Abstract
A novel strain of Saccaropolyspora hirsuta was isolated from an insect Tapinoma simrothi for the first time and was morphologically and physiologically characterized. It was genetically identified using 16S rRNA and sequence similarity percentage in genbank with closely related species as strain ess_amA6 of Saccaropolyspora hirsuta. The accession number of strain ess_amA6 is KF996506. Antagonistic activity of strain ess_amA6 against some pathogenic Gram positive and negative bacteria, and unicellular fungus Candida albicans was studied. In addition, star shaped silver nanoparticles were biosynthesized using strain ess_amA6. The silver Nano stars were characterized by UV-us spectrophotometer. Fourier transform infrared spectroscopy analysis confirmed the conversion of Ag+ ions to Nano silver due to the reduction by capping material of extract. Transmission electron microscopically studies of biosynthesized Nano silver particles showed that they are spherical ranging from 10 nm to 30 nm in size. Silver atoms were checked in Nano sample by Energy Dispersive X-ray spectroscopy. Bioactivity of biosynthesized Nano silver was observed against some pathogenic microorganisms such as Staphylococcus aureus, Streptococcus pyogenes, Salmonella typhi, pseudomonas aeruginosa, Klebsiella pneumonia and Candida albicans. These tested microbes were highly sensitive to Nano silver. This study recommended that strain ess_amA6 can be used to effectively biosynthesize bioactive Nano silver compounds.
Keywords: Saccharopolyspora hirsute, Nano silver, Antimicrobial, Pathogenic microbes
1. Introduction
Insects have symbiotic relationships with various microorganisms for the purposes of nutrition and protection from natural risks (Klepzig et al., 2009). In the center of symbiotic relations, microorganism -insect associations that involve actinomycetes could be of specific interest to microbiologists looking for antimicrobials and ways to biosynthesize nanomaterials. One such relationship is attine ants with actinomycetes genera Streptomyces and Amycolatopsis from which Mueller et al. (2008) extracted antimicrobial active compounds. Nanotechnology involves the synthesis of nanoparticles (i.e., materials no more than 100 nm in diameter) via physical, chemical, and biological approaches (Ebrahiminezhad et al., 2016, Kalimuthu et al., 2008), as well as the integration of resulting nanostructures into diverse applications. Nanomaterials vary by size, distribution, and particle morphology (Service, 2000). Among nanometals, silver nanoparticles (AgNPs) are the most commonly used, given their range of biological activities and good chemical stability (Vennila et al., 2018, Prabhu and Poulose, 2012,). In the present work, we aimed to isolate and identify actinobacterial isolate from an insect Tapinoma simrothi. The study also aimed ability of this isolate to synthesize Nano silver as anti-microbiological agents
2. Materials and methods
2.1. Isolation of Saccharopolyspora hirsuta
The insect Tapinoma simrothi collected from Eldrieh, Riyadh, Saudi Arabia (24.7N, 46.7E) to isolate actinomycetes and suspension of T. simrothi was prepared in normal saline solution (NSS) for isolation process. Starch casein agar (SCA) medium (pH 7.2 ± 0.2) was used in the isolation Cochrane, 1961, Srinivasan et al., 1991.
2.2. Morphological characterization
The color of aerial mycelium was determined from mature, sporulating aerial mycelia of colonies of actinomycetes on different media, including ISP-2, ISP-4, ISP-6, ISP-7, Czapek–Dox agar, and SCA. Color was determined visually by observing color changes in the medium due to diffusing pigments produced by strain Ess_amA6 (Shirling and Gottlieb, 1966) and with lists of color names (Pridham, 1965).
2.3. Physiological characterization
Carbohydrates and nitrogen sources were identified and physiological investigations performed with media and methods recommended by Gorajana et al., 2007, Luedemann and Brodsky, 1964, Pridham and Gottlieb, 1948. All cultures were incubated at 30 °C for 7 d. The assay for enzymatic activity was achieved according to Bibb et al.’s (1977) procedure.
2.4. Extraction and purification of genomic DNA
The total genomic DNA of strain Ess_amA6 was extracted by crushing of mycelium biomass (0.1 g) with liquid nitrogen and the grinded biomass was mixed with 500 μL lysis buffer containing 50 mM of Tris-HCl (pH 8.0), 5 mM of EDTA (pH 8.0), 50 mM of NaCl, and 20 μL of lysozyme 10 mg/mL. The mixture was vigorously vortexed, and incubated at 37 °C for 30 min. Consequently, 20 μL of proteinase K (10 mg/mL) and 20 μL SDS (10% w/v) were added to the Eppendorf tube and incubated at 55 °C for 30 min. The cell lysate was exposed to very low temperature and extracted once in phenol: chloroform: isoamyl alcohol mixture (25:24:1 v/v) and followed by centrifugation at 10,000 rpm for 5 min to collect aqueous phase. The total genomic DNA was precipitated from the obtained aqueous phase by adding 600 μL of chilled isopropanol, and the precipitate was pelletized by centrifugation at 13,000 rpm for 30 min. The pelletized DNA genome was washed with 100% ethanol, air dried under laminar flow, and dissolved in 50 μL TE buffer
2.5. Molecular characterization of actinomycete isolate by amplification of 16S rRNA gene and it sequencing
To molecularly identify actinomycete isolate, the 16S rRNA gene was amplified using primer set Star-F (5′-GAGTTTGATCMTGGCTCTG-3′) and 1387-R (5′-CGGGCGGTGTGTACAACG-3′). Amplification was done on a thermalcycler machine (D-37085, SensoQuest GmbH, Göttingen, Germany). The PCR products was sequenced and analyzed by GATC Biotech at the European Custom Sequencing Centre in Cologne, Germany. PCR is programmed denaturation at 94 °C for 1 min, annealing at 51 °C for 1 min, extension at 72 °C for 1 min with an initial denaturation at 94 °C for 3 min, and a final extension at 72 °C for 7 min. number of cycles was 35.
2.6. Phylogenetic construction of actinomycete isolate
The 16S rRNA gene sequence obtained was compared with homologous sequences retrieved from GenBank using BLASTN per the method of Altschul et al. (1997). Multiple sequence alignments with the sequences of different actinomycetes groups was performed using ClustalW with default parameters (Thompson et al., 1994). A phylogenetic tree was constructed by the neighbor-joining method with nucleotide pairwise genetic distances modified by the Kimura two-parameter method using TreeCon.
2.7. Nano silver synthesis method by actinomycete isolate
Following the method of Li et al. (2011) to biosynthesize nanoparticles, the isolate was further cultured in ISP-2 liquid medium, under growth conditions at 30 °C, pH 7.2 and 220 rpm for 72 h. Separation of biomass and Cell filtrate (CF) was occurred by centrifugation at 4 °C, 10,000 rpm for 10 min. For bioreduction, AgNO3 (Qualigens 99.8%) was added to 100 mL of CF at 1 mM and incubated at 30 °C in the dark for 48 h.
2.8. Characterization of biosynthesized Nano silver
Morphological, elemental, functional and structural properties of the silver Nano stars were implemented using TEM (JEM-1010, JEOL) at an accelerating voltage of 80 kV, energy-dispersive X-ray spectrometer (EDS) with a scanning electron microscope (JSM-6380 LA, JEOL, Japan), U/V-vis spectrophotometer (Lambda 35, Hitachi, Tokyo, Japan) and FTIR spectrophotometer (Nicolet 6700, Madison, USA)
2.9. Test pathogens
The bioactivity of biosynthesized nanosilver particles was tested against Gram-negative (i.e., Salmonella typhi ATCC 6539, Pseudomonas aeruginosa ATTC 27853, and Klebsiella pneumonia ATCC 700603), Gram-positive (i.e., Streptococcus pyogenes ATCC 19615 and Staphylococcus aureus ATCC 25923) bacterial species and Candida albicans strain ATCC 90028 as antifungal activity. We procured on Test pathogenic microbes from Khalid Hospital in Riyadh. All test pathogenic microbes were preserved in brain heart infusion medium (BHI) at −20 °C. Later, 300 μL of each stock culture was inoculated in 3 mL of BHI broth. Overnight cultures were kept for 24 h at 37 ± 1 °C, and the purity of cultures was checked after 8 h of incubation.
2.10. Antimicrobial activity of Nano silver
The bioactivity of the biosynthesized nanosilver particles were performed by agar diffusion method. The nutrient agar medium was inoculated with freshly prepared cells of each bacterium and C. albicans on Sabouraud agar. After the solidification of the agar, several sterilized disks were dipped into CF as positive control and biosynthesized Nano silver at concentrations of 3, 8, 16, and 30 μg and were transferred on the plates. Bioactivity of biosynthesized Nano silver was measured as the diameter of the inhibition zone after incubation at 37 °C for 24 h.
3. Results and discussion
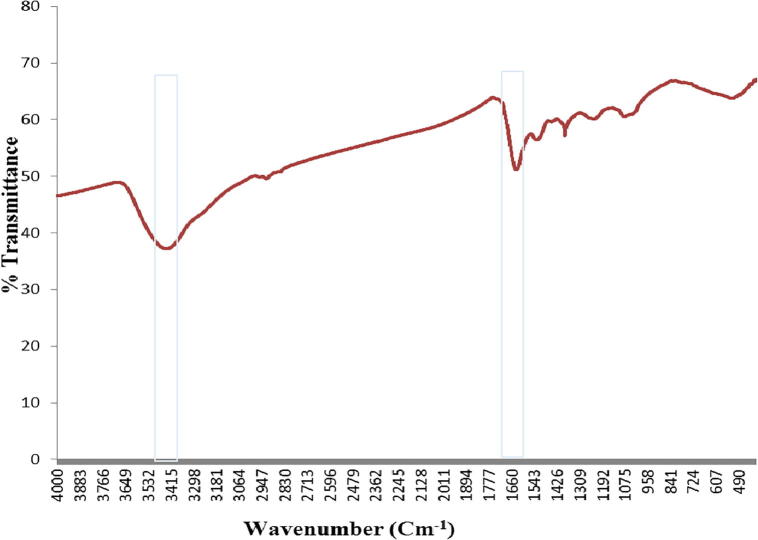
The strain ess_amA6 isolated from an insect Tapinoma simrothi which was collected from Riyadh, Saudi Arabia is a Gram-positive filamentous bacterium. The culture, when examining by light microscope (100X) has straight to hairy surfaced spore chains arising from the aerial mycelium and may be placed in Retinaculum Apertum (RA) group of Saccharopolyspora species (Figs. 1and 2). The strains were characterized by both biochemical and physiological methods and as well as molecular methods. The aerial mycelium displayed white color, while the substrate mycelium showed brown to whitish grey color (Table 1). These characteristic morphological properties strongly suggested that the isolate belonged to genus Saccharopolyspora. The biochemical and physiological properties of the isolated actinomycete are exposed in Table 2. The growth temperature is ranged from 26 to 37 °C with optimum of 30 °C and it grew up to 4% NaCl concentration with neutral pH 7. The strain could utilize Rhaffinose, Fructose, Sucrose, Arabinose, lactose, d-galactose, d-xylose, Citrate along with amino acids l-Histidine, l-Phenylalanine, l-Valine, l-Cysteine (Table 2). The strain ess_amA6 showed resistance to the entire antibiotics tested expects Erythromycin, Chloramphenicol and Imipenem (Table 2). The partial sequence of 16S rRNA gene of the actinomycete isolate was compared with the nucleotide sequences of other Saccharopolyspora strains and retrieved from the NCBI database using the neighbour-joining method. The strain had very highly 16S rRNA gene sequence similarity (98%) with Saccharopolyspora hirsuta. Hence, on the basis of cultural, biochemical, physiological and 16S rRNA gene sequence, the novel isolate ess_amA6 was designated as Saccharopolyspora hirsuta strain ess_amA6. The DNA sequences were aligned and phylogenetic tree was constructed using CLUSTALW (Fig. 3). The sequences were deposited in GenBank (NCBI) with accession number KF996506.1 (Kumar et al., 2015). Nanotechnology is the best promising area of research in modern biomedical applications. Nanomaterials are the leading substances in the nanobiomedical field (Kumar et al., 2015). In this context Saccharopolyspora hirsuta strain ess_amA6 was further selected for the biosynthesis of nanosilver. To the best of our knowledge, this is the first report on extracellular biosynthesis of nanosilver by the Saccharopolyspora hirsuta strain ess_amA6. The formation of biosynthesized silver nanoparticles was observed by the color changes (Fig. 4). Unreduced silver nitrate was colorless whereas silver nanoparticles were dark brown in color due to their specific surface plasmon resonance property (Ebrahiminezhad et al., 2016, Kalishwaralal et al., 2008). These nanoparticles were star in shape and ranging from 10 nm to 30 nm in size with polydispersity (Fig. 5). Fig. 6 clearly shows that the biosynthesized silver nanoparticles surface plasmon resonance occurs at 420 nm using UV-Visible spectrophotometer, suggesting that the particles are well dispersed in the aqueous solution (Kumar et al., 2015). To explore the reduction process of silver nitrate by the extracellular supernatant of S. hirsuta strain ess_amA6 and to detect possible interfaces between silver metals and protein molecules, Fourier transform infrared spectroscopy measurements were approved peak at 1660, 1426 (C–N stretching vibrations) and 3415 cm−1 which might reason in the reduction of silver ions and stabilization of Nano silver (Fig. 7). the EDX measurements of nanomaterials sample were carried out To identify presence of Ag+ ions, The optical absorption band of peak was in the range of 0.392–2.984 keV (Table 3) and this is typical for the absorption of Ag Nano metals (Ebrahiminezhad et al., 2016, Sadhasivam et al., 2010).
Fig. 1.
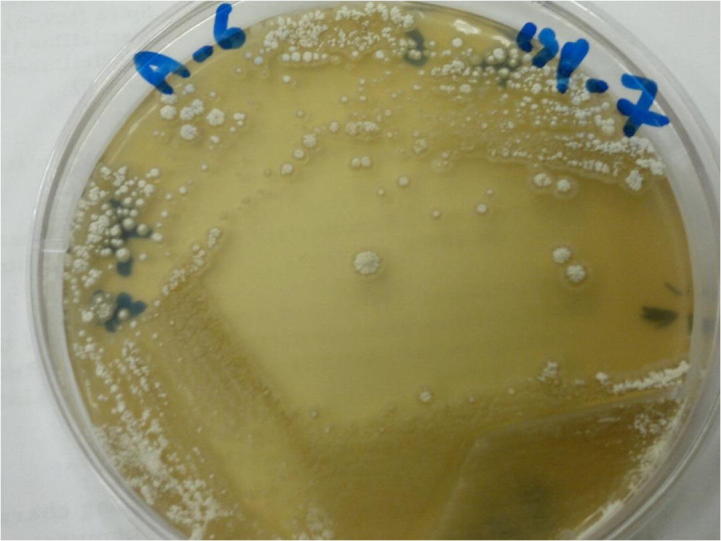
Colonies of S. hirsuta on international Streptomyces project 7 medium.
Fig. 2.
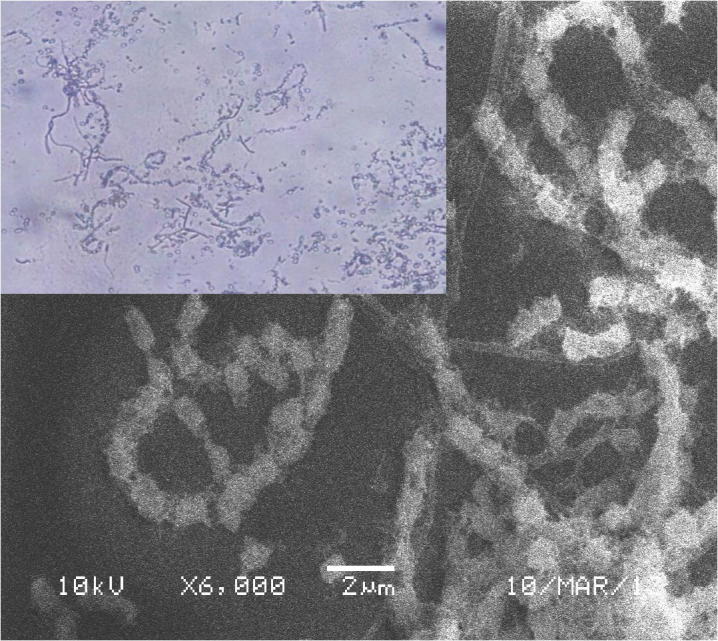
Morphological shape of the strain ess_amA6 of S. hirsuta under simple and scanning electron microscope.
Table 1.
Cultural characterization of Saccharopolyspora hirsuta Strain ess_amA6 on different culture media.
| Parameters | Media |
||||||
|---|---|---|---|---|---|---|---|
| ISP-2 | ISP-4 | ISP-5 | ISP-6 | ISP-7 | Czapex Dox | Starch casein agar | |
| Color of aerial mycelium | White | White | White | Creamy White | White | White | White |
| Color of substrate mycelium | Brown | Light white | Light grey | Light white | Light white | Light white | Light white |
| AM/SM | AM | AM | AM | Am | Am | Am | Am |
| Pigmentation | Brown | – | – | Brownish black | Violet | – | Red |
| Melanin production | – | – | – | +ve | – | – | – |
| Growth | Good | Good | Moderate | Good | Good | Poor | Good |
| Form of spore chain | Retinaculum apertum | Retinaculum apertum | Retinaculum apertum | Retinaculum apertum | Retinaculum apertum | Retinaculum apertum | Retinaculum apertum |
Table 2.
Physiological and biochemical properties of Saccharopolyspora hirsuta Strain ess_amA6.
| Carbon sources | Nitrogen sources | Growth at different conc. NaCl | Growth at different pH | Growth at different temperatures °C | Enzyme activity | Antibiotic resistance | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rhaffinose | + | l-Cysteine | + | 1% | ++ | 5 | − | 4 | − | Amylase | + | Erythromycin (15 µg) | − |
| Fructose | + | l-Valine | + | 4% | + | 7 | + | 26 | ++ | Protease | + | Gentamamicin (10 µg) | + |
| Sucrose | ++ | l-Phenylalanine | + | 7% | − | 10 | − | 30 | +++ | Chitinase | + | Penicillin G (10 µg) | + |
| Arabinose | + | l-Histidine | + | 10% | − | 37 | ++ | Catalase | + | Rifampicin (30 µg) | + | ||
| Lactose | ++ | 14% | − | 45 | − | DNase | + | Kanamycin (1000 µg) | + | ||||
| d-galactose | + | Hydrolysis of esculin | + | Vancomycin (5 µg) | + | ||||||||
| d-xylose | + | Lecithin hydrolysis | + | Colisttin sulphate (10 µg) | + | ||||||||
| Citrate | + | H2S production | − | Amikacin (30 µg) | + | ||||||||
| Nitrate reduction | − | Aztreonam (30 µg) | + | ||||||||||
| Urea hydrolysis | + | Chloramphenicol (30 µg) | − | ||||||||||
| Lipid hydrolysis | + | Ceftazidime (30 µg) | + | ||||||||||
| Imipenem (10 µg) | − | ||||||||||||
| Ciprofloxacin (1 µg) | + | ||||||||||||
| Piperacillin (100 µg) | + | ||||||||||||
| Tetracycline (30 µg) | + | ||||||||||||
Fig. 3.
Phylogenetic tree of strain ess_amA6 of S. hirsuta (accession No. KF996506.1).
Fig. 4.

Reduction of silver nitrate to Nano silver and color change to brown color by S. hirsuta strain ess_amA6.
Fig. 5.
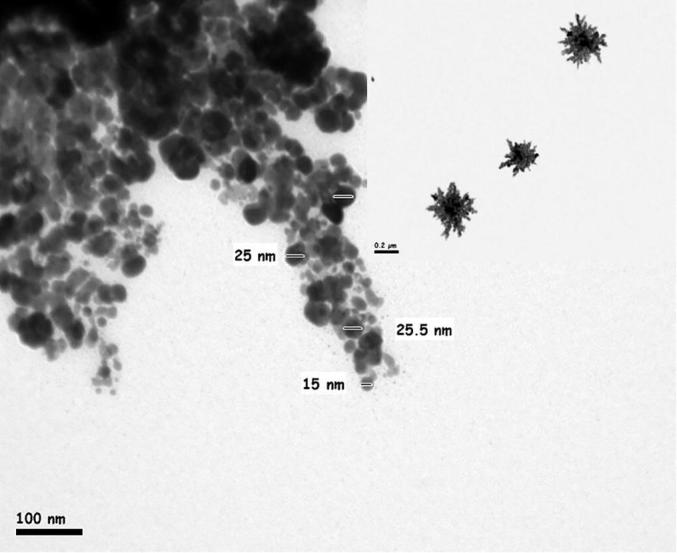
Image of star shaped Nano silver under transmission electron microscope.
Fig. 6.
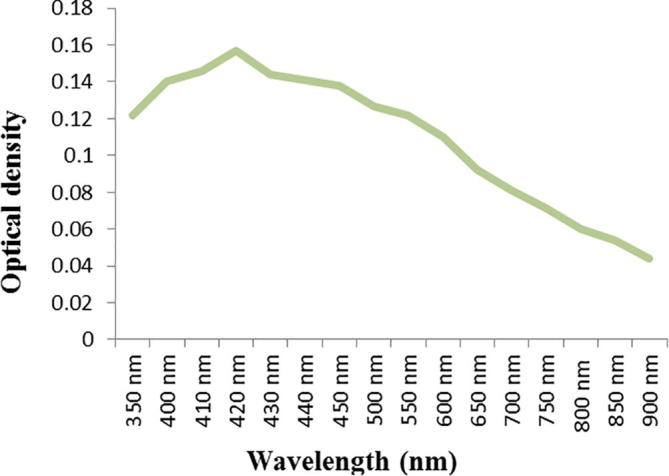
Degree of UV Absorbance of biosynthesized Nano silver.
Fig. 7.
FTIR analysis of Nano silver showing properties peak at 1660 and 3415 cm−1.
Table 3.
EDS analysis of biosynthesized star shaped Nano silver.
| Element | (keV) | Mass % | Error % | Atomic % | Compound |
|---|---|---|---|---|---|
| AgNPs A-6 | |||||
| N K | 0.392 | 0.94 | 0.15 | 5.76 | |
| O K | 0.525 | 3.48 | 0.04 | 18.59 | 100% Ag O |
| Ag L | 2.984 | 95.57 | 0.01 | 75.65 | |
| Total | 100 | 100 | 100 | ||
The produced nanoparticles were tested against Gram-positive, Gram-negative and yeast microorganism. Different concentrations of silver-nanoparticles were prepared and tested (0, 3, 8, 16, 30 µg/disc). In this study, we showed that the biosynthesized nanosilver were more effectively activity at the concentrations of 8 µg/disc to 30 µg/disc. The highest inhibition activities were observed at 30 µg/disc against Staphylococcus aureus (13 mm), Klebsiella pneumonia (13 mm), Salmonella typhi (12 mm), Streptococcus pyogenes (12 mm), Pseudomonas aeruginosa (13 mm) and Candida albicans (11 mm) (Fig. 8, Fig. 9) (Vennila et al. 2018).
Fig. 8.
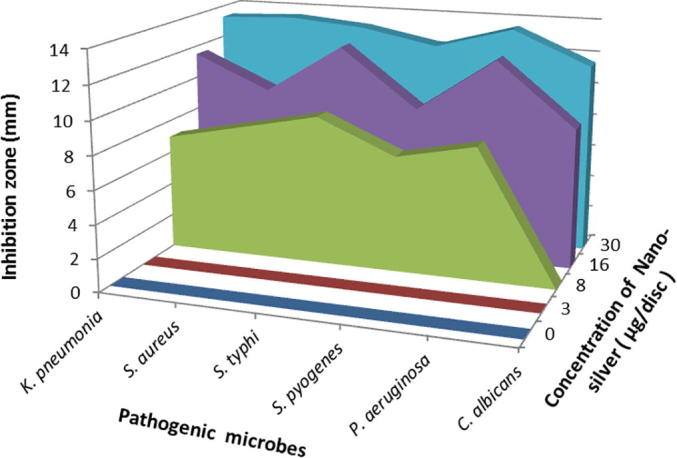
Antimicrobial activity of biosynthesized star shaped Nano silver.
Fig. 9.
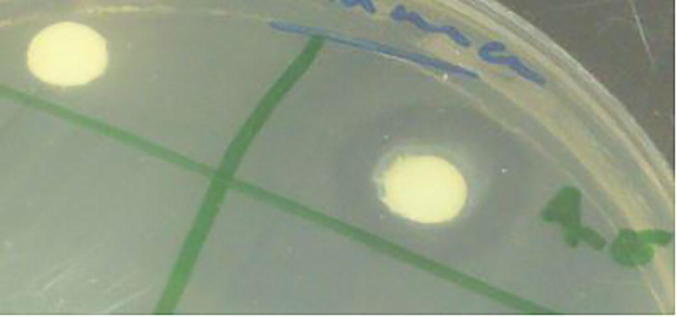
Antimicrobial activity of biosynthesized star shaped Nano silver against Pseudomonas aeruginosa.
4. Conclusion
In the current study we isolated ess_amA6 strain of Saccharopolyspora hirsuta from an insect Tapinoma simrothi. It was used to synthesize Nano silver which showed good more effectively antimicrobial characteristics.
Acknowledgments
Acknowledgement
The authors thank the Deanship of Scientific Research at King Saud University for funding this work through research group RG-1439-025.
Conflict of interest
No conflict of interest was declared.
Footnotes
Peer review under responsibility of King Saud University.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M.J., Freeman R.F., Hopwood D.A. Physical and genetical characterisation of a second sex factor, SCP2, for Streptomyces coelicolor A3 (2) Mol. Gen. Genet. 1977;154(2):155–166. [Google Scholar]
- Cochrane V. Physiology of actinomycetes. Ann. Rev. Microbiol. 1961;15(1):1–24. [Google Scholar]
- Ebrahiminezhad A., Bagheri M., Taghizadeh S.M., Berenjian A., Ghasemi Y. Biomimetic synthesis of silver nanoparticles using microalgal secretory carbohydrates as a novel anticancer and antimicrobial. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016;7(1):015018. [Google Scholar]
- Gorajana A., Venkatesan M., Vinjamuri S., Kurada B.V., Peela S., Jangam P., Poluri E., Zeeck A. Resistoflavine, cytotoxic compound from a marine actinomycete, Streptomyces chibaensis AUBN 1/7. Microbiol. Res. 2007;162(4):322–327. doi: 10.1016/j.micres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Kalimuthu K., Babu R.S., Venkataraman D., Bilal M., Gurunathan S. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloid Surf. B. 2008;65(1):150–153. doi: 10.1016/j.colsurfb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Kalishwaralal K., Deepak V., Ramkumarpandian S., Nellaiah H., Sangiliyandi G. Extracellular biosynthesis of silver nanoparticles by the culture supernatant of Bacillus licheniformis. Mat. Lett. 2008;62(29):4411–4413. [Google Scholar]
- Klepzig K.D., Adams A., Handelsman J., Raffa K. Symbioses: a key driver of insect physiological processes, ecological interactions, evolutionary diversification, and impacts on humans. Environ. Entomol. 2009;38(1):67–77. doi: 10.1603/022.038.0109. [DOI] [PubMed] [Google Scholar]
- Kumar P.S., Balachandran C., Duraipandiyan V., Ramasamy D., Ignacimuthu S., Al-Dhabi N.A. Extracellular biosynthesis of silver nanoparticle using Streptomyces sp. 09 PBT 005 and its antibacterial and cytotoxic properties. Appl. Nanosci. 2015;5(2):169–180. [Google Scholar]
- Li G., He D., Qian Y., Guan B., Gao S., Cui Y., Yokoyama K., Wang L. Fungus-mediated green synthesis of silver nanoparticles using Aspergillus terreus. Int. J. mol. Sci. 2011;13(1):466–476. doi: 10.3390/ijms13010466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann G.M., Brodsky B.C. Taxonomy of gentamicin-producing Micromonospora. Antimicrob. Agents Chemother. 1964;1963:116–124. [PubMed] [Google Scholar]
- Mueller U.G., Dash D., Rabeling C., Rodrigues A. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution. 2008;62(11):2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- Prabhu S., Poulose E.K. Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int. Nano Lett. 2012;2(1):32. [Google Scholar]
- Pridham T., Gottlieb D. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. J. Bacteriol. 1948;56(1):107. doi: 10.1128/jb.56.1.107-114.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridham T.G. Color and streptomycetes report of an international workshop on determination of color of streptomycetes. Appl. Microbiol. 1965;13(1):43–61. doi: 10.1128/am.13.1.43-61.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhasivam S., Shanmugam P., Yun K. Biosynthesis of silver nanoparticles by Streptomyces hygroscopicus and antimicrobial activity against medically important pathogenic microorganisms. Colloid Surface B. 2010;81(1):358–362. doi: 10.1016/j.colsurfb.2010.07.036. [DOI] [PubMed] [Google Scholar]
- Service R.F. Atom-scale research gets real. Science. 2000:1524–1531. [Google Scholar]
- Shirling E.T., Gottlieb D. Methods for characterization of Streptomyces species1. Int. J. Syst. Evol. Microbiol. 1966;16(3):313–340. [Google Scholar]
- Srinivasan M., Laxman R., Deshpande M. Physiology and nutritional aspects of actinomycetes: an overview. World J. Microbiol. Biotechnol. 1991;7(2):171–184. doi: 10.1007/BF00328987. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennila K., Chitra L., Balagurunathan R., Palvannan T. Comparison of biological activities of selenium and silver nanoparticles attached with bioactive phytoconstituents: green synthesized using Spermacoce hispida extract. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018;9(1):015005. [Google Scholar]