Summary
Antigen presentation to T cells in major histocompatibility complex class II (MHC class II) requires the conversion of early endo/phagosomes into lysosomes by a process called maturation. Maturation is driven by the phosphoinositide kinase PIKfyve. Blocking PIKfyve activity by small molecule inhibitors caused a delay in the conversion of phagosomes into lysosomes and in phagosomal acidification, whereas production of reactive oxygen species (ROS) increased. Elevated ROS resulted in reduced activity of cathepsin S and B, but not X, causing a proteolytic defect of MHC class II chaperone invariant chain Ii processing. We developed a novel universal MHC class II presentation assay based on a bio-orthogonal “clickable” antigen and showed that MHC class II presentation was disrupted by the inhibition of PIKfyve, which in turn resulted in reduced activation of CD4+ T cells. Our results demonstrate a key role of PIKfyve in the processing and presentation of antigens, which should be taken into consideration when targeting PIKfyve in autoimmune disease and cancer.
Subject Areas: Molecular Mechanism of Gene Regulation, Immunology, Immune Response
Graphical Abstract
Highlights
-
•
PIKfyve converts PI(3)P into PI(3,5)P2 on the surface of endosomes and phagosomes
-
•
PIKfyve of DCs modulates phagosomal maturation, acidification, and ROS production
-
•
PIKfyve defects disrupt cathepsin S trafficking and activity
-
•
Inactive cathepin S slows Ii processing for MHC class II trafficking and antigen loading
Molecular Mechanism of Gene Regulation; Immunology; Immune Response
Introduction
Endocytosis and phagocytosis of microbial pathogens, virus-infected cells, and cancer cells is essential for their immune clearance by phagocytes of the immune system (Fairn and Grinstein, 2012, Neefjes et al., 2017). In addition, dendritic cells present peptides derived from ingested antigens on major histocompatibility complex (MHC) classes I and II to T lymphocytes and are thereby responsible for the initiation of adaptive immune responses (Banchereau and Steinman, 1998). There are various types of endocytosis and phagocytosis, which differ in the receptors engaged, the role of the cytoskeleton, cage proteins, and lipid composition (Fairn and Grinstein, 2012, Levin et al., 2015, Nunes et al., 2013). Following uptake, endosomes and phagosomes undergo a transition from early to late endo/phagosomes and eventually to lysosomes. This process is mediated by membrane remodeling called maturation, where endosomes and phagosomes fuse with other endosomes and lysosomes and recruit cytosolic proteins to their membranes, such as Rab-GTPases and tethering factors (Egami, 2016, Fairn and Grinstein, 2012, Flannagan et al., 2012, Naufer et al., 2018, Neefjes et al., 2017, Uribe-Querol and Rosales, 2017, Vieira et al., 2003). Early endosomes and phagosomes are only mildly acidic or even basic (Flannagan et al., 2009, Mantegazza et al., 2008, Savina et al., 2006), whereas lysosomes and phagolysosomes are very acidic due to activation of vacuolar proton pumps (pH < 5; Trombetta et al., 2003) and have increased presence and activity of metabolic enzymes for the degradation of ingested material (Kinchen and Ravichandran, 2008).
The MHC class II loading of proteolytically derived peptides occurs within specialized lysosome-like compartments called MHC class II compartments (MIIC) (Amigorena et al., 1994, Calafat et al., 1994, Castellino and Germain, 1995, Sadegh-Nasseri, 2016, Tulp et al., 1994, West et al., 1994). Within these acidic luminal compartments, the non-polymorphic chain of MHC class II Ii is proteolytically activated by its cleavage, reducing it to the 24-amino acid class II-associated Ii peptide (CLIP) (Jasanoff et al., 1999). Ii is a chaperone carrying sorting signals on its cytoplasmic tail and interacting with MHC class II thereby controlling its trafficking from the endoplasmic reticulum to the trans-Golgi network (TGN), endosomal compartments, and eventually to the cell surface (Jones et al., 1978, Teyton et al., 1990). Ii prevents premature antigen loading into the cleft of MHC class II, whereas CLIP can be exchanged in MIIC for an antigen peptide by the chaperone human leukocyte antigen (HLA)-DM. The protease cathepsin S (CatS) plays an essential role for Ii processing to CLIP and for the proteolytic processing of ingested antigen (Honey and Rudensky, 2003, Riese et al., 1996, Villadangos and Ploegh, 2000), and only three cathepsins (CatB, CatH, and CatS) suffice for generating essential immunodominant epitopes in vitro (Kim et al., 2014a).
A central hallmark of endo/phagosomal maturation is the progressive change in the content of phosphoinositide lipids on the endosomal and phagosomal membrane (Levin et al., 2015, Shisheva, 2001). These phosphoinositides are important for endo/phagosomal maturation, as they are major determinants of organellar identity and form anchor points for numerous proteins involved in membrane trafficking or cytoskeletal tethering (Baranov et al., 2016, Baranov et al., 2017, Levin et al., 2015). Within minutes after uptake, phosphatidylinositol 4,5-bisphosphate, which is the main phosphoinositide species at the plasma membrane, is converted into phosphatidylinositol 3-phosphate (PI(3)P) by various kinases and phosphatases. PI(3)P is the main phosphoinositide species at early endosomes and phagosomes and resides there for 5–10 min (Ellson et al., 2001a, Fratti et al., 2001, Gillooly et al., 2000, Vieira et al., 2003) before the acidification of these organelles (Naufer et al., 2018). The maturation of early into late endo/phagosomes and lysosomes requires the conversion of PI(3)P into phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) (Ho et al., 2012, Sbrissa et al., 2002). PI(3,5)P2 is a low-abundance phosphoinositide amounting to ∼0.04%–0.1% of the total phosphoinositide pool (Ho et al., 2012, McCartney et al., 2014). Phosphoinositide 5-kinase (PIKfyve; yeast ortholog Fab1p) is the sole enzyme capable of producing PI(3,5)P2 from PI(3)P, which it targets via its FYVE domain (Ho et al., 2012, Sbrissa et al., 2002, Shisheva et al., 2015). Blocking of PIKfyve activity delays phagosomal maturation, leading to the buildup of cellular PI(3)P and a drastic reduction of cellular levels of PI(3,5)P2 (Hazeki et al., 2012, Kim et al., 2014b). Given the late endosome/lysosome-like nature of MIIC, a role of PI(3,5)P2 and PIKfyve in MHC class II presentation can be expected, but has not been shown.
PIKfyve can be inhibited by two small molecule inhibitors: apilimod (STA-5,326) (Cai et al., 2013) and YM201636 (Hazeki et al., 2012, Jefferies et al., 2008). Apilimod was originally developed to treat inflammatory diseases (Burakoff et al., 2006, Krausz et al., 2012, Sands et al., 2010, Wada et al., 2007, Wada et al., 2012) and was later found to specifically target and inhibit PIKfyve (Cai et al., 2013). YM201636 is a specific antagonist of PIKfyve (Jefferies et al., 2008), and although it has not been tested in clinical trials, it is commonly used to inhibit PIKfyve (Compton et al., 2016, Gomez et al., 2018, Hazeki et al., 2012, Ikonomov et al., 2009, Kerr et al., 2010, Kim et al., 2014b, Krishna et al., 2016, Sbrissa et al., 2012). Pharmacological inhibition of PIKfyve or expression of a dominant negative form of PIKfyve (Ikonomov et al., 2001, Jefferies et al., 2008, Shisheva, 2001) causes the formation of enlarged (“foam-like”) vacuoles (Cai et al., 2013, Dong et al., 2010, Jefferies et al., 2008, Min et al., 2014), likely because of osmotic differences caused by the reduced activity of PI(3,5)P2-dependent cation channels such as the lysosomal cation channel TRPML1/MCOLN1 (Compton et al., 2016). These enlarged vacuoles are likely endosomes that cannot be reformed into lysosomes (Bissig et al., 2017), and their formation can be used as a readout of depletion of cellular PI(3,5)P2 (Kim et al., 2014b).
Given the role of late endosome/lysosome-related MIIC in the proteolytic processing of antigen and subsequent MHC class II loading, we hypothesized that interfering with PIKfyve activity would inhibit the degradation and presentation of antigen by dendritic cells. To address this hypothesis, we studied the effects of PIKfyve inhibition by apilimod and YM201636 on endo/phagosomal maturation, protease activity, and antigen presentation. As reported (Cai et al., 2013, Cai et al., 2014), PIKfyve inhibition blocked interleukin (IL)-12 secretion in monocyte-derived dendritic cells. PIKfyve inhibition also affected the maturation of phagosomes, with impaired acidification and lower recruitment of the lysosomal proteins lysosome-associated membrane protein 1 (LAMP1) and mannose 6-phosphate receptor (M6PR) to phagosomes, whereas the early phagosomal markers PI(3)P and EEA1 were more abundant (Compton et al., 2016, Dayam et al., 2017, Dove et al., 2009, Gayle et al., 2017a, Gomez et al., 2018, Hazeki et al., 2012, Ikonomov et al., 2009, Kerr et al., 2010, Kim et al., 2014b, Krishna et al., 2016, Sbrissa et al., 2007, Sbrissa et al., 2012). Moreover, inhibition of PIKfyve selectively blocked the activity of cathepsin B and S, and this resulted in an increased presence of Ii-bound MHC class II within the cell, but not on the cell surface. PIKfyve inhibitors caused prolonged presence of the NADPH oxidase NOX2, which is present on early, but not late, endosomes and phagosomes (Dingjan et al., 2017a, Dingjan et al., 2017b). This resulted in increased reactive oxygen species (ROS) production, leading to lower activity of CatS. We show that the impaired processing of Ii and MHC class II trafficking defects upon blockage of endo/phagosomal maturation with PIKfyve inhibitors results in lower MHC class II antigen presentation. This was demonstrated with a novel assay based on a viral antigen carrying an unnatural amino acid amendable to bio-orthogonal labeling of the MHC-class-II-presented epitope. The combined effects of reduced proteolytic processing and reduced MHC class II presentation upon blocking PIKfyve result in an impaired activation of antigen-specific T cells. These results show a key role for PIKfyve in T cell activation, and this should be taken into consideration for the use of PIKfyve-targeting drugs in clinical trials.
Results
PIKfyve Inhibition Delays Phagosomal Maturation
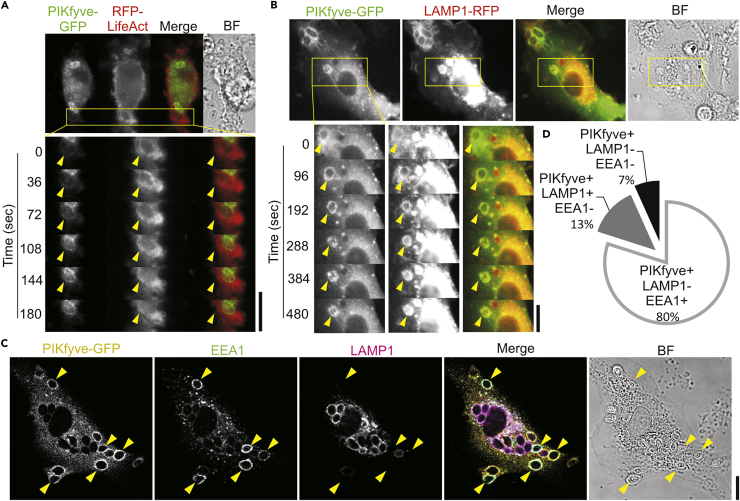
We started by determining whether PIKfyve would localize to antigen-containing compartments. To visualize the localization of PIKfyve in live cells, we transfected dendritic cells differentiated from human-blood-derived monocytes with plasmids coding for GFP-tagged PIKfyve (PIKfyve-GFP) (Figure 1A; Video S1). The actin cytoskeleton was visualized by simultaneous transfection with a plasmid coding for the F-actin-binding probe LifeAct fused to the red-shifted fluorescent protein mRFPruby (Riedl et al., 2008). Late endo/phagosomes and lysosomes were identified with LAMP1 fused to red fluorescent protein (RFP) (Figure 1B; Video S2) (Sherer et al., 2003). The dendritic cells were pulsed with the model pathogen zymosan, relatively large yeast-derived particles (∼4–5 μm diameter) that allow for the unequivocal assignment of phagosomal localization of candidate proteins by microscopy and result in an increased expression of PIKfyve (Figure S1A). We observed that PIKfyve-GFP was recruited within 2 min after the formation of the phagosomal F-actin cytoskeleton (Figure 1A), which we recently showed is formed at the nascent cup of emerging phagosomes by tethering to phosphatidylinositol 3,4-bisphosphate (PI(3,4)P2) and is subsequently removed after closure of the phagocytic cup (Baranov et al., 2016). At later time points after uptake (>7 min), the signal of PIKfyve-GFP gradually decreased at the phagosomes, whereas the signal of LAMP1-RFP increased (Figure 1B). We also immunostained zymosan-pulsed dendritic cells expressing PIKfyve-GFP for endogenous LAMP1 as well as for the early endosomal marker EEA1 (Figure 1C). EEA1 is known to bind to early endo/phagosomes by interactions of its FYVE domain with PI(3)P (Ho et al., 2012, Sbrissa et al., 2002), which is the substrate for PIKfyve. PIKfyve-GFP was partially co-residing with EEA1 on phagosomes, whereas overlap with LAMP1 was lower (Figure 1D). Together, these results show that PIKfyve is recruited to phagosomes within minutes after their formation and remains present at early phagosomes before their conversion into late phagosomes.
Figure 1.
PIKfyve Is Present Early during Phagosomal Maturation
(A) Live cell imaging of a human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with the F-actin-binding probe RFP-LifeAct (red) and pulsed with IgG-opsonized zymosan particles. The insets show snapshots at the indicated time points. Yellow arrowheads, time series of phagosome. See also Video S1. BF, brightfield.
(B) Same as (A), but now with PIKfyve-GFP (green) and LAMP1-RFP (red). See also Video S2.
(C) Confocal micrograph of representative zymosan-pulsed (1 hr) dendritic cell overexpressing PIKfyve-GFP (yellow in merge) with immunostaining for EEA1 (green) and LAMP1 (magenta). Yellow arrowheads, PIKfyve-GFP and EEA1 double-positive phagosomes.
(D) Quantification of (C) (∼150 phagosomes per donor; three donors).
Scale bars, 10 μm. See also Figure S1.
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with the F-actin-binding probe LifeAct-RFP (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with LAMP1-RFP (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.
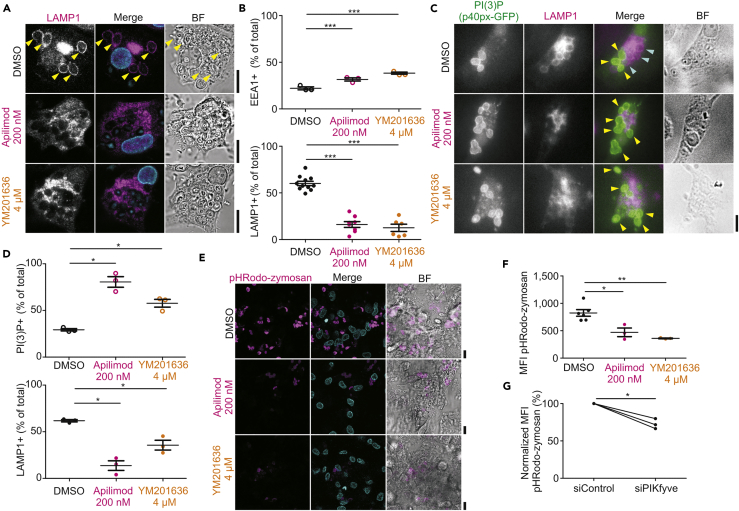
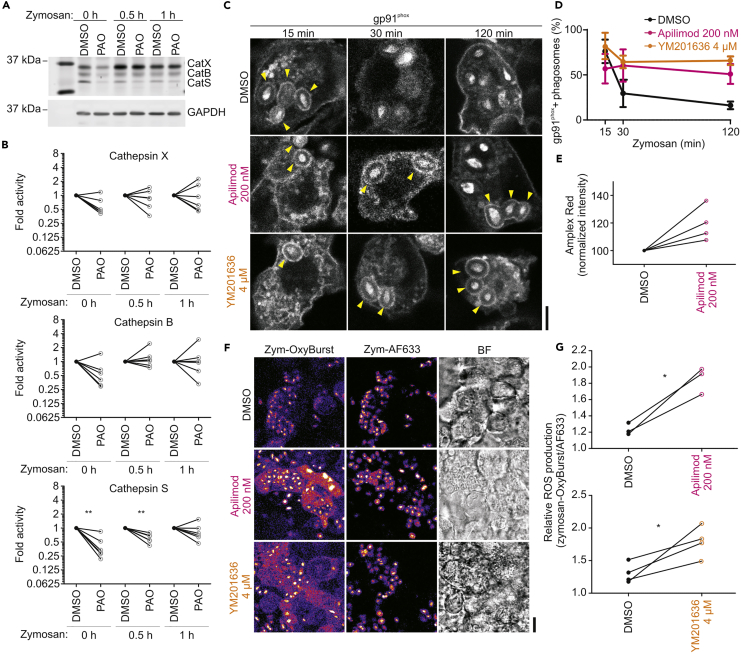
Next, we validated the reported inhibition of endosome and phagosome maturation by blockage of PIKfyve activity with apilimod and YM201636 (Compton et al., 2016, Dayam et al., 2017, Dove et al., 2009, Gayle et al., 2017a, Gomez et al., 2018, Hazeki et al., 2012, Ikonomov et al., 2009, Kerr et al., 2010, Kim et al., 2014b, Krishna et al., 2016, Sbrissa et al., 2007, Sbrissa et al., 2012). We selected concentrations of 200 nM apilimod (Cai et al., 2013, Compton et al., 2016, Terajima et al., 2016, Wada et al., 2012, Wong et al., 2017) and 4 μM YM201636 (Currinn et al., 2016, Ikonomov et al., 2009), which did not significantly affect the viability of the dendritic cells, although metabolic activity was somewhat affected by apilimod (by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay; Baranov et al., 2016, Baranov et al., 2017, Dingjan et al., 2016) and YM201636 treatment non-significantly increased the fraction of early apoptotic cells (by annexin V staining; Figures S1B–S1D). These concentrations were used in previous studies and resulted in inhibition of IL-12 production by the dendritic cells (Cai et al., 2013, Wada et al., 2012) (Figure S1E). Treatment of dendritic cells with apilimod or YM201636 resulted in massive cellular vacuolization (Figure S1F), a well-known effect of PIKfyve inhibition (Cai et al., 2013, Dayam et al., 2017, Krishna et al., 2016). This vacuolization was also observed when a dominant negative form of PIKfyve was expressed (PIKfyve-GFP with K1831E [Cai et al., 2013, Ikonomov et al., 2001, Sbrissa et al., 2000]) (Figure S1G) and with small interfering RNA (siRNA) knockdown (Figure S1H). Moreover, treatment of dendritic cells by apilimod or YM201636 compromised phagocytosis of the zymosan particles as well as endocytosis of fluorescently labeled albumin by almost half (Figures S1I and S1J). Previously reported PIKfyve perturbations led to inhibition of IgG-latex bead uptake by neutrophils (Dayam et al., 2017) and mouse RAW 264.7 macrophages (Kim et al., 2014b) and to lower Escherichia coli uptake by RAW 264.7 macrophages (Wong et al., 2017). To account for this reduced antigen uptake, we present our results relative to the total number of phagosomes per cell. Both apilimod and YM201636 treatment resulted in blockage of phagosome maturation, as apparent from a reduced fraction of LAMP1 and increased fractions of EEA1-positive (Figures 2A and 2B) and PI(3)P-positive phagosomes (Figures 2C and 2D). In addition, phagosomal recruitment of M6PR, another late endosomal marker that is involved in trafficking from the TGN to lysosomes, was also reduced upon apilimod and YM201636 treatment (Figures S1K and S1L). To exclude off-target effects of the drugs, we performed siRNA knockdown of PIKfyve. Even though we only reached an ∼50% knockdown efficiency of PIKfyve by qPCR (Figure S1M), this led to a reduced recruitment of LAMP1 to phagosomes (Figure S1N) similar to the small molecule inhibitors. Finally, acidification of the phagosomal lumen was reduced by apilimod or YM201636 treatment, as shown by experiments with zymosan conjugated to pHRodo, a pH-sensitive dye with increased fluorescence at low pH (Figures 2E and 2F). Similar blockage of acidification was observed with PIKfyve siRNA knockdown (Figure 2G).
Figure 2.
PIKfyve Controls Phagosomal Maturation
(A) Confocal images of representative dendritic cells treated with apilimod (200 nM) or YM201636 (4 μM) for 3 hr and pulsed with zymosan 1 hr before fixation. DMSO, vehicle control. Cells were immunolabeled for LAMP1 (magenta in merge). Blue, DAPI staining. BF, brightfield.
(B) Quantification of (A) normalized to the total number of phagosomes per cell (∼150 phagosomes per condition per donor; mean ± SEM of three donors for EEA1 and six donors for LAMP1).
(C) Epifluorescence microscopy of representative dendritic cells expressing GFP-tagged phosphoinositide-probe for PI(3)P based on PX domain of NCF4 (PI(3)P; green in merge) and immunolabeled for LAMP1 (magenta). The cells were treated as in (A). Yellow arrowheads, PI(3)P-positive phagosomes; cyan arrowheads, LAMP1-positive phagosomes.
(D) Quantification of (C) normalized to the total number of phagosomes per cell (∼116 phagosomes per condition per donor; mean ± SEM of three donors).
(E) Confocal imaging of dendritic cells treated with DMSO, apilimod, or YM201636 for 3 hr before addition of pHRodo-labeled zymosan 1 hr before live imaging. The color intensity of pHRodo (magenta) scales with acidic pH. Cyan in merge, Hoechst.
(F) Quantification of (E); MFI, mean fluorescence intensity (mean ± SEM for three donors, ∼1,000 phagosomes per condition per donor).
(G) Same as (F), but now with siPIKfyve (∼400 phagosomes per condition per donor; see also Figure S1M for knockdown levels).
Scale bars: 10 μm. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. See also Figure S1.
Cathepsin B and S Activities and Proteolytic Activation of MHC Class II Are Reduced upon PIKfyve Inhibition
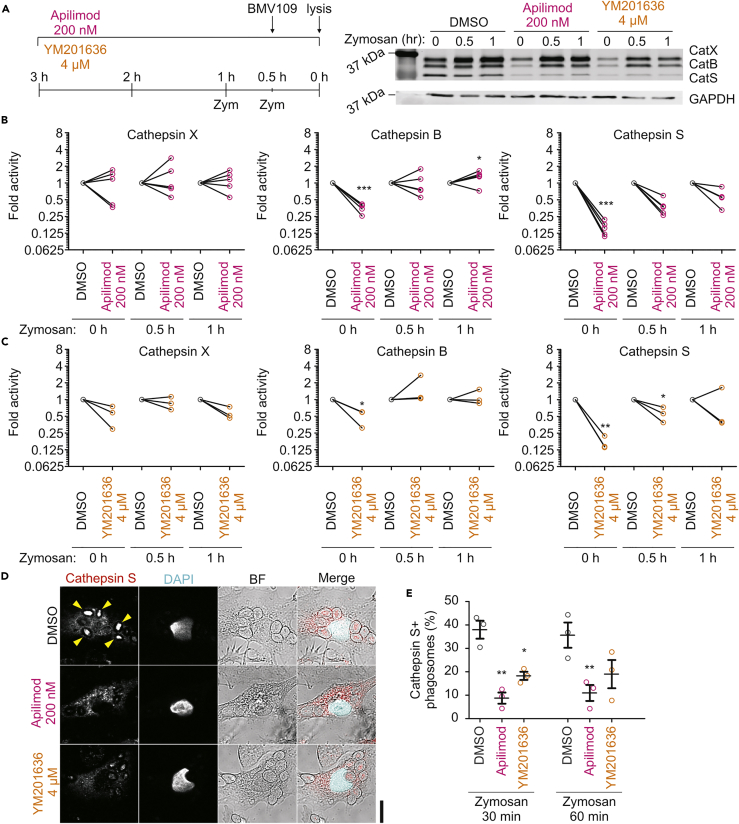
Phagosomal maturation and acidification are necessary to trigger the activity of hydrolases involved in the degradation of ingested antigen, and thereby for deriving antigenic peptides for presentation in MHC class II (Blum et al., 2013, ten Broeke et al., 2013). A major class of proteases are cysteine cathepsins, named after the catalytic cysteine located within their active sites (Verma et al., 2016) and consisting of 11 family members present in endosomes, phagosomes, and lysosomes (Turk et al., 2012). CatS is predominantly expressed by professional antigen-presenting cells, such as dendritic cells and B cells (Blum et al., 2013) and, unlike other cathepsins, is active not only in acidic but also in neutral and slightly alkaline environments (Jancic et al., 2007, Kirschke et al., 1989). As we observed that inhibition of PIKfyve by apilimod or YM201636 impaired lysosomal acidification and affected trafficking to late endosomes/lysosomes, we hypothesized that this would alter the activity of cathepsins. We applied the activity-based probe BMV109 to living cells before lysis (Verdoes et al., 2013). The BMV109 probe results in covalent attachment of a fluorophore to the catalytic sites of active cysteine cathepsins (Verdoes et al., 2013), thereby allowing for the quantitative assessment of their activities by in-gel fluorescence (Figure S2A). We compared unstimulated and zymosan-pulsed dendritic cells treated with apilimod or YM201636 (Figure 3A). Analysis of protein levels by western blot showed that PIKfyve inhibitors did not cause changes of total cathepsin levels (Figure S2A). For unstimulated cells, we observed reduced cathepsin B and S activities by treatment with apilimod or YM201636, whereas the activity of cathepsin X was not altered (Figures 3B, 3C, and S2B). However, stimulation of the cells with zymosan diminished these effects, and significant differences in cathepsin activity could no longer be detected after 1-hr incubation. Immunolabeling of endogenous CatS in phagocytic dendritic cells was visible at the core of the zymosan particles (Figure 3D). Microscopy experiments with the activity-based probe BMV109 confirmed that this is the site of cathepsin activity (Figures S2C and S2D). BMV109 signal became apparent at phagosomes 30–60 min after zymosan uptake and was present in LAMP1-positive phagosomes (Figure S2C). Immunofluorescence microscopy revealed that the majority of CatS-positive phagosomes are also LAMP1 positive (Figure S2E). Treatment with apilimod or YM201636 reduced the signals of both CatS immunostaining (Figures 3D–3E) and BMV109 (Figures S2D and S2F) at the phagosomes, indicating that trafficking of CatS to phagosomes is reduced by PIKfyve inhibitors. To determine whether the reduced CatS activity was solely attributable to its reduced recruitment to phagosomes upon PIKfyve inhibition, we performed experiments with BMV109 in lysates of dendritic cells (Figure S3A). For some conditions, apilimod and YM201636 still reduced the signal of BMV109 for CatS, whereas the signals for cathepsins B and X were unaltered (Figures S3B and S3C). We conclude that the reduced CatS activity upon PIKfyve inhibition is not solely due to altered trafficking, because CatS activity is also reduced in cell lysates.
Figure 3.
Pharmacological Inhibition of PIKfyve Blocks Cathepsin S Activity and Trafficking
(A) SDS-PAGE with in-gel fluorescence of the BMV109-Cy5 activity-based probe labeling cathepsin X (CatX; top band), cathepsin B (CatB; middle band), and cathepsin S (CatS; lower band) in resting dendritic cells (0 hr) or after stimulation with zymosan for 0.5 or 1 hr. Cells were treated with DMSO (vehicle control), apilimod, or YM201636 for 3 hr before lysis according to the left-hand scheme. GAPDH, loading control by western blot. Only part of the SDS PAGE/ polyvinylidene fluoride blot is shown; the rest of the image carried no information.
(B and C) Quantification of (A) (individual donors shown; normalized to DMSO controls).
(D) Confocal images of representative dendritic cells treated with PIKfyve inhibitors and stimulated with zymosan as in (A) before fixation and staining for CatS (red in merge). Yellow arrowheads, CatS-positive phagosomes. Scale bar, 10 μm.
(E) Quantification of (D) for cells stimulated with zymosan for 0.5 or 1 hr (>133 phagosomes per condition per donor; mean ± SEM of three donors; individual donors shown; normalized to the total number of phagosomes per cell).
∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. See also Figures S2 and S3.
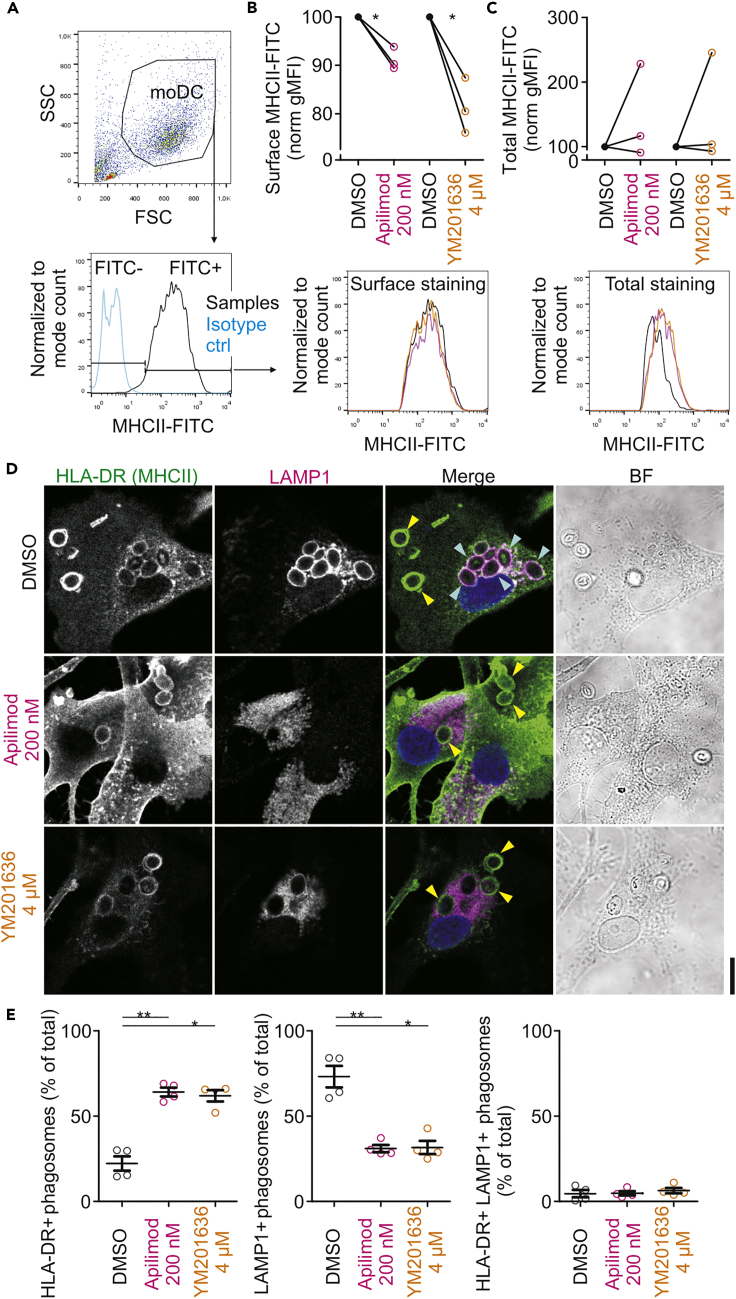
CatS is expressed at high levels by dendritic cells and has a strong immunological relevance, because it controls the generation of CLIP (Honey and Rudensky, 2003, Riese et al., 1996, Villadangos and Ploegh, 2000). Within endolysosomes, CLIP is derived from the invariant chain Ii via the intermediate fragments Iip23 and Iip10 (Amigorena et al., 1995, Neefjes and Ploegh, 1992, Villadangos et al., 2000). The final processing of Iip10 into CLIP is mediated by CatS. The inhibition or loss of CatS in mice (Nakagawa et al., 1999, Shi et al., 1999) causes accumulation of Iip10-bound MHC class II in endo/lysosomal compartments, impeding MHC class II trafficking to the cell surface (Brachet et al., 1997, Riese and Chapman, 2000). As inhibition of PIKfyve results in reduced activity of CatS, we hypothesized that apilimod and YM201636 would cause a defect of Ii processing and MHC class II trafficking. To address this hypothesis, we first assessed the effects of apilimod or YM201636 treatment on plasma membrane localization of MHC class II. Flow cytometry experiments with an antibody recognizing the extracellular domain of MHC class II revealed that apilimod or YM201636 treatment caused a small (∼10%–20%) but consistent reduction of the presence of MHC class II at the plasma membrane (Figures 4A and 4B). In contrast, total cellular levels of MHC class II, measured by immunolabeling in the presence of a detergent, were unaltered or even (inconsistently) increased (Figure 4C). Similar observations were made with dendritic cells derived from mouse bone marrow (Figures S4A–S4C). We also performed flow cytometry experiments with an antibody recognizing Ii or CLIP-bound to αβMHC class II (clone CerCLIP.1; Denzin et al., 1994), revealing that the presence of this inactive form of MHC class II was also reduced at the plasma membrane, whereas the total cellular levels were increased (Figures S4D–S4F). The defect in processing of CLIP upon PIKfyve inhibition was also observed by western blot with mouse bone-marrow-derived dendritic cells using an antibody recognizing the Iip10 fragment (Figures S4G and S4H). Here, we used the CatS inhibitor LHVS (morpholinurea-leucine-homophenylalanin-vinylsulfone-phenyl) as a positive control (Palmer et al., 1995, Riese et al., 1998). A direct inhibition of CatS with LHVS also had a similar effect on MHC class II trafficking as PIKfyve inhibition with apilimod or YM201636 (Figures S4I and S4J). Microscopy imaging of phagocytic cells overexpressing fluorescently tagged MHC class II (Zwart et al., 2005) showed that MHC class II is recruited to phagosomes after zymosan uptake (Video S3). Immunofluorescence microscopy showed that treatment with PIKfyve inhibitors showed increased accumulation of endogenous MHC class II on the phagosomes (Figures 4D and 4E), suggesting a trafficking defect. Together, these results show that PIKfyve supports CatS activity and thereby regulates the proteolytic activation of MHC class II.
Figure 4.
Pharmacological Inhibition of PIKfyve Impairs Proteolytic Activation and Trafficking of MHC Class II
(A) Gating strategy for flow cytometry of human-monocyte-derived dendritic cells immunolabeled for MHC class II (MHCII-FITC; HLA-DR, DQ). Light blue in histogram, isotype control; SSC, side scatter; FSC, forward scatter; FITC, fluorescein isothiocyanate.
(B) Representative histograms and quantification of surface MHC class II with flow cytometry experiments of dendritic cells treated for 3 hr with apilimod (200 nM) or YM201636 (4 μM) (geometric mean fluorescence intensities [gMFI]; individual donors shown; normalized to DMSO control).
(C) Same as (B), but now for total MHC class II with detergent permeabilization.
(D) Confocal images of representative dendritic cells treated with apilimod (200 nM) or YM201636 (4 μM) for 3 hr and pulsed with zymosan 1 hr before fixation. DMSO, vehicle control. Cells were immunolabeled for HLA-DR (green in merge) and LAMP1 (magenta in merge). Blue, DAPI staining. BF, brightfield. Scale bar, 10 μm. Yellow arrowheads, HLA-DR-positive phagosomes; cyan arrowheads, LAMP1-positive phagosomes.
(E) Quantification of percentages of HLA-DR-positive and LAMP1-positive phagosomes from (D) normalized to the total number of phagosomes per cell (∼280 phagosomes per condition per donor; mean ± SEM of four donors).
∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. See also Figures S4–S6.
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with MHC-II-mCherry (HLA-DRA-IRES-HLA-DRB-mCherry) (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.
The Inactivation of Cathepsin S upon PIKfyve Inhibition Is due to Oxidative Modifications
In dendritic cells, NOX2 produces large amounts of ROS within the lumen of endosomes and phagosomes, and this ROS production is sustained for hours after uptake (Dingjan et al., 2016, Dingjan et al., 2017a, Dingjan et al., 2017b, Jancic et al., 2007, Joffre et al., 2012, Kotsias et al., 2013, Mantegazza et al., 2008, Nunes et al., 2013, Savina et al., 2006, Vulcano et al., 2004). ROS alter the processing of the ingested antigen for MHC classes I and II presentation (Hari et al., 2015, Mantegazza et al., 2008, Rybicka et al., 2012), with one of the mechanisms being the oxidative modification of a cysteine located within the catalytic site of CatS (Allan et al., 2014, Hari et al., 2015, Jancic et al., 2007, Kanai et al., 2001, Rybicka et al., 2012). In line with this, treatment of cells with the membrane-permeable organometallic compound phenylarsine oxide, which modifies cysteines, resulted in an inhibition of CatS, but not or less of cathepsins B and X (Figures 5A and 5B), indicating that the activity of CatS is particularly sensitive to cysteine modifications. We recently showed that NOX2 is already present at nascent phagosomes, because the catalytic subunit gp91phox of NOX2 is co-invaginated from the plasma membrane together with the antigen during phagocytosis (Dingjan et al., 2017a). Moreover, NOX2 requires the presence of PI(3,4)P2 and/or PI(3)P for its activity and is active on early phagosomes (Anderson et al., 2010, Ellson et al., 2001b, Groemping and Rittinger, 2005, Kanai et al., 2001). We also showed that even though NOX2 resides in lysosomal compartments in naive dendritic cells, it is gradually removed from phagosomes during their conversion into LAMP1-positive phagolysosomes (Dingjan et al., 2017a, Dingjan et al., 2017b). As our data show that blockage of PIKfyve led to an accumulation of PI(3)P on phagosomes (Figures 2C and 2D) and delayed the conversion of early into late endo/phagosomes, we hypothesized that the reduced CatS activity was caused by an increased ROS production resulting from a prolonged presence and increased activity of NOX2. The presence of phagosomal NOX2 upon PIKfyve inhibition by apilimod and YM201636 was assessed using immunofluorescence staining for its main catalytic subunit gp91phox (Dingjan et al., 2017a, Dingjan et al., 2017b). Treatment with apilimod and YM201636 resulted in a prolonged presence of gp91phox at zymosan-containing phagosomes compared with the DMSO control (Figures 5C and 5D). We also measured ROS production in cells pulsed for 1 hr with zymosan using two different assays. First, apilimod treatment resulted in an increased extracellular presence of H2O2 as measured with the Amplex Red assay (Dingjan et al., 2016) (Figure 5E). Second, intra-phagosomal ROS production was measured with zymosan particles conjugated to the ROS-sensitive probe OxyBURST (Dingjan et al., 2017a), and this also showed an increased ROS production upon PIKfyve inhibition with apilimod or YM201636 (Figures 5F and 5G). Our data suggest that CatS is more sensitive to oxidative modifications than other cathepsins, and the increased ROS production upon PIKfyve inhibition can thereby explain the reduced activity of CatS (but not B and X) in cell lysates (Figure S3). Together, we conclude that PIKfyve inhibition results in an increased production of ROS within endo/phagosomes and that this leads to a reduced activity of CatS, which is more prone to oxidative modifications than cathepsins B and X.
Figure 5.
Pharmacological Inhibition of PIKfyve Promotes NOX2-Mediated ROS Production
(A) SDS-PAGE with in-gel fluorescence for the cathepsin-activity-based probe BMV109-Cy5 for dendritic cells pulsed with zymosan for 0.5 or 1 hr and untreated (0 hr). PAO, cysteine-modifying agent phenylarsine oxide; DMSO, vehicle control; GAPDH, loading control by western blot.
(B) Quantification of (A) (individual donors shown; normalized to DMSO controls).
(C) Confocal images of representative dendritic cells pulsed with zymosan for 15, 30, or 120 min in the presence of apilimod (200 nM) or YM201636 (4 μM) for 3 hr and immunostained for gp91phox. Yellow arrowheads, gp91phox-positive phagosomes. Scale bar, 5 μm.
(D) Quantification of (C); Gp91phox-positive phagosomes were counted and normalized to the total number of phagosomes per cell (mean ± SEM for three donors, ∼300 phagosomes per condition).
(E) Extracellular H2O2 measurements with Amplex Red assay for dendritic cells treated for 3 hr with apilimod and 1 hr with zymosan.
(F and G) Phagosomal ROS production determined with zymosan labeled to both OxyBURST and Alexa Fluor 633 (AF633) and confocal live imaging. Cells were treated for 3 hr with apilimod (F) or YM201636 (G) and incubated for 1 hr with zymosan-OxyBURST/AF633. Signals from OxyBURST signal were normalized to that of AF633 (∼3,000 phagosomes per condition; individual donors shown). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
PIKfyve Inhibition Impairs MHC Class II Presentation
In the final set of experiments, we tested whether inhibition of PIKfyve would lead to impaired MHC class II antigen presentation. We first performed a classical T cell activation assay with T cells isolated from transgenic OT-II mice (Barnden et al., 1998). CD4+ T cells from these mice carry a T cell receptor specifically recognizing ovalbumin residues 323–339 (OVA323-339) presented on H-2IAb MHC class II. Apilimod treatment resulted in an ∼25% reduction of production of interferon (IFN) γ by the OT-II T cells over time, whereas YM201636 even completely blocked IFNγ release (Figures S5A and S5B). However, the long incubation times with apilimod or YM201636 required for T cell activation, reduced the viabilities of both the dendritic cells and OT-II T cells by 20%–40% (Figures S5C and S5D). Moreover, as expected, surface presentation of MHC class II in those experiments was reduced by ∼10-30% upon PIKfyve inhibition (Figures S5E and S5F). Because of these effects, the impaired activation of T cells upon PIKfyve inhibition cannot be solely attributed to lower antigen presentation. Indeed, control experiments showed that both apilimod and especially YM201636 blocked T cell activation independent of antigen presentation, as these compounds resulted in reduced production of IFNγ and lower surface expression of the activation markers CD25 and CD69 (Figure S6). Thus, although we observed a reduction of T cell activation by inhibition of PIKfyve, this is probably mainly caused by direct effects of apilimod and YM201636 on the T cells and the T cell activation assay does not allow to discern direct effects on MHC class II presentation.
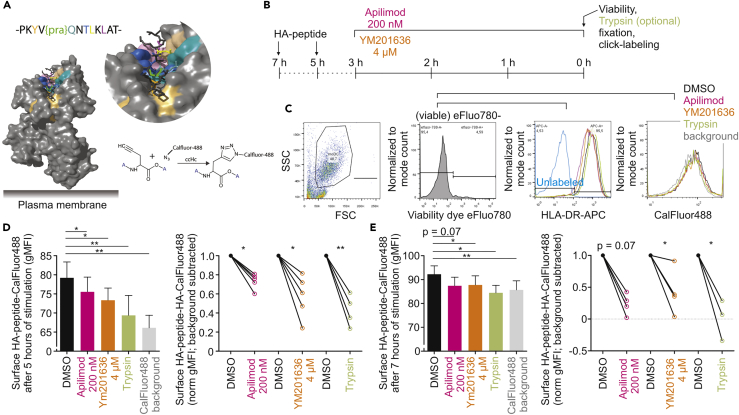
To overcome this problem, we developed a new method allowing for direct visualization of MHC class II presentation. This is based on a recently developed assay for visualization of MHC class I presentation (Pawlak et al., 2015, Pawlak et al., 2016), and employs synthetic peptides as antigen carrying a non-natural amino acid amendable to bio-orthogonal ligation with fluorescent dyes. Based on the crystal structure of a complex of human HLA-DR1 MHC class II with influenza A virus hemagglutinin (HA) (strain A/Aichi/2/1968 H3N2) peptide (322–334 amino acid positions; PKYVKQNTLKLAT; HA322-334) (Stern et al., 1994, Zavala-Ruiz et al., 2004), we synthesized a peptide consisting of the MHC class II epitope of influenza extended both N and C terminally with four natural amino acids (YGACPKYVKQNTLKLATGMRN; HA318-338) (Figure S7, related to Table S1). This extended peptide needs to be processed and loaded by HLA-DM in MIIC to be presented on MHC class II. The central lysine (K326) was converted into a propargylglycine. Modeling indicated that the alkyne moiety of propargylglycine is solvent accessible when this peptide is bound to MHC class II (Figure 6A). This alkyne moiety can be conjugated to CalFluor-488, which increases the fluorescence signal by two orders of magnitude, resulting in a low signal-to-noise ratio (Shieh et al., 2015). Moreover, the alkyne moiety is only three carbon atoms in size and does not (or less) perturb the molecular mechanisms of antigen processing and presentation, in contrast to large fluorophores, and survives the harsh environment in lysosomes (Bakkum et al., 2018). By incubating human dendritic cells with bio-orthogonal functionalized peptide HA318-338, we could show increased MHC class II presentation over time for at least up to 5-hr incubation (Figures 6B–6D and S8). Moreover, no increase in CalFluor-488 signal was observed with negative control experiments with a wild-type HA318-338 peptide (without propargylglycine) or the HA322-334 epitope, which cannot be processed for HLA-DM loading onto MHC class II, or by proteolytically cleaving surface-exposed MHC class II with trypsin. Compared with the 5-hr time point, longer incubation (18 hr) led to reduction of the CalFluor-488 signal, suggesting that the peptide was internalized and/or degraded by the cells (Figure S8). When apilimod or YM201636 was present during the last 3 hr of 5- or 7-hr incubation with the bio-orthogonally functionalized peptide HA318-338, the signals from CalFluor-488 were consistently reduced by 20%–80% (depending on the donor) (Figures 6C–6E), demonstrating that these drugs directly blocked presentation of the HA antigen. Thus using bio-orthogonal functionalized antigenic peptide, we could demonstrate that blockage of PIKfyve leads to impaired MHC class II antigen presentation.
Figure 6.
PIKfyve Inhibition Impairs MHC Class II Presentation
(A) Crystal structure of human HLA-DR1 (DRA, DRB1*0101; PDB: 1DLH) in complex with a virus HA (strain A/Aichi/2/1968 H3N2) peptide residues 322–334. The position of a clickable non-naturalized amino acid L-C-propargylglycine {pra} for bio-orthogonal labeling with CalFluor-488 is indicated (bright green); pockets for peptide binding at the HLA-DR1 cleft are highlighted in different colors. A scheme of the bio-orthogonal reaction is also shown.
(B) Time line of stimulation with HA peptide residues 318–338 for MHC class II presentation by human dendritic cells.
(C) Flow cytometry gating strategy for quantification of HLA-DR1 presented HA by labeling with CalFluor-488 as depicted in (A and B). Cell viability was assessed with fixable viability dye eFluo780. Surface levels of HLA-DR1 were assessed with HLA-DR-APC.
(D) Quantification of HA-CalFluor-488 signals from (C) after 5 hr of stimulation with HA-peptide (bar graphs). Gray, non-specific background from CalFluor labeling of dendritic cells that were not incubated with HA-peptide; green, trypsin-positive control for HA removal from the cell surface; line graphs, individual donors with background subtracted and normalized to DMSO control.
(E) The same as (D), but now for 7 hr of stimulation with HA peptide.
∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. See also Figures S7 and S8 and Table S1.
Discussion
In this study, we studied the role of PIKfyve in MHC class II antigen presentation by dendritic cells. Our data confirm the key role of PIKfyve in phagosome maturation (Compton et al., 2016, Dayam et al., 2017, Dove et al., 2009, Gayle et al., 2017a, Gomez et al., 2018, Hazeki et al., 2012, Ikonomov et al., 2009, Kerr et al., 2010, Kim et al., 2014b, Krishna et al., 2016, Sbrissa et al., 2007, Sbrissa et al., 2012) and treatment of dendritic cells with apilimod and YM201636 blocked recruitment of LAMP1, M6PR, and CatS to phagosomes. A still open question is via which mechanisms the reduced levels of cellular PI(3,5)P2 cause these trafficking defects. Endosomal maturation via the conversion of Rab5- to Rab7-positive membranes by the endosomal tethering complexes CORVET (class C core vacuole/endosome tethering) and HOPS (homotypic fusion and protein sorting) is well understood (Balderhaar and Ungermann, 2013). The role of PI(3,5)P2 and downstream effectors of PI(3,5)P2 in endosomal maturation is uncertain, and PI(3,5)P2 might govern trafficking events via several possible mechanisms. The best characterized downstream effector of PI(3,5)P2 is the cation channel TRPML1; its activity directly depends on PI(3,5)P2. However, it is also not known if and how TRPML1 affects endo/phagosomal maturation. As TRPML1 triggers the efflux of Ca2+ from lysosomes to the cytosol, one possibility is that it might be able to activate ALG-2, a dynein-interacting protein sensitive to Ca2+. Dyneins promote retrograde vesicular migration along the tubular tracks, and this might somehow facilitate endosome-lysosome fusion (Li et al., 2016). Alternatively or additionally, the increased Ca2+ efflux might directly promote intracellular fusion events via Ca2+-sensing proteins (Hay, 2007). Another downstream effector of PI(3,5)P2 is the V-ATPase, as yeast strains with mutations in the PIKfyve ortholog Fab1p showed impaired vacuolar acidification (Li et al., 2014), and this might explain the reduced acidification upon pharmacological inhibition or siRNA knockdown of PIKfyve.
Our results showed that the blockage of phagosome maturation by PIKfyve inhibition resulted in reduced activity of CatS and B, but not of cathepsin X. Our data suggest that the reduced activity of CatS upon apilimod or YM201636 treatment might be attributed to both a reduced trafficking to phagosomes and an increased ROS production by NOX2. Our microscopy data show that CatS trafficking to LAMP1-positive phagosome relies on PIKfyve activity. Moreover, we showed that NOX2-produced ROS affect cysteine cathepsins, including CatS, because the ROS can modify cysteines within their catalytic cores (Allan et al., 2014, Hari et al., 2015, Jancic et al., 2007, Rybicka et al., 2012). The increased ROS production caused by a prolonged presence of NOX2 at phagosomes upon apilimod or YM201636 treatment might even be amplified by higher phagosomal levels of PI(3)P, which might enhance NOX2 activity through NOX2 subunit p40phox binding to PI(3)P by its PX domain (Anderson et al., 2010, Ellson et al., 2001b, Groemping and Rittinger, 2005, Kanai et al., 2001). An open question is why addition of zymosan reduced the inactivation of cathepsin B and S. As we also observe this effect in our lysate controls, perhaps the zymosan particles sequester ROS species and thereby prevent the oxidative modification of CatS. This suggests that the NOX2-mediated reduction of CatS activity will mainly have physiological relevance in the absence of phagocytic cargo, and thereby might be involved in the maintenance of self-tolerance (Steinman and Nussenzweig, 2002), for instance, for the induction of Th2 responses by immature dendritic cells (Na et al., 2016). In any case, our data show that PIKfyve-mediated phagosomal maturation affects the activity of cysteine cathepsins via NOX-produced ROS, and this can modulate the epitope repertoire for MHC presentation (Balce et al., 2011, Rybicka et al., 2010, Rybicka et al., 2012). Because NOX2 mediates the disulfide reduction of protein antigens (Ewanchuk and Yates, 2018), the effects of NOX2-produced ROS on antigen processing will be even more pronounced. Only linear peptide/protein stretches are accessible to proteases for cleavage, hence all inter- and intra-disulfide bonds have to be reduced before antigen processing (Bogunovic et al., 2010), as has been also demonstrated for MHC class II with model antigens (Jensen, 1991). γ-Interferon-inducible lysosomal thiol reductase activity can be inhibited by ROS, and this may cause different antigen processing patterns leading to altered repertoires of peptides for presentation, because some of the proteins cannot be unfolded and are therefore not accessible to proteinases for degradation (Ewanchuk and Yates, 2018).
In addition to the proteolytic processing of ingested antigens, CatS mediates the activation of MHC class II in MIIC via the processing of Ii to CLIP and is thereby required for antigen presentation (Nakagawa et al., 1999, Riese and Chapman, 2000, Shi et al., 1999). Our data show that inhibition of PIKfyve by apilimod or YM201636 results in an accumulation of Iip10, the precursor of CLIP, and in reduced trafficking of MHC class II to the cell surface. This is in line with the finding that the inhibitor of PIKfyve AS2795440 caused a marked reduction of surface MHC class II expression on CD45R+ B cells in anti-IgM-stimulated whole blood from rats (Terajima et al., 2016). These findings can be explained by the decreased activity of CatS, as incomplete invariant chain degradation is known to result in disturbance of MHC class II presentation and blockage of MHC class II transfer from endocytic compartments to the cell surface (Brachet et al., 1997, Riese and Chapman, 2000), as confirmed by our experiments with the CatS inhibitor LHVS. The proteolytic truncation of Ii to MHC-class-II-bound CLIP peptide is a prerequisite for antigen loading and thereby for MHC class II antigen presentation (Roche and Furuta, 2015), and surface trafficking of MHC class II is important for successful antigen presentation (Rocha and Neefjes, 2008). This might explain the reduced presentation of influenza virus HA322–334 on HLA-DR1 by human dendritic cells upon YM201636 or apilimod treatment. Unsurprisingly, we also observed reduced activation of CD4+ T cells upon PIKfyve inhibition, although this is probably caused by the combined effects of (1) reduced uptake of antigen, (2) blocked production of IL-12/IL-23 and type I IFN (Cai et al., 2013, Cai et al., 2014), (3) reduced MHC class II presentation, (4) reduced cell viability of both the dendritic cells and T cells, and (5) other direct effects of apilimod and YM201636 on T cells.
To overcome this, we developed a new universal assay allowing for the direct visualization of MHC class II antigen presentation. This assay is an adaptation of a recent assay to measure antigen presentation on MHC class I (Pawlak et al., 2015, Pawlak et al., 2016) and is based on synthetic peptides containing an MHC class-II-compatible epitope (in this case from HA residues 318–338 containing epitope residues 322–334). One residue on the epitope, which remains solvent exposed when bound to MHC class II, is converted to a non-natural propargylglycine, which contains an alkyne moiety of only three atoms in size. In contrast to large fluorescent labels, peptides carrying propargylglycine can still be proteolytically processed and loaded onto MHC class II. After fixation of the cells, the peptide bound to MHC class II at the surface of the dendritic cells can be labeled with CalFluor dyes that increase in fluorescence signal by orders of magnitude (Shieh et al., 2015). As previously shown for MHC class I (Pawlak et al., 2015, Pawlak et al., 2016), this signal increase allows for the detection of the limited amounts of MHC class-II-presented epitopes by flow cytometry. In contrast to assays that assess T cell priming and rely on co-stimulatory factors, such as the OT-II assay, the assay developed in this study provides a direct and quantitative readout of MHC class II presentation and is easily adaptable for different antigens and MHC haplotypes.
In conclusion, we showed that the phagosomal maturation driven by phosphoinositide kinase PIKfyve is essential for MHC class II antigen presentation (Figure 7). Disruption of this process by the PIKfyve inhibitors apilimod and YM201636 delays the maturation process of the phagosome. Delayed phagosomal maturation, in turn, results in elevated levels of ROS thereby deactivating CatS, which plays a key role in the proteolytic cleavage of Ii to CLIP and is essential for MHC class II antigen presentation. PIKfyve inhibition results in an incomplete processing of CLIP, and Ii-bound MHC class II is retained in phagosomes. Apilimod is proposed as a promising treatment for multiple variations of B cell non-Hodgkin lymphoma (NHL) due to its cytotoxic effect on B-NHL cancer cells lines (Gayle et al., 2017b) and was used in clinical trials for blocking IL-12/IL-23-mediated Th1/17 pro-inflammatory response in Crohn disease, psoriasis, and rheumatoid arthritis (Burakoff et al., 2006, Krausz et al., 2012, Sands et al., 2010, Wada et al., 2007, Wada et al., 2012). As concluded from NHL cancer cell line experiments, these cells have maximal sensitivity to apilimod, due to non-understood modulatory effects of PI(3,5)P2 on the transcription factor TFEB and apilimod leading to elevated expression of genes coding for lysosomal proteins, resulting in lysosomal swelling and disruption of lysosomal homeostasis and maturation, proliferation block, and cell death. In addition, PIKfyve might be a potential target for targeting solid tumors, as it plays a role in metastasis and promotes cancer cell migration and invasion together with the phosphoinositide phosphatase myotubularin-related protein 3 (MTMR3), which converses PI(3,5)P2 to PI(5)P (Oppelt et al., 2014). Depletion of PIKfyve and MTMR3, as well as inhibition of PIKfyve alone by YM201636, resulted in decreased in vitro migration of cancer cell lines of lung, rhabdomyosarcoma, and osteosarcoma origins (Oppelt et al., 2014), indicating that PIKfyve could be a novel therapeutic target in cancer migration. However, our results show that inhibition of PIKfyve impairs antigen presentation by dendritic cells and reduces activation of T cells. These effects on immune activation must be taken into account in clinical trials targeting PIKfyve.
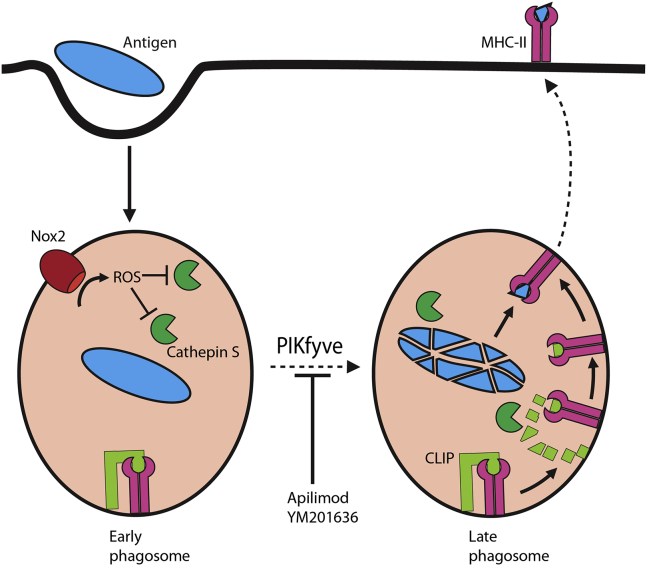
Figure 7.
Delayed Phagosomal Maturation Results in a Reduced MHC Class II Antigen Presentation
Model figure showing antigen uptake by a dendritic cell and subsequent processing within phagosomes by ROS and cathepsins. Upon phagosomal maturation, MHC class II bound to Ii is trafficked to the phagosome where cathepsin S is involved in the processing of Ii to CLIP, which can then be exchanged by an antigen to be presented by MHC class II on the cell surface. Inhibiting PIKfyve delays phagosomal maturation, and due to prolonged ROS formation, cathepsin S becomes oxidized and deactivated, thereby blocking CLIP-to-antigen exchange and eventually resulting in impaired antigen presentation.
Limitations of the Study
In this study, we addressed the role of phagosomal and endosomal maturation of MHC class II presentation with a focus on the phosphoinositide kinase PIKfyve. Most of our results are obtained using the small molecule inhibitors that are shown to be specific for PIKfyve: apilimod (Cai et al., 2013) and YM201636 (Jefferies et al., 2008). We confirmed that PIKfyve inhibition with these compounds was similar to PIKfyve siRNA knock down in primary human monocyte-derived dendritic cells, and both resulted in formation of enlarged endosomes, delayed phagosomal acidification, and delayed LAMP1 recruitment. However, the efficiency of our siRNA knockdown was incomplete, leading to less pronounced functional effects compared with the inhibitors. Our work could benefit from stable knockdown/out of PIKfyve to demonstrate that all experiments with drug inhibitors are comparable to knockout. However, the primary human monocyte-derived dendritic cells used in our study are terminally differentiated and not amenable to CRISPR/Cas knockout.
A potential caveat of our study is that we only managed to immunolabel endogenous CatS, but not the other two cathepsins X and B. Therefore we cannot draw clear conclusions on the effects of PIKfyve inhibition on the trafficking of those cathepsin species.
We admit that MHC class II labeling with CalFluor-488 method has still some limitations, mainly a low signal-to-noise ratio, which is likely caused by a low number of MHC-class-II-presented epitopes. Additional optimizations of this method will be required in future.
In our live cell microscopy experiments we followed the trafficking of MHC class II to phagosomes, revealing that a large portion of it is recruited from the cell surface during formation of the phagosome. However, we only followed the fate of MHC class II during PIKfyve inhibition for a short time and never studied longer time points after zymosan uptake. It would be interesting to determine whether MHC class II also reaches phagosomes from an intracellular pool, as we recently showed for NOX2 (Dingjan et al., 2017a).
Our study raises the key open question as to which effectors of PI(3,5)P2 mediate phagosomal maturation, as explained in the Discussion section. It would be interesting to identify those in the future.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Michael Sixt for LifeAct-RFP construct, Assia Shisheva for pEGFP-HA-PIKfyve and pEGFP-HA-PIKfyveK1831E, Jacques Neefjes for the MHC-II-YFP construct, and Matthew Bogyo for the LHVS. The authors express their gratitude to Yusuf Dölen for providing OT-II cells and PLGA-OVA, and Olga Ilina for OT-I cells. This work was supported by a Starting Grant from the European Research Council (ERC) under the European Union's Seventh Framework Program (Grant Agreement 336479). G.v.d.B. is funded by a Hypatia fellowship from the Radboud University Medical Center, a Career Development Award from the Human Frontier Science Program, the NWO Gravitation Program 2013 (ICI-024.002.009), and a Vidi grant from the Netherlands Organisation for Scientific Research (NWO-ALW VIDI 864.14.001). F.B. is funded by an EMBO Short-Term fellowship (7280) and a Veni grant from the Netherlands Organization for Scientific Research (016.Veni.192.026). Work from C.R.B.'s laboratory at Stanford University was supported by the NIH grant R37 GM058867.
Author Contributions
Conceptualization: M.V.B., F.B., and G.v.d.B.; Methodology: F.B., S.M., E.M.M., M.V.B., and G.v.d.B.; Investigation: M.V.B., M.A.C.v.A, F.B., S.M., A.M., I.D., and M.t.B; Writing – Original Draft: M.V.B and G.v.d.B; Writing – Review & Editing: F.B., M.V. and M.t.B.; Visualization: M.V.B., F.B, G.v.d.B.; CalFluor labeling: C.R.B. and S.G.L.K; peptide synthesis: A.S. and U.D.; activity-based probes: M.V.; Resources: G.v.d.B., M.V., U.D., and C.R.B; Supervision: G.v.d.B.; Funding Acquisition: G.v.d.B.
Declaration of Interests
The authors declare no competing interests.
Published: January 25, 2019
Footnotes
Supplemental Information includes Transparent Methods, eight figures, one table, and three videos and can be found with this article online at https://doi.org/10.1016/j.isci.2018.12.015.
Supplemental Information
References
- Allan E.R., Tailor P., Balce D.R., Pirzadeh P., McKenna N.T., Renaux B., Warren A.L., Jirik F.R., Yates R.M. NADPH oxidase modifies patterns of MHC class II-restricted epitopic repertoires through redox control of antigen processing. J. Immunol. 2014;192:4989–5001. doi: 10.4049/jimmunol.1302896. [DOI] [PubMed] [Google Scholar]
- Amigorena S., Drake J.R., Webster P., Mellman I. Transient accumulation of new class II MHC molecules in a novel endocytic compartment in B lymphocytes. Nature. 1994;369:113–120. doi: 10.1038/369113a0. [DOI] [PubMed] [Google Scholar]
- Amigorena S., Webster P., Drake J., Newcomb J., Cresswell P., Mellman I. Invariant chain cleavage and peptide loading in major histocompatibility complex class II vesicles. J. Exp. Med. 1995;181:1729–1741. doi: 10.1084/jem.181.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.E., Chessa T.A., Davidson K., Henderson R.B., Walker S., Tolmachova T., Grys K., Rausch O., Seabra M.C., Tybulewicz V.L. PtdIns3P and Rac direct the assembly of the NADPH oxidase on a novel, pre-phagosomal compartment during FcR-mediated phagocytosis in primary mouse neutrophils. Blood. 2010;116:4978–4989. doi: 10.1182/blood-2010-03-275602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkum T., van Leeuwen T., Sarris A.J.C., van Elsland D.M., Poulcharidis D., Overkleeft H.S., van Kasteren S.I. Quantification of bioorthogonal stability in immune phagocytes using flow cytometry reveals rapid degradation of strained alkynes. ACS Chem. Biol. 2018;13:1173–1179. doi: 10.1021/acschembio.8b00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balce D.R., Li B., Allan E.R., Rybicka J.M., Krohn R.M., Yates R.M. Alternative activation of macrophages by IL-4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood. 2011;118:4199–4208. doi: 10.1182/blood-2011-01-328906. [DOI] [PubMed] [Google Scholar]
- Balderhaar H.J., Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Baranov M.V., Revelo N.H., Dingjan I., Maraspini R., Ter Beest M., Honigmann A., van den Bogaart G. SWAP70 organizes the actin cytoskeleton and is essential for phagocytosis. Cell Rep. 2016;17:1518–1531. doi: 10.1016/j.celrep.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov M.V., Revelo N.H., Verboogen D.R.J., Ter Beest M., van den Bogaart G. SWAP70 is a universal GEF-like adaptor for tethering actin to phagosomes. Small GTPases. 2017 doi: 10.1080/21541248.2017.1328302. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Bissig C., Hurbain I., Raposo G., van Niel G. PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic. 2017;18:747–757. doi: 10.1111/tra.12525. [DOI] [PubMed] [Google Scholar]
- Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic B., Srinivasan P., Ueda Y., Tomita Y., Maric M. Comparative quantitative mass spectrometry analysis of MHC class II-associated peptides reveals a role of GILT in formation of self-peptide repertoire. PLoS One. 2010;5:e10599. doi: 10.1371/journal.pone.0010599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachet V., Raposo G., Amigorena S., Mellman I. Ii chain controls the transport of major histocompatibility complex class II molecules to and from lysosomes. J. Cell Biol. 1997;137:51–65. doi: 10.1083/jcb.137.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakoff R., Barish C.F., Riff D., Pruitt R., Chey W.Y., Farraye F.A., Shafran I., Katz S., Krone C.L., Vander Vliet M. A phase 1/2A trial of STA 5326, an oral interleukin-12/23 inhibitor, in patients with active moderate to severe Crohn's disease. Inflamm. Bowel Dis. 2006;12:558–565. doi: 10.1097/01.ibd.0000225337.14356.31. [DOI] [PubMed] [Google Scholar]
- Cai X., Xu Y., Cheung A.K., Tomlinson R.C., Alcazar-Roman A., Murphy L., Billich A., Zhang B., Feng Y., Klumpp M. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem. Biol. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Xu Y., Kim Y.M., Loureiro J., Huang Q. PIKfyve, a class III lipid kinase, is required for TLR-induced type I IFN production via modulation of ATF3. J. Immunol. 2014;192:3383–3389. doi: 10.4049/jimmunol.1302411. [DOI] [PubMed] [Google Scholar]
- Calafat J., Nijenhuis M., Janssen H., Tulp A., Dusseljee S., Wubbolts R., Neefjes J. Major histocompatibility complex class II molecules induce the formation of endocytic MIIC-like structures. J. Cell Biol. 1994;126:967–977. doi: 10.1083/jcb.126.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino F., Germain R.N. Extensive trafficking of MHC class II-invariant chain complexes in the endocytic pathway and appearance of peptide-loaded class II in multiple compartments. Immunity. 1995;2:73–88. doi: 10.1016/1074-7613(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Compton L.M., Ikonomov O.C., Sbrissa D., Garg P., Shisheva A. Active vacuolar H+ ATPase and functional cycle of Rab5 are required for the vacuolation defect triggered by PtdIns(3,5)P2 loss under PIKfyve or Vps34 deficiency. Am. J. Physiol. Cell Physiol. 2016;311:C366–C377. doi: 10.1152/ajpcell.00104.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H., Guscott B., Balklava Z., Rothnie A., Wassmer T. APP controls the formation of PI(3,5)P(2) vesicles through its binding of the PIKfyve complex. Cellular and molecular life sciences. Cell. Mol. Life Sci. 2016;73:393–408. doi: 10.1007/s00018-015-1993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayam R.M., Sun C.X., Choy C.H., Mancuso G., Glogauer M., Botelho R.J. The lipid kinase PIKfyve coordinates the neutrophil immune response through the activation of the Rac GTPase. J. Immunol. 2017;199:2096–2105. doi: 10.4049/jimmunol.1601466. [DOI] [PubMed] [Google Scholar]
- Denzin L.K., Robbins N.F., Carboy-Newcomb C., Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Dingjan I., Linders P.T., van den Bekerom L., Baranov M.V., Halder P., Ter Beest M., van den Bogaart G. Oxidized phagosomal NOX2 complex is replenished from lysosomes. J. Cell Sci. 2017;130:1285–1298. doi: 10.1242/jcs.196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingjan I., Paardekooper L.M., Verboogen D.R.J., von Mollard G.F., Ter Beest M., van den Bogaart G. VAMP8-mediated NOX2 recruitment to endosomes is necessary for antigen release. Eur. J. Cell Biol. 2017;96:705–714. doi: 10.1016/j.ejcb.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingjan I., Verboogen D.R., Paardekooper L.M., Revelo N.H., Sittig S.P., Visser L.J., Mollard G.F., Henriet S.S., Figdor C.G., Ter Beest M. Lipid peroxidation causes endosomal antigen release for cross-presentation. Sci. Rep. 2016;6:22064. doi: 10.1038/srep22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S.K., Dong K., Kobayashi T., Williams F.K., Michell R.H. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem. J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- Egami Y. Molecular imaging analysis of Rab GTPases in the regulation of phagocytosis and macropinocytosis. Anat. Sci. Int. 2016;91:35–42. doi: 10.1007/s12565-015-0313-y. [DOI] [PubMed] [Google Scholar]
- Ellson C.D., Anderson K.E., Morgan G., Chilvers E.R., Lipp P., Stephens L.R., Hawkins P.T. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr. Biol. 2001;11:1631–1635. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- Ellson C.D., Gobert-Gosse S., Anderson K.E., Davidson K., Erdjument-Bromage H., Tempst P., Thuring J.W., Cooper M.A., Lim Z.Y., Holmes A.B. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat. Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- Ewanchuk B.W., Yates R.M. The phagosome and redox control of antigen processing. Free Rad. Biol. Med. 2018;125:53–61. doi: 10.1016/j.freeradbiomed.2018.03.040. [DOI] [PubMed] [Google Scholar]
- Fairn G.D., Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33:397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Cosio G., Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009;7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Jaumouille V., Grinstein S. The cell biology of phagocytosis. Annu. Rev. Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- Fratti R.A., Backer J.M., Gruenberg J., Corvera S., Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle S., Landrette S., Beeharry N., Conrad C., Hernandez M., Beckett P., Ferguson S.M., Mandelkern T., Zheng M., Xu T. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129:1768–1778. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle S., Landrette S., Beeharry N., Conrad C., Hernandez M., Beckett P., Ferguson S.M., Xu T., Rothberg J., Lichenstein H. B-cell non-Hodgkin lymphoma: selective vulnerability to PIKFYVE inhibition. Autophagy. 2017;13:1082–1083. doi: 10.1080/15548627.2017.1304871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez N.M., Lu W., Lim J.C., Kiselyov K., Campagno K.E., Grishchuk Y., Slaugenhaupt S.A., Pfeffer B.A., Fliesler S.J., Mitchell C.H. Robust lysosomal calcium signaling through channel TRPML1 is impaired by lysosomal lipid accumulation. FASEB J. 2018;32:782–794. doi: 10.1096/fj.201700220RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groemping Y., Rittinger K. Activation and assembly of the NADPH oxidase: a structural perspective. Biochem. J. 2005;386:401–416. doi: 10.1042/BJ20041835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari A., Ganguly A., Mu L., Davis S.P., Stenner M.D., Lam R., Munro F., Namet I., Alghamdi E., Furstenhaupt T. Redirecting soluble antigen for MHC class I cross-presentation during phagocytosis. Eur. J. Immunol. 2015;45:383–395. doi: 10.1002/eji.201445156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J.C. Calcium: a fundamental regulator of intracellular membrane fusion? EMBO Rep. 2007;8:236–240. doi: 10.1038/sj.embor.7400921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeki K., Nigorikawa K., Takaba Y., Segawa T., Nukuda A., Masuda A., Ishikawa Y., Kubota K., Takasuga S., Hazeki O. Essential roles of PIKfyve and PTEN on phagosomal phosphatidylinositol 3-phosphate dynamics. FEBS Lett. 2012;586:4010–4015. doi: 10.1016/j.febslet.2012.09.043. [DOI] [PubMed] [Google Scholar]
- Ho C.Y., Alghamdi T.A., Botelho R.J. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 2012;13:1–8. doi: 10.1111/j.1600-0854.2011.01246.x. [DOI] [PubMed] [Google Scholar]
- Honey K., Rudensky A.Y. Lysosomal cysteine proteases regulate antigen presentation. Nat. Rev. Immunol. 2003;3:472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- Ikonomov O.C., Sbrissa D., Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- Ikonomov O.C., Sbrissa D., Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem. Biophys. Res. Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic C., Savina A., Wasmeier C., Tolmachova T., El-Benna J., Dang P.M., Pascolo S., Gougerot-Pocidalo M.A., Raposo G., Seabra M.C. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat. Cell Biol. 2007;9:367–378. doi: 10.1038/ncb1552. [DOI] [PubMed] [Google Scholar]
- Jasanoff A., Song S., Dinner A.R., Wagner G., Wiley D.C. One of two unstructured domains of Ii becomes ordered in complexes with MHC class II molecules. Immunity. 1999;10:761–768. doi: 10.1016/s1074-7613(00)80075-8. [DOI] [PubMed] [Google Scholar]
- Jefferies H.B., Cooke F.T., Jat P., Boucheron C., Koizumi T., Hayakawa M., Kaizawa H., Ohishi T., Workman P., Waterfield M.D. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.E. Reduction of disulfide bonds during antigen processing: evidence from a thiol-dependent insulin determinant. J. Exp. Med. 1991;174:1121–1130. doi: 10.1084/jem.174.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- Jones P.P., Murphy D.B., McDevitt H.O. Two-gene control of the expression of a murine Ia antigen. J. Exp. Med. 1978;148:925–939. doi: 10.1084/jem.148.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F., Liu H., Field S.J., Akbary H., Matsuo T., Brown G.E., Cantley L.C., Yaffe M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- Kerr M.C., Wang J.T., Castro N.A., Hamilton N.A., Town L., Brown D.L., Meunier F.A., Brown N.F., Stow J.L., Teasdale R.D. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J. 2010;29:1331–1347. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A., Hartman I.Z., Poore B., Boronina T., Cole R.N., Song N., Ciudad M.T., Caspi R.R., Jaraquemada D., Sadegh-Nasseri S. Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources. Nat. Commun. 2014;5:5369. doi: 10.1038/ncomms6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.H., Dayam R.M., Prashar A., Terebiznik M., Botelho R.J. PIKfyve inhibition interferes with phagosome and endosome maturation in macrophages. Traffic. 2014;15:1143–1163. doi: 10.1111/tra.12199. [DOI] [PubMed] [Google Scholar]
- Kinchen J.M., Ravichandran K.S. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke H., Wiederanders B., Bromme D., Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem. J. 1989;264:467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsias F., Hoffmann E., Amigorena S., Savina A. Reactive oxygen species production in the phagosome: impact on antigen presentation in dendritic cells. Antioxid. Redox Signal. 2013;18:714–729. doi: 10.1089/ars.2012.4557. [DOI] [PubMed] [Google Scholar]
- Krausz S., Boumans M.J., Gerlag D.M., Lufkin J., van Kuijk A.W., Bakker A., de Boer M., Lodde B.M., Reedquist K.A., Jacobson E.W. Brief report: a phase IIa, randomized, double-blind, placebo-controlled trial of apilimod mesylate, an interleukin-12/interleukin-23 inhibitor, in patients with rheumatoid arthritis. Arthritis Rheum. 2012;64:1750–1755. doi: 10.1002/art.34339. [DOI] [PubMed] [Google Scholar]
- Krishna S., Palm W., Lee Y., Yang W., Bandyopadhyay U., Xu H., Florey O., Thompson C.B., Overholtzer M. PIKfyve regulates vacuole maturation and nutrient recovery following engulfment. Dev. Cell. 2016;38:536–547. doi: 10.1016/j.devcel.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin R., Grinstein S., Schlam D. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta. 2015;1851:805–823. doi: 10.1016/j.bbalip.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Li S.C., Diakov T.T., Xu T., Tarsio M., Zhu W., Couoh-Cardel S., Weisman L.S., Kane P.M. The signaling lipid PI(3,5)P(2) stabilizes V(1)-V(o) sector interactions and activates the V-ATPase. Mol. Biol. Cell. 2014;25:1251–1262. doi: 10.1091/mbc.E13-10-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X., Xu H. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 2016;18:404–417. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza A.R., Savina A., Vermeulen M., Perez L., Geffner J., Hermine O., Rosenzweig S.D., Faure F., Amigorena S. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood. 2008;112:4712–4722. doi: 10.1182/blood-2008-01-134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney A.J., Zhang Y., Weisman L.S. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. Bioessays. 2014;36:52–64. doi: 10.1002/bies.201300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.H., Suzuki A., Stalker T.J., Zhao L., Wang Y., McKennan C., Riese M.J., Guzman J.F., Zhang S., Lian L. Loss of PIKfyve in platelets causes a lysosomal disease leading to inflammation and thrombosis in mice. Nat. Commun. 2014;5:4691. doi: 10.1038/ncomms5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H., Cho M., Chung Y. Regulation of Th2 cell immunity by dendritic cells. Immune Netw. 2016;16:1–12. doi: 10.4110/in.2016.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T.Y., Brissette W.H., Lira P.D., Griffiths R.J., Petrushova N., Stock J., McNeish J.D., Eastman S.E., Howard E.D., Clarke S.R. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity. 1999;10:207–217. doi: 10.1016/s1074-7613(00)80021-7. [DOI] [PubMed] [Google Scholar]
- Naufer A., Hipolito V.E.B., Ganesan S. pH of endophagosomes controls association of their membranes with Vps34 and PtdIns(3)P levels. J. Cell Biol. 2018;217:329–346. doi: 10.1083/jcb.201702179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J., Jongsma M.M.L., Berlin I. Stop or go? endosome positioning in the establishment of compartment architecture, dynamics, and function. Trends Cell Biol. 2017;27:580–594. doi: 10.1016/j.tcb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Neefjes J.J., Ploegh H.L. Inhibition of endosomal proteolytic activity by leupeptin blocks surface expression of MHC class II molecules and their conversion to SDS resistance alpha beta heterodimers in endosomes. EMBO J. 1992;11:411–416. doi: 10.1002/j.1460-2075.1992.tb05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes P., Demaurex N., Dinauer M.C. Regulation of the NADPH oxidase and associated ion fluxes during phagocytosis. Traffic. 2013;14:1118–1131. doi: 10.1111/tra.12115. [DOI] [PubMed] [Google Scholar]
- Oppelt A., Haugsten E.M., Zech T., Danielsen H.E., Sveen A., Lobert V.H., Skotheim R.I., Wesche J. PIKfyve, MTMR3 and their product PtdIns5P regulate cancer cell migration and invasion through activation of Rac1. Biochem. J. 2014;461:383–390. doi: 10.1042/BJ20140132. [DOI] [PubMed] [Google Scholar]
- Palmer J.T., Rasnick D., Klaus J.L., Bromme D. Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem. 1995;38:3193–3196. doi: 10.1021/jm00017a002. [DOI] [PubMed] [Google Scholar]
- Pawlak J.B., Gential G.P., Ruckwardt T.J., Bremmers J.S., Meeuwenoord N.J., Ossendorp F.A., Overkleeft H.S., Filippov D.V., van Kasteren S.I. Bioorthogonal deprotection on the dendritic cell surface for chemical control of antigen cross-presentation. Angew. Chem. Int. Ed. 2015;54:5628–5631. doi: 10.1002/anie.201500301. [DOI] [PubMed] [Google Scholar]
- Pawlak J.B., Hos B.J., van de Graaff M.J., Megantari O.A., Meeuwenoord N., Overkleeft H.S., Filippov D.V., Ossendorp F., van Kasteren S.I. The optimization of bioorthogonal epitope ligation within MHC-I complexes. ACS Chem. Biol. 2016;11:3172–3178. doi: 10.1021/acschembio.6b00498. [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese R.J., Chapman H.A. Cathepsins and compartmentalization in antigen presentation. Curr. Opin. Immunol. 2000;12:107–113. doi: 10.1016/s0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- Riese R.J., Mitchell R.N., Villadangos J.A., Shi G.P., Palmer J.T., Karp E.R., De Sanctis G.T., Ploegh H.L., Chapman H.A. Cathepsin S activity regulates antigen presentation and immunity. J. Clin. Invest. 1998;101:2351–2363. doi: 10.1172/JCI1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese R.J., Wolf P.R., Bromme D., Natkin L.R., Villadangos J.A., Ploegh H.L., Chapman H.A. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- Rocha N., Neefjes J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche P.A., Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicka J.M., Balce D.R., Chaudhuri S., Allan E.R., Yates R.M. Phagosomal proteolysis in dendritic cells is modulated by NADPH oxidase in a pH-independent manner. EMBO J. 2012;31:932–944. doi: 10.1038/emboj.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybicka J.M., Balce D.R., Khan M.F., Krohn R.M., Yates R.M. NADPH oxidase activity controls phagosomal proteolysis in macrophages through modulation of the lumenal redox environment of phagosomes. Proc. Natl. Acad. Sci. U S A. 2010;107:10496–10501. doi: 10.1073/pnas.0914867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh-Nasseri S. A step-by-step overview of the dynamic process of epitope selection by major histocompatibility complex class II for presentation to helper T cells. F1000Res. 2016;5:1305. doi: 10.12688/f1000research.7664.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands B.E., Jacobson E.W., Sylwestrowicz T., Younes Z., Dryden G., Fedorak R., Greenbloom S. Randomized, double-blind, placebo-controlled trial of the oral interleukin-12/23 inhibitor apilimod mesylate for treatment of active Crohn's disease. Inflamm. Bowel Dis. 2010;16:1209–1218. doi: 10.1002/ibd.21159. [DOI] [PubMed] [Google Scholar]
- Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Dumenil A.M., Seabra M.C., Raposo G., Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., Filios C., Delvecchio K., Shisheva A. Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636. Am. J. Physiol. Cell Physiol. 2012;303:C436–C446. doi: 10.1152/ajpcell.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., Shisheva A. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., Shisheva A. PIKfyve lipid kinase is a protein kinase: downregulation of 5'-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., Shisheva A. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization. J. Biol. Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- Sherer N.M., Lehmann M.J., Jimenez-Soto L.F., Ingmundson A., Horner S.M., Cicchetti G., Allen P.G., Pypaert M., Cunningham J.M., Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- Shi G.P., Villadangos J.A., Dranoff G., Small C., Gu L., Haley K.J., Riese R., Ploegh H.L., Chapman H.A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- Shieh P., Dien V.T., Beahm B.J., Castellano J.M., Wyss-Coray T., Bertozzi C.R. CalFluors: a universal motif for fluorogenic azide probes across the visible spectrum. J. Am. Chem. Soc. 2015;137:7145–7151. doi: 10.1021/jacs.5b02383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisheva A. PIKfyve: the road to PtdIns 5-P and PtdIns 3,5-P(2) Cell Biol. Int. 2001;25:1201–1206. doi: 10.1006/cbir.2001.0803. [DOI] [PubMed] [Google Scholar]
- Shisheva A., Sbrissa D., Ikonomov O. Plentiful PtdIns5P from scanty PtdIns(3,5)P2 or from ample PtdIns? PIKfyve-dependent models: evidence and speculation (response to: DOI 10.1002/bies.201300012) Bioessays. 2015;37:267–277. doi: 10.1002/bies.201400129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M., Nussenzweig M.C. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc. Natl. Acad. Sci. U S A. 2002;99:351–358. doi: 10.1073/pnas.231606698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L.J., Brown J.H., Jardetzky T.S., Gorga J.C., Urban R.G., Strominger J.L., Wiley D.C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- ten Broeke T., Wubbolts R., Stoorvogel W. MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harb. Perspect. Biol. 2013;5:a016873. doi: 10.1101/cshperspect.a016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima M., Kaneko-Kobayashi Y., Nakamura N., Yuri M., Hiramoto M., Naitou M., Hattori K., Yokota H., Mizuhara H., Higashi Y. Inhibition of c-Rel DNA binding is critical for the anti-inflammatory effects of novel PIKfyve inhibitor. Eur. J. Pharmacol. 2016;780:93–105. doi: 10.1016/j.ejphar.2016.03.036. [DOI] [PubMed] [Google Scholar]
- Teyton L., O'Sullivan D., Dickson P.W., Lotteau V., Sette A., Fink P., Peterson P.A. Invariant chain distinguishes between the exogenous and endogenous antigen presentation pathways. Nature. 1990;348:39–44. doi: 10.1038/348039a0. [DOI] [PubMed] [Google Scholar]
- Trombetta E.S., Ebersold M., Garrett W., Pypaert M., Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- Tulp A., Verwoerd D., Dobberstein B., Ploegh H.L., Pieters J. Isolation and characterization of the intracellular MHC class II compartment. Nature. 1994;369:120–126. doi: 10.1038/369120a0. [DOI] [PubMed] [Google Scholar]
- Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Querol E., Rosales C. Control of phagocytosis by microbial pathogens. Front. Immunol. 2017;8:1368. doi: 10.3389/fimmu.2017.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoes M., Oresic Bender K., Segal E., van der Linden W.A., Syed S., Withana N.P., Sanman L.E., Bogyo M. Improved quenched fluorescent probe for imaging of cysteine cathepsin activity. J. Am. Chem. Soc. 2013;135:14726–14730. doi: 10.1021/ja4056068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Dixit R., Pandey K.C. Cysteine proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 2016;7:107. doi: 10.3389/fphar.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O.V., Bucci C., Harrison R.E., Trimble W.S., Lanzetti L., Gruenberg J., Schreiber A.D., Stahl P.D., Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol. Cell. Biol. 2003;23:2501–2514. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos J.A., Driessen C., Shi G.P., Chapman H.A., Ploegh H.L. Early endosomal maturation of MHC class II molecules independently of cysteine proteases and H-2DM. EMBO J. 2000;19:882–891. doi: 10.1093/emboj/19.5.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos J.A., Ploegh H.L. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 2000;12:233–239. doi: 10.1016/s1074-7613(00)80176-4. [DOI] [PubMed] [Google Scholar]
- Vulcano M., Dusi S., Lissandrini D., Badolato R., Mazzi P., Riboldi E., Borroni E., Calleri A., Donini M., Plebani A. Toll receptor-mediated regulation of NADPH oxidase in human dendritic cells. J. Immunol. 2004;173:5749–5756. doi: 10.4049/jimmunol.173.9.5749. [DOI] [PubMed] [Google Scholar]
- Wada Y., Cardinale I., Khatcherian A., Chu J., Kantor A.B., Gottlieb A.B., Tatsuta N., Jacobson E., Barsoum J., Krueger J.G. Apilimod inhibits the production of IL-12 and IL-23 and reduces dendritic cell infiltration in psoriasis. PLoS One. 2012;7:e35069. doi: 10.1371/journal.pone.0035069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y., Lu R., Zhou D., Chu J., Przewloka T., Zhang S., Li L., Wu Y., Qin J., Balasubramanyam V. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood. 2007;109:1156–1164. doi: 10.1182/blood-2006-04-019398. [DOI] [PubMed] [Google Scholar]
- West M.A., Lucocq J.M., Watts C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature. 1994;369:147–151. doi: 10.1038/369147a0. [DOI] [PubMed] [Google Scholar]
- Wong C.O., Gregory S., Hu H., Chao Y., Sepulveda V.E., He Y., Li-Kroeger D., Goldman W.E., Bellen H.J., Venkatachalam K. Lysosomal degradation is required for sustained phagocytosis of bacteria by macrophages. Cell Host Microbe. 2017;21:719–730.e6. doi: 10.1016/j.chom.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala-Ruiz Z., Strug I., Anderson M.W., Gorski J., Stern L.J. A polymorphic pocket at the P10 position contributes to peptide binding specificity in class II MHC proteins. Chem. Biol. 2004;11:1395–1402. doi: 10.1016/j.chembiol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Zwart W., Griekspoor A., Kuijl C., Marsman M., van Rheenen J., Janssen H., Calafat J., van Ham M., Janssen L., van Lith M. Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity. 2005;22:221–233. doi: 10.1016/j.immuni.2005.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with the F-actin-binding probe LifeAct-RFP (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with LAMP1-RFP (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.
Live cell imaging of human monocyte-derived dendritic cell overexpressing human PIKfyve-GFP (green in merge) together with MHC-II-mCherry (HLA-DRA-IRES-HLA-DRB-mCherry) (red) and pulsed with IgG-opsonized zymosan particles. BF, brightfield.