with the enzyme-labeling solution provided with the kit (Roche) for 1 hr at 37°C. Cells were then incubated with DAPI for 15 min. Three times washing was done at every step to remove residual substances. In the case of neonatal rat cardiomyocytes, cells were additionally stained with sarcomeric alpha-actinin (Sigma-Aldrich, #A7811) after TUNEL staining, to facilitate specific counting of TUNEL-positive cardiomyocytes. Images were acquired using the Nikon Eclipse Ti microscope and analyzed with NIS Elements.
Cell Size Measurements
Neonatal rat cardiomyocytes or human iPSC-derived cardiomyocytes were fixed, permeabilized, and stained with sarcomeric alpha-actinin (Sigma-Aldrich, #A7811) and DAPI to measure cell size. Images were taken with a Nikon Eclipse Ti microscope and were analyzed with NIS Elements. Several images were taken from different regions of the well, and the area of more than 200 cardiomyocytes was calculated and averaged.
Paraffin-embedded heart sections were fixed and stained with Alexa Fluor 488-labeled wheat germ agglutinin (Invitrogen) and DAPI. Images were taken with a Nikon Eclipse Ti microscope and were analyzed with NIS Elements. Five to six different regions of the heart were imaged, and the area of nearly 300 cardiomyocytes was measured and averaged per animal.
Doxorubicin-Induced Cardiotoxicity Animal Model
Permission was granted for all the animal experiments with local animal authorities at Hannover Medical School and Niedersachsen Landesamt für Verbraucherschutz (TVA 14/1665). A previously described mouse model of doxorubicin-induced cardiotoxicity was used.4 Adult C57BL/6N male mice were given doxorubicin at a dose of 5 mg/kg weekly for 5 consecutive weeks to establish doxorubicin-induced cardiotoxicity. 2 × 1012 viral particles of AAV9-control or AAV9-miR-212/132 were administered a day before starting the doxorubicin treatment. Control mice received AAV9-control virus together with saline injections. One week after the last doxorubicin injection (total, 5 weeks), echocardiographic data were acquired to assess the cardiac function, and animals were sacrificed. Echocardiography data were recorded using the Vevo 2100 system (FUJIFILM VisualSonics), and analysis was performed with standard imaging protocols (M-mode and B-mode) using Vevostrain software (FUJIFILM VisualSonics). All the animal experiments were randomized and blinded.
Electron Microscopy
Small pieces of explanted heart tissues were immersion-fixed in 150 mM HEPES (pH 7.35), containing 1.5% formaldehyde and 1.5% glutaraldehyde. Tissue embedding and heart section preparation were done as previously mentioned at the Institute of Functional and Applied Anatomy, Hannover Medical School.29 Images were taken at Transmission Electron Microscopy Morgagni 268, located in the Central Electron Microscopy Facility, Hannover Medical School.
RNA and Real-Time PCR
RNA was isolated using TriFast (Peqlab) per the manufacturer’s instructions. Concentration of the isolated RNA was measured using Take3 Plates on a Bio-Tek plate reader (Synergy HT). For mRNA measurements, RNA (500–1,000 ng) was reverse transcribed using the iScript Select cDNA Synthesis Kit (Bio-Rad), and SYBR Green-based real-time PCR was performed with iQ SYBR Green mix (Bio-Rad) on a C1000 Touch Thermocycler (Bio-Rad) using specific primer pairs listed in Table S1. For microRNA quantification, mature microRNA TaqMan assays (Applied Biosystems) for miR-132, miR-212, small nucleolar RNA (snoRNA), and sno202 were used. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems), and real-time PCR was conducted on a ViiA7 (Applied Biosystems) real-time PCR machine using ABsolute Blue QPCR Mix (Thermo Fisher Scientific).
mRNA Array
An array was performed on n = 3 heart samples each from the AAV9-control and AAV9-miR-212/132 groups treated with doxorubicin. RNA isolation was performed as mentioned earlier using TriFast (Peqlab), and RNA quality was evaluated by using RIN numbers measured on the Bioanalyzer System (Agilent). An array was conducted and analyzed as described previously.4 Differentially expressed genes with a cutoff of 1.25-fold were selected. A heatmap showing differentially regulated genes was generated using the Clustvis online tool. Microarray data has been submitted to the GEO database with the accession number GEO: GSE121965.
Gene Ontology Analysis
The gene IDs of 245 differentially regulated genes were uploaded on the PANTHER Gene Ontology website http://www.pantherdb.org/. A PANTHER overrepresentation test was conducted using PANTHER v13.1 with Mus musculus organism selection. The PANTHER GO-slim Biological Process was selected from an annotation dataset, a binomial test was conducted, and all the processes with p < 0.05 were selected.
AAV Production
Four hundred eighty-six base pairs (486 bp) encoding for miR-212/132 were cloned in an AAV backbone. A Fitm2 coding sequence of 786 bp was synthesized as a gene string from Thermo Fisher Scientific and cloned into a separate AAV backbone. AAV9 and AAV6 were produced as previously described.30 AAV9 was injected intravenously at a dose of 2 × 1012 viral particles. AAV6 was used at an MOI of 104 for primary rat cardiomyocytes.
Western Blot
Cell pellets were lysed in 1× lysis buffer (Cell Signaling Technology) together with Pefabloc (Sigma-Aldrich). Protein lysate was centrifuged at 8,000 × g for 5 min at 4°C to separate insoluble pellets. Protein concentration of the soluble fraction was measured by using Bradford reagent (Roti Quant, Carl Roth) on a spectrophotometer (Bio-Rad). Protein (50–30 μg) was run on an SDS gel, and wet blotting was performed to transfer protein on a polyvinylidene fluoride (PVDF) membrane (Bio-Rad). Specific proteins were detected with their respective antibodies (p62, Abcam, #ab56416; vinculin, Sigma-Aldrich #V9131; MnSOD, BD Biosciences, #611580; and Foxo3, Cell Signaling #2497).
Statistics
All the data were analyzed with GraphPad Prism 6 and are presented as mean ± SEM. An unpaired two-tailed t test was conducted to compare two groups wherever required. For comparison of ≥3 groups, a one-way-ANOVA was conducted with a post hoc Tukey test. For one-way ANOVA, Brown-Forsythe and Bartlett’s tests were conducted to check for significantly different SDs. In case of significant difference among SDs, a one-way ANOVA with Welch’s correction and a Games-Howell post hoc test were conducted to compare two groups in Excel.
Author Contributions
S.K.G. developed the concept, designed the study, received funding, performed most of the experiments, analyzed the data, and wrote the manuscript. A.G. helped with preparation of AAV6 viruses. P.A. and S.E. provided AAV9 viruses. K.S.-B. provided the human-iPSC-derived cardiomyocytes. S.B. wrote the animal permission for the experiments and helped in study design. T.T. helped in developing the concept, provided guidance during the study, and helped with manuscript writing.
Conflicts of Interest
S.K.G. and T.T. hold and licensed patent rights for the use of the miR-212/132 family as a therapeutic target. TT. and S.B. are co-founders and hold shares of Cardior Pharmaceuticals. The other authors report no conflict of interest.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG) GU 1664-1-1 to S.K.G. T.T. received funding from the REBIRTH Excellence Cluster, Fondation Leducq (MIRVAD [MicroRNA-Based Therapeutic Strategies in Vascular Disease]) and the European Union-funded ERC (European Research Council) Consolidator Grant LongHeart. K.S-B. is funded by the BMBF grant e:Bio—Modul II—Verbundprojekt: CaRNAtion (031L0075C).
We would like to thank Karina Zimmer and Ariana Foinquinos from the Institute of Molecular and Translational Therapeutic Strategies, Hannover Medical School, for their help with the animal handling. We would also like to thank Dr. Oliver Dittrich-Breiholz, Transcriptomics Facility, Hannover Medical School, for performing the mRNA arrays. We would also like to thank Dr. Jan Hegermann and Gerhard Preiss, Institute of Functional and Applied Anatomy, Hannover Medical School, for their help with transmission electron microscopy.
Footnotes
Supplemental Information includes four figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.11.004.
Contributor Information
Shashi Kumar Gupta, Email: gupta.shashi@mh-hannover.de.
Thomas Thum, Email: thum.thomas@mh-hannover.de.
Supplemental Information
References
- 1.GBD 2016 Causes of Death Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Ewer M.S., Ewer S.M. Cardiotoxicity of anticancer treatments. Nat. Rev. Cardiol. 2015;12:620. doi: 10.1038/nrcardio.2015.133. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S.K., Garg A., Bar C., Chatterjee S., Foinquinos A., Milting H., Streckfuss-Bomeke K., Fiedler J., Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neilan T.G., Coelho-Filho O.R., Pena-Herrera D., Shah R.V., Jerosch-Herold M., Francis S.A., Moslehi J., Kwong R.Y. Left ventricular mass in patients with a cardiomyopathy after treatment with anthracyclines. Am. J. Cardiol. 2012;110:1679–1686. doi: 10.1016/j.amjcard.2012.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vejpongsa P., Yeh E.T. Prevention of anthracycline-induced cardiotoxicity: challenges and opportunities. J. Am. Coll. Cardiol. 2014;64:938–945. doi: 10.1016/j.jacc.2014.06.1167. [DOI] [PubMed] [Google Scholar]
- 7.Cardinale D., Colombo A., Bacchiani G., Tedeschi I., Meroni C.A., Veglia F., Civelli M., Lamantia G., Colombo N., Curigliano G. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. 2015;131:1981–1988. doi: 10.1161/CIRCULATIONAHA.114.013777. [DOI] [PubMed] [Google Scholar]
- 8.Li M., Sala V., De Santis M.C., Cimino J., Cappello P., Pianca N., Di Bona A., Margaria J.P., Martini M., Lazzarini E. Phosphoinositide 3-kinase gamma inhibition protects from anthracycline cardiotoxicity and reduces tumor growth. Circulation. 2018;138:696–711. doi: 10.1161/CIRCULATIONAHA.117.030352. [DOI] [PubMed] [Google Scholar]
- 9.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ucar A., Gupta S.K., Fiedler J., Erikci E., Kardasinski M., Batkai S., Dangwal S., Kumarswamy R., Bang C., Holzmann A. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jentzsch C., Leierseder S., Loyer X., Flohrschütz I., Sassi Y., Hartmann D., Thum T., Laggerbauer B., Engelhardt S. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J. Mol. Cell. Cardiol. 2012;52:13–20. doi: 10.1016/j.yjmcc.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Dirks-Naylor A.J. The role of autophagy in doxorubicin-induced cardiotoxicity. Life Sci. 2013;93:913–916. [PubMed] [Google Scholar]
- 13.Berthiaume J.M., Wallace K.B. Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol. 2007;23:15–25. doi: 10.1007/s10565-006-0140-y. [DOI] [PubMed] [Google Scholar]
- 14.Candas D., Li J.J. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid. Redox Signal. 2014;20:1599–1617. doi: 10.1089/ars.2013.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanujam D., Sassi Y., Laggerbauer B., Engelhardt S. Viral vector-based targeting of miR-21 in cardiac nonmyocyte cells reduces pathologic remodeling of the heart. Mol. Ther. 2016;24:1939–1948. doi: 10.1038/mt.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadereit B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., Silver D.L. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA. 2008;105:94–99. doi: 10.1073/pnas.0708579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipshultz S.E., Colan S.D., Gelber R.D., Perez-Atayde A.R., Sallan S.E., Sanders S.P. Late cardiac effects of doxorubicin therapy for acute lymphoblastic leukemia in childhood. N. Engl. J. Med. 1991;324:808–815. doi: 10.1056/NEJM199103213241205. [DOI] [PubMed] [Google Scholar]
- 18.Miranda D.A., Koves T.R., Gross D.A., Chadt A., Al-Hasani H., Cline G.W., Schwartz G.J., Muoio D.M., Silver D.L. Re-patterning of skeletal muscle energy metabolism by fat storage-inducing transmembrane protein 2. J. Biol. Chem. 2011;286:42188–42199. doi: 10.1074/jbc.M111.297127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiedler J., Gupta S.K., Thum T. Identification of cardiovascular microRNA targetomes. J. Mol. Cell. Cardiol. 2011;51:674–681. doi: 10.1016/j.yjmcc.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Gao C., Ren S., Lee J.H., Qiu J., Chapski D.J., Rau C.D., Zhou Y., Abdellatif M., Nakano A., Vondriska T.M. RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure. J. Clin. Invest. 2016;126:195–206. doi: 10.1172/JCI84015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchlis G., Podsakoff G.M., Radu A., Hawk S.M., Flake A.W., Mingozzi F., High K.A. Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood. 2012;119:3038–3041. doi: 10.1182/blood-2011-09-382317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sehara Y., Fujimoto K.I., Ikeguchi K., Katakai Y., Ono F., Takino N., Ito M., Ozawa K., Muramatsu S.I. Persistent expression of dopamine-synthesizing enzymes 15 years after gene transfer in a primate model of Parkinson’s disease. Hum. Gene Ther. Clin. Dev. 2017;28:74–79. doi: 10.1089/humc.2017.010. [DOI] [PubMed] [Google Scholar]
- 23.Lesizza P., Prosdocimo G., Martinelli V., Sinagra G., Zacchigna S., Giacca M. Single-dose intracardiac injection of pro-regenerative microRNAs improves cardiac function after myocardial infarction. Circ. Res. 2017;120:1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Searle A.K., Hohmann J.D., Liu A.L., Abraham M.K., Palasubramaniam J., Lim B., Yao Y., Wallert M., Yu E. Dual-targeted theranostic delivery of miRs arrests abdominal aortic aneurysm development. Mol. Ther. 2018;26:1056–1065. doi: 10.1016/j.ymthe.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 26.Gupta S.K., Foinquinos A., Thum S., Remke J., Zimmer K., Bauters C., de Groote P., Boon R.A., de Windt L.J., Preissl S. Preclinical development of a microRNA-based therapy for elderly patients with myocardial infarction. J. Am. Coll. Cardiol. 2016;68:1557–1571. doi: 10.1016/j.jacc.2016.07.739. [DOI] [PubMed] [Google Scholar]
- 27.Streckfuss-Bömeke K., Wolf F., Azizian A., Stauske M., Tiburcy M., Wagner S., Hübscher D., Dressel R., Chen S., Jende J. Comparative study of human-induced pluripotent stem cells derived from bone marrow cells, hair keratinocytes, and skin fibroblasts. Eur. Heart J. 2013;34:2618–2629. doi: 10.1093/eurheartj/ehs203. [DOI] [PubMed] [Google Scholar]
- 28.Borchert T., Hübscher D., Guessoum C.I., Lam T.D., Ghadri J.R., Schellinger I.N., Tiburcy M., Liaw N.Y., Li Y., Haas J. Catecholamine-dependent β-adrenergic signaling in a pluripotent stem cell model of Takotsubo cardiomyopathy. J. Am. Coll. Cardiol. 2017;70:975–991. doi: 10.1016/j.jacc.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 29.Rudat C., Grieskamp T., Röhr C., Airik R., Wrede C., Hegermann J., Herrmann B.G., Schuster-Gossler K., Kispert A. Upk3b is dispensable for development and integrity of urothelium and mesothelium. PLoS ONE. 2014;9:e112112. doi: 10.1371/journal.pone.0112112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., Dittrich M., Maetzig T., Zimmer K., Remke J. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016;8:326ra22. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.