Abstract
The CRISPR-Cas9-based multiplexed gene editing (MGE) provides a powerful method to modify multiple genomic regions simultaneously controlling different agronomic traits in crops. We applied the MGE construct built by combining the tandemly arrayed tRNA–gRNA units to generate heritable mutations in the TaGW2, TaLpx-1, and TaMLO genes of hexaploid wheat. The knockout mutations generated by this construct in all three homoeologous copies of one of the target genes, TaGW2, resulted in a substantial increase in seed size and thousand grain weight. We showed that the non-modified gRNA targets in the early generation plants can be edited by CRISPR-Cas9 in the following generations. Our results demonstrate that transgenerational gene editing activity can serve as the source of novel variation in the progeny of CRISPR-Cas9-expressing plants and suggest that the Cas9-inducible trait transfer for crop improvement can be achieved by crossing the plants expressing the gene editing constructs with the lines of interest.
Introduction
Wheat is the second most important food crop in the developing world. Currently, the genetic diversity of the natural1,2 or chemically mutagenized populations3 of wheat and its wild ancestors4 are the main sources of useful allelic variation for wheat improvement.5 However, the deployment of these alleles in breeding is affected by the distribution of genetic diversity across genes controlling a trait, as well as the local recombination rate around these genes that can be severely suppressed in the peri-centromeric, highly divergent, or structurally rearranged genomic regions. CRISPR-Cas9* technology, with its multiplex genome editing capacity,6–14 holds great promise in overcoming some of these limitations and provides a tool for inducing beneficial modifications in multiple genes controlling major agronomic traits.
Since the introduction of CRISPR-Cas9 for editing mammalian genomes,8,9,15 it has been applied to modify the genomes of a number of model and crop plants, including tobacco,16,17 tomato,6,18 barley,19 Arabidopsis,16 wheat,10,20–25 rice,11,14 and maize.7,26 Following the first reports of CRISPR-Cas9-based genome editing in wheat protoplasts,22,25 gene edited plants have been regenerated from the wheat immature embryos transformed with CRISPR-Cas9 in different forms, including plasmids, linear DNA fragments, linear RNA, and ribonuleoprotein complexes.20,21,23
Several strategies have been developed for multiplexed gene editing (MGE). The first MGE reported in human cells mimicked the natural architecture of the CRISPR-Cas9 locus; protospacers targeting different genes were included into a single pre-crRNA array, which was then processed into individual crRNAs by Cas9, tracrRNA, and endogenous RNase III. The resulting ribonucleoprotein complexes, including Cas9, tracrRNA, and crRNA, were capable of inducing mutations at multiple target sites.9 Another MGE strategy based on combining multiple gRNAs under the control of their own promoters into a single construct was also successfully applied in wheat, rice, tomato, maize, and Arabidopsis.7,11–13,16 However, due to the size of the individual promoter-gRNA units (450 bp), the level of multiplexing achievable using this approach is limited by the insert size capacity of plasmid vectors. The third strategy that used the Csy4 nuclease and its 28 bp recognition site to generate functional gRNAs from the tandemly arrayed units was successfully tested in human cells12 and several plant species.10 The small size of the Csy4 recognition site allowed a higher level of multiplexing to be achieved but required the simultaneous expression of Csy4. Finally, effective MGE in rice was accomplished by spacing multiple gRNAs with tRNA under the control of a single promoter, where functional gRNAs are generated by processing a polycistronic gene transcript through the endogenous tRNA processing system.14 The functionality of gRNAs produced by either Csy4 ribonuclease or endogenous tRNA processing systems was successfully demonstrated in immature wheat embryos.10
Here, we investigated the frequency and heritability of the mutations generated by the MGE construct expressing the gRNA–tRNA units in allopolyploid wheat. The MGE construct included gRNAs targeting the TaGW2, TaLpx-1, and TaMLO genes. The A genome homoeolog of the TaGW2 gene was previously shown to be negatively associated with the thousand grain weight (TGW), grain area, grain width, and grain length.27–31 Silencing the TaLpx-1 gene, which encodes 9-lipoxygenase, rendered wheat resistant to Fusarium graminearum.32 Knockout mutations in all three homoeologs of TaMLO provided resistance against powdery mildew in wheat.21 To test the feasibility of the selective editing of all three, two, or only one of the gene homoeologs in the allopolyploid wheat genome, the gRNAs were designed to target the TaGW2 gene in the region conserved in all three genomes, the TaLpx-1 gene in the region conserved in only two genomes, and the TaMLO gene in the A-genome specific region. The MGE construct was assembled using the gRNAs whose activity was validated in the wheat protoplasts. The ability of the MGE construct to affect agronomic traits was investigated by studying the effects of mutations in the TaGW2 gene on easily scorable seed morphology traits controlled by this gene. By analyzing the transmission of Cas9-induced mutations in the progenies of the MGE construct-expressing wheat lines, we showed that the transgenerational CRISPR-Cas9 activity is an important source of novel variation that can be utilized to obtain mutations across multiple sites targeted by the gRNAs from the MGE construct. This transgenerational gene editing activity can also be exploited for modifying the genomes of wheat breeding lines by crossing them with the plants expressing the CRISPR-Cas9 constructs.
Materials and Methods
Plasmids and vector construction
The wheat codon-optimized Cas9 fused with the nuclear localization signals and wheat U6 promoter-gRNA was synthesized by Integrated DNA Technology. Plasmid pA9mRFP, pA9Cas9, and pU6sg were constructed as shown in Supplementary Fig. S1 and Supplementary File 1 (Supplementary Data are available online at www.liebertpub.com/crispr). Plasmid pBUN4217 with the maize codon-optimized Cas9 gene (Supplementary Fig. S1) and pGTR14 containing gRNA–tRNA units were ordered from Addgene.
To design gRNAs targeting the TaGW2 and TaLpx-1 genes, the CDSs of these genes were submitted to the sgRNA Scorer 1.0.33 The highest scoring gRNAs were compared against the genomic sequence of cultivar Chinese Spring using the URGI website (https://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST). Only those gRNAs that did not have possible off-target matches in the genome were selected to create the MGE construct. The A genome copy of the TaMLO gene was targeted using the previously reported gRNA.21 The guide sequence oligonucleotides were subcloned into pU6sg or pBUN421 (Supplementary Fig. S1 and Supplementary File 1).12 The editing capability of different gRNAs was evaluated by co-transforming wheat protoplasts with the pU6sg constructs in combination with the pBUN421 (maize-optimized Cas9) or pA9Cas9 (wheat-optimized Cas9) plasmids. To compare the editing efficiency of single and multiple gRNA:CRISPR-Cas9 constructs, the annealed guide sequence oligonucleotides were subcloned into pBUN421. The MGE construct pBUN421-GLM was built by combining multiple polymerase chain reaction (PCR) fragments, each including a gRNA–tRNA block, using a single-step Golden Gate reaction (Supplementary Fig. S2).14
Protoplast transformation and DNA isolation
The seedlings of wheat cultivar Bobwhite were grown in the dark for 2 weeks. Shoot tissues were fine-sliced and digested for 2.5 h in an enzymatic solution containing 1.5% Cellulase R10 (from Trichoderma viride, 7.5 IU/mg) and 0.75% Macerozyme R10 (from Rhizopus sp.). The protoplast transformation was performed following the previously reported protocol.34 The molar amount of plasmid DNA for each transformation reaction was adjusted to be the same. The transformation efficiency was assessed by counting the fraction of fluorescent-positive protoplasts transformed with pA9mRFP. Protoplasts were collected 48 h after transformation, and DNAs were isolated with PureLink Genomic DNA Mini Kit (cat. no. K182002; Thermo Fisher Scientific) following the manufacturer's protocol.
Next-generation sequencing of PCR amplicons and estimation of gene editing efficiency
To detect CRISPR-Cas9-induced mutations, genomic regions harboring the gRNA targets were PCR amplified and sequenced. To facilitate simultaneous next-generation sequencing (NGS) analysis of multiple genomic regions, Illumina's TruSeq adaptors were added to the both ends of amplicons using two rounds of PCR (Supplementary Fig. S3A). PCR products were purified and sequenced at the K-State Integrated Genomics Facility on the MiSeq instrument (Supplementary Fig. S3B and C). All reads passing quality control were aligned to the wild-type reference sequences. The genome editing frequency at each target site was calculated by dividing the number of mutated reads to the total number of all aligned reads.
Regeneration and genotyping of transgenic plants
Wheat immature embryo transformation and plant regeneration were performed, as previously described.35 To isolate DNA, leaf tissues were sampled and homogenized in 500 μL of TPS buffer, then incubated for 20 min at 75°C. After centrifugation, 150 μL of the supernatant was mixed with 150 μL of isopropanol and incubated for 20 min at room temperature. DNA was precipitated, washed with 70% ethanol, and re-suspended in 100 μL of deionized water.
The presence of CRISPR-Cas9 constructs in the transgenic plants was validated by PCR using three pairs of primers amplifying different regions of the Cas9 and gRNA expression cassettes (Supplementary File 2). The CRISPR-Cas9-induced mutations were examined only in the plants showing the presence of all three PCR products. The detection of mutations was performed using the NGS-based procedure. Four bar-coding bases between the target-specific primer and Illumina TruSeq adaptors (Supplementary File 2) were added to index multiple plants under each of the Illumina TruSeq barcodes.
Screening of gw2 knockout mutants
To screen the gw2 knockout mutants, the GW2T2 target region from all three homoeologs was amplified, and PCR products were digested with XmaI (NEB). If all three copies of the GW2T2 target site are mutated by CRISPR-Cas9, the PCR products should not be digested. The genome specificity of the primers flanking the GW2T2 target sites (Supplementary file 2) was validated by performing PCR with DNA of Chinese Spring nullisomic-tetrasomic lines (Supplementary Fig. S4). For plants having nondigestible PCR products, the mutations at the GW2T2 target sites were confirmed by the Sanger sequencing of genome-specific PCR products.
Screening of the TaLpx-1 gene mutants
The progenies of T1 plant GLM-2-5 were screened for the presence of mutations in the TaLpx-1 gene. The flanking regions of the LPX1T2 target site were amplified using DNA isolated from the T2 generation wheat lines. PCR products were digested with SalI-HF (NEB). It was expected that the PCR amplicons carrying mutations in the LPX1T2 target site would be resistant to the SalI-HF digestion. Plants having strong nondigestible bands were subjected to NGS analysis.
Plant growth and phenotypic analysis of gw2 plants
Plants were grown in the greenhouse under 12 h of light for 1 month, and then grown until seed harvesting under 16 h of light at a temperature of 24°C during the day and 21°C at night. The MARVIN seed analyzer (GTA Sensorik GmbH) was used to estimate the TGW and grain width, length, and area of the wild-type plants (genotype AABBDD) and gw2 knockout mutants (genotype aabbdd). The mean phenotypic values were estimated for each plant and used for further analyses. Box and whisker plots were generated with the “boxplot” package implemented in R.
Statistical analysis
To evaluate the segregation ratio of mutated alleles in T1 plants, a chi-square test was performed using the CHITEST function implemented in Microsoft® Excel. A two-tailed Student's t-test was applied to assess the significance of the phenotypic differences between the wild-type and gene edited plants.
Results
Multiplex genome editing in the wheat protoplasts
The editing efficiency of the designed gRNAs targeting the TaGW2 and TaLpx-1 genes was assessed using a protoplast expression assay and the NGS of target sites (Supplementary Fig. S3B and C). The target sites GW2T2 and LPX1T2, which showed the highest editing efficiency (Supplementary File 3), together with the previously reported target site MLOT1 in the TaMLO gene, were used for further analysis. The gene editing efficiency of the wheat codon-optimized Cas9 construct pA9Cas9, and the maize codon-optimized Cas9 construct pBUN421 were compared. The gene editing efficiency of wheat (pA9Cas9) and maize (pBUN421) codon-optimized Cas9 were comparable for all three gRNA targets (p > 0.05; Supplementary File 4) likely due to either similar codon usage for this gene construct in wheat and maize and/or similar Cas9 translocation efficiency to nucleus in these species. All three gRNAs were combined into a tRNA-spaced polycistronic gene and subcloned into the pBUN421 plasmid (henceforth, pBUN421-GLM; Fig. 1A). The editing efficiency of the individual gRNAs from the pBUN421-GLM construct (Fig. 1A and B) was comparable to that of the single gRNA constructs estimated in the same transformation experiment (Table 1). This result is different from the one reported for maize where higher editing efficiency was found for a multiplex gRNA construct.36 It is difficult to discern specific reasons for such a discrepancy, which could be caused by either the differences in the amounts of gRNA produced from single and multiple gRNA constructs in maize, or by the efficiency of mutated DNA repair between maize and wheat, which has at least three copies for the targeted genes.
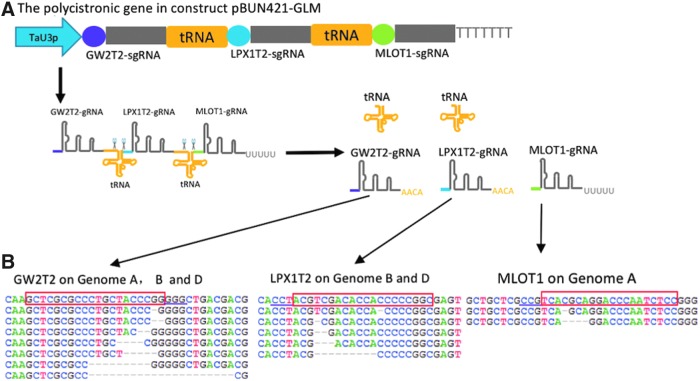
FIG. 1.
CRISPR-Cas9-based multiplex editing in hexaploid wheat using gRNAs processed through the endogenous tRNA-processing system. (A) Schematic of tRNA-based processing of a polycistronic gene transcript. The polycistronic gene containing three tRNA–gRNA blocks was driven by a TaU3 promoter in a MGE construct (pBUN421-GLM). Guide sequences GW2T2, LPX1T2, and MLOT1 are shown in purple, blue, and green, respectively. The GW2T2-gRNA, LPX1T2-gRNA, and MLOT1-gRNA are released after the tRNA processing. (B) The representative next-generation sequencing (NGS) results obtained for three genomic regions targeted by the pBUN421-GLM construct. The wild-type sequences are shown on the top. The target sequences are shown in the red rectangles; the PAM sequences are underlined; the deletions are shown by dashed lines.
Table 1.
Efficiency of multiplex gene editing in the protoplasts of hexaploid wheat
| Constructs | Spacer | Total reads analyzed | Mutated readsa | Proportion of mutated readsb |
|---|---|---|---|---|
| pBUN421-GLM | GW2T2 | 103,076 | 3,545 | 0.17 |
| LPX1T2 | 63,133 | 377 | 0.03 | |
| MLOT1 | 110,290 | 7,047 | 0.32 | |
| pBUN421-GW2T2 | GW2T2 | 77,731 | 6,210 | 0.40 |
| pBUN421-LPX1T2 | LPX1T2 | 49,108 | 888 | 0.09 |
| pBUN421-MLOT1 | MLOT1 | 111,379 | 5,965 | 0.27 |
The examples of mutated reads are shown in Supplementary Figures S5 and S6.
Proportion of mutated reads was normalized using the 20% protoplast transformation efficiency estimated for this experiment.
Transmission and phenotypic effects of the MGE construct-induced mutations
The transgenic plants were regenerated from the wheat immature embryos transformed with the MGE construct pBUN421-GLM. The single gRNA construct pBUN421-GW2T2 was used as a control. In total, 102 plants were regenerated for the pBUN421-GLM construct (henceforth GLM plants), and 61 plants were regenerated for the pBUN421-GW2T2 construct (henceforth GT2 plants). Based on the PCR results, 22 GLM plants and 17 GT2 plants had the intact CRISPR-Cas9 expression cassette. Six GLM plants and six GT2 plants carried mutated alleles at the GW2T2 target site, and nine GLM plants carried mutated alleles at the MLOT1 target site (Fig. 2A and B, Supplementary file 5). Five GLM plants carried mutated alleles at the GW2T2 and MLOT1 target sites simultaneously. While mutations at the LPX1T2 target site were detected by NGS in several GLM plants, within the analyzed set of lines, we did not recover fixed heritable mutations at this site (Supplementary File 5). Among the 12 plants that had mutations at the GW2T2 target site, only one plant, GLM-1, carried the mutated version of the TaGW2 gene in the homozygous state in all three wheat genomes (Fig. 2 and Supplementary File 5).
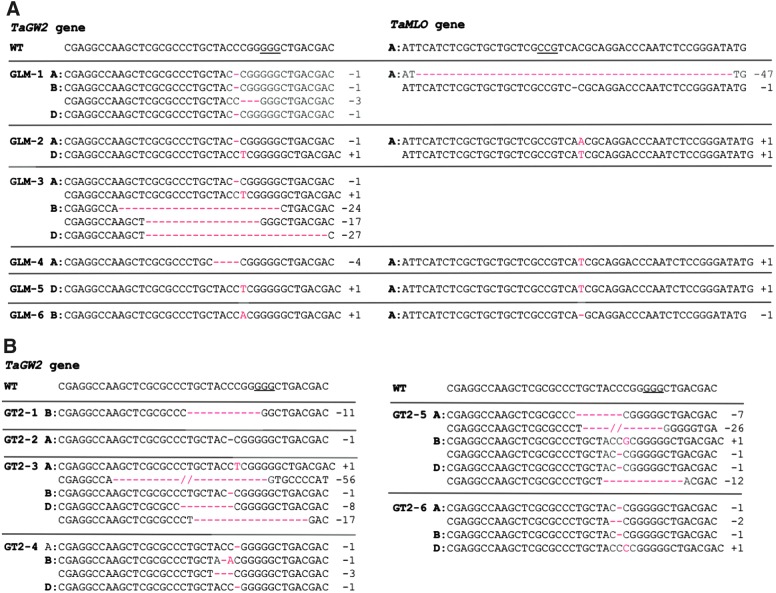
FIG. 2.
CRISPR-Cas9-induced mutations in the T0 transgenic plants identified by NGS. (A) CRISPR-Cas9-induced mutations in the homoeologs of the TaGW2 and TaMLO genes in the T0 plants expressing MGE construct pBUN421-GLM. (B) CRISPR-Cas9-induced mutations in the homoeologs of the TaGW2 gene in the T0 plants expressing single gRNA construct pBUN421-GT2. Only mutated reads with a frequency >30% were shown in (A) and (B) (see Supplementary File 5 for details). WT, wild-type alleles in wheat cultivar Bobwhite; “–” and “+” signs and numbers after them, nucleotides deleted and inserted, respectively. The PAM sequences are underlined; the mutated nucleotides are highlighted in red.
The transmission of CRISPR-Cas9-induced mutations to the next generation was investigated in the self-pollinated progeny of two T0 GLM plants (GLM-1 and GLM-2) with the mutant alleles in both the TaGW2 and TaMLO genes (Supplementary File 5). For line GLM-1, 100% transmission of the homozygous mutated alleles from T0 to T1 generation was detected, as expected. In the progeny of line GLM-2, carrying both mutated and wild-type alleles at the GW2T2 target site in the A and D genome copies of the TaGW2 gene, the proportion of mutated alleles was consistent with the Mendelian segregation ratio (p > 0.05). Interestingly, for the MLOT1 target site, no homozygous plants carrying the wild-type allele in the A genome of the TaMLO gene was detected (Table 2 and Supplementary File 5). One of the possible factors contributing to the observed non-Mendelian segregation at this locus could be the activity of CRIRSP/Cas9 in the progeny of the GLM-2 plant.
Table 2.
Transmission of CRISPR-Cas9-induced mutations in the progenies of MGE construct-expressing wheat lines
| T0 plants | Genome | TaGW2 genotypea | TaMLO genotypea | N.O. of T1 progenies | N.O. of TaGW2 genotypesa | N.O. of TaMLO genotypesa | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| GLM-1 | A | aa | aa | 28 | 0 AA | 0 Aa | 28 aa | 0 AA | 0 Aa | 28 aac |
| B | bb | BB | 0 BB | 0 Bb | 28 bbb | — | — | — | ||
| D | dd | DD | 0 DD | 0 Dd | 28 dd | — | — | — | ||
| GLM-2 | A | Aa | Aad | 31 | 7 AA | 17 Aa | 7 aa | 0 AAd | 10 Aa | 21 aa |
| B | BB | BB | 30 BB | 1 Bbe | 0 bb | — | — | — | ||
| D | Dd | DD | 10 DD | 15 Dd | 6 dd | — | — | — | ||
A/a, B/b, and D/d represent loci in different genomes; the uppercase and lowercase stand for wild-type and mutated alleles, respectively.
In GLM-1, the B genome copy of the TaGW2 gene has two different mutation types: 1 bp deletion (b1) and 3 bp deletion (b2; Fig. 2A). The segregation ratio of these two mutations is 10:14:4 (b1b1:b1b2:b2b2).
In GLM-1, the A genome copy of the TaMLO gene has two different mutation types: 1 bp deletion (a1) and 47 bp deletion (a2; Fig. 2A). The segregation ratio of these two mutations is 4:14:10 (a1a1:a1a2:a2a2).
Plant GLM-2 has wild-type reads and two different types of mutated reads (Fig. 2A) in the A genome copy of the TaMLO gene. However, its T1 progenies do not have homozygous wild-type plants.
New TaGW2 gene alleles in the B genome were induced by the transgenerationally active CRISPR-Cas9.
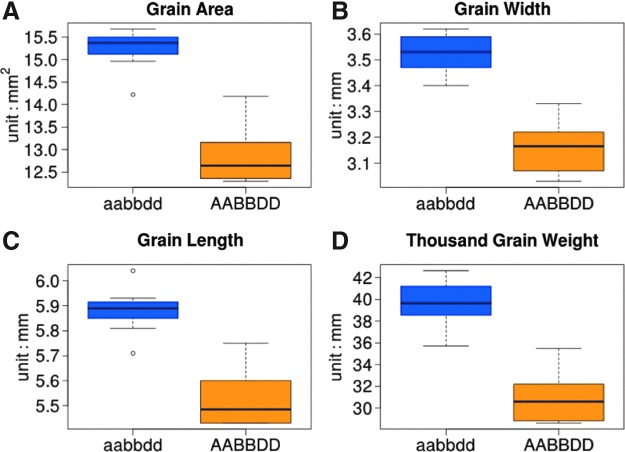
We investigated the effects of MGE construct-induced mutations on the phenotype by measuring easily scorable seed morphology traits in the T1 progeny of GLM-1. T1 plants carrying knockout mutations in all three copies of the TaGW2 gene (genotype aabbdd) showed significantly increased TGW (27.7%), grain area (17.0%), grain width (10.9%), and grain length (6.1%) compared to wild-type cultivar Bobwhite (Fig. 3 and Supplementary File 6).
FIG. 3.
Phenotypic effects of CRISPR-Cas9-induced mutations in the TaGW2 gene. Box and whisker plots are used to show (A) grain area, (B) grain width, (C) grain length, and (D) thousand grain weight (TGW) of gw2 knockout (aabbdd) and wild-type plants (AABBDD).
Generation of novel mutations in the TaGW2 and TaLpx-1 genes by transgenerationally active CRISPR-Cas9
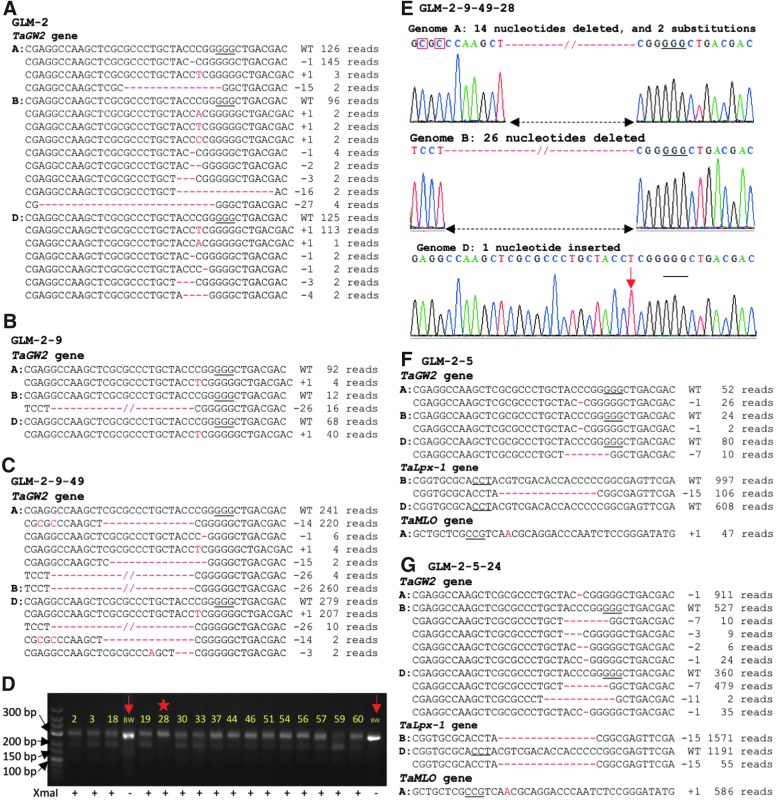
Among the 39 CRISPR-Cas9-positive plants, we detected only one gw2 knockout mutant that carried mutations in all three homoeologous copies of the gene. The rarity of this editing event suggests its dependence on the number of gene copies in the genome, and indicates that the recovery of mutant lines carrying editing events in all homoeologs or multiple targets can be challenging for polyploid crops. However, it is possible that the ongoing CRISPR-Cas9 activity in the somatic cells can serve as a source of novel heritable mutations in the next generations of CRISPR-Cas9-expressing plants, thereby reducing the need in performing additional plant transformation experiments. The ongoing CRISPR-Cas9 activity in somatic cells is evidenced by the discovery of different types of editing events at the GW2T2, LPX1T2, and MLOT1 target sites of the plants from different generations (Supplementary File 5, Fig. 4, and Supplementary Fig. S7). For instance, eight different types of mutations were found in the B genome copy of the TaGW2 gene in line GLM-2 (Fig. 4A).
FIG. 4.
Transgenerational CRISPR-Cas9 activity induces new mutations in the TaGW2 and TaLpx-1 genes. NGS reads flanking the GW2T2 target site and their frequencies in (A) T0 line GLM-2, (B) T1 line GLM-2-9, and (C) T2 line GLM-2-9-49 are shown. (D) Restriction enzyme digestion of polymerase chain reaction (PCR) amplicons to screen gw2 knockout mutations in the T3 progenies of line GLM-2-9-49. The GW2T2 flanking region was amplified by PCR and digested with XmaI; non-digested PCR amplicons correspond to mutated GW2T2 target sites. The numbers on the gel image are identifiers of the GLM-2-9-49 progenies. Lanes marked with arrows are PCR products from wild-type plant not digested with XmaI and loaded as controls; the knockout mutant plant was marked with a star. BW, wild-type cultivar Bobwhite. (E) Sanger sequencing of PCR-amplified GW2T2 target sites of T3 line GLM-2-9-49-28. Genome specific primers were used to amplify regions flanking the GW2T2 target sites. Nucleotide substitutions are marked with red rectangles, and the inserted nucleotide is shown by the red arrow. Types and frequencies of mutations at the GW2T2, LPX1T2, and MLOT1 target sites in (F) T1 line GLM-2-5, and (G) T2 line GLM-2-5-24 are shown. WT, wild-type alleles in wheat cultivar Bobwhite; “–” and “+” signs and numbers after them, nucleotides deleted and inserted, respectively. The frequency of each mutation type is shown on the right. The PAM sequences are underlined; the deleted nucleotides are shown with red dashed lines; the insertions and deletions are highlighted in red.
To investigate the process of gene editing across generations, we screened the progenies of several plants lacking mutated alleles in at least one of the homoeologous genomes at the target sites (Fig. 4 and Supplementary Figs. S7–S9). For the GLM-2 plant that carried the TaGW2 mutated alleles in the heterozygous states in the A and D genomes and had no mutated allele in the B genome (genotype AaBBDd), new edited alleles in the B and A genomes were detected in the T1 and T2 generations, respectively (Fig. 4B and C, Table 2, and Supplementary File 5). One gw2 knockout mutant (genotype aabbdd) was detected among 60 plants in the T3 generation of GLM-2 (Fig. 4D and E, and Supplementary Fig. S8). The analysis of the progenies of the T0 line GLM-7, which had no mutations in the TaGW2 gene (Supplementary Fig. S7A), revealed a mutated allele in the A genome in the T1 generation (Supplementary Fig. S7B), and mutated alleles in both the B and D genomes in the T2 generation (Supplementary Fig. S7C–E). Among the 26 plants from progenies of the T1 line GLM-2-5, which carried mutated alleles in both the TaGW2 and TaMLO genes but not in the TaLpx-1 gene (Fig. 4F), we discovered two plants that acquired mutated variants of the TaLpx-1 gene in the B genome along with the mutated variants of the TaGW2 and TaMLO genes being inherited (Fig. 4G, Supplementary Fig. S9, and Supplementary File 5). These results suggest that the transgenerational CRISPR-Cas9 activity can significantly facilitate the recovery of mutations across the multiple gene targets.
Discussion
Consistent with the results previously reported in rice and maize,14, 36 we demonstrated that heritable mutations in multiple gene targets in the wheat genome can be induced using a construct with an array of the gRNA–tRNA units driven by a single promoter. The phenotypic effects of the MGE construct-induced mutations were confirmed by measuring the seed morphology traits of the gw2 knockout mutants carrying mutations in all three copies of the TaGW2 gene. Several studies previously demonstrated that the functionally active A genome homoeolog of the TaGW2 gene is negatively associated with grain weight and grain width.28–31 Two other studies also reported that the A genome homoeolog is negatively associated with the grain length.29,31 In our study, the loss-of-function mutations in all three copies of the TaGW2 gene resulted in a substantial increase of the TGW, grain area, grain width, and grain length. The observed increase was higher than that previously reported, suggesting that the homoeologs of the TaGW2 gene in the B and D genomes might also have the same function as the A genome homoeolog, and have additive effects on the seed phenotypes.
The MGE approach provides a flexible tool for implementing complex gene editing strategies in wheat where the majority of genes have at least three homoeologous copies. By designing gRNAs to the targets carrying homoeolog-specific sites or to the conserved targets identical in all three wheat genomes, it should be possible either to edit multiple genes from a specific genome selectively, or to introduce modifications into all duplicated gene copies. Thus, multiple beneficial allelic changes across the wheat genome can theoretically be induced by transforming plants with a single polycistronic tRNA-gRNA/Cas9 construct. However, our results suggest that the recovery of T0 lines carrying mutations in all copies of multiple genes for polyploid plants, due to whole genome duplication, could be more challenging than for diploid plants.
The NGS of CRISPR-Cas9-expressing plants allowed us to detect rare somatic gene editing events present in only part of the cells in the leaf tissues, indicative of ongoing CRISPR-Cas9 activity. The successful recovery of novel gene editing events in the progenies of transgenic plants suggested that this CRISPR-Cas9 activity can serve as the source of novel heritable variation. Consistent with this conclusion, we successfully recovered plants with multiple edited targets in the progenies derived from a single plant carrying the active CRISPR-Cas9 MGE construct. Another advantage of the transgenerational CRISPR-Cas9 activity in the MGE construct-expressing plants is that it provides the possibility for obtaining lines carrying different combinations of mutations across multiple targets. For traits with a complex genetic basis, these lines can be used to study epistatic and/or additive interactions among multiple edited gene variants and to select the most optimal combinations of novel alleles.
Recently, the transgenerational CRISPR-Cas9 activity has been shown to induce new functional variants at multiple target sites in tomato F1 plants.6 This result suggests that beneficial mutations in the wheat breeding lines not amenable to transformation could be induced by crossing these lines with the wheat lines carrying the CRISPR-Cas9 constructs. This strategy provides an effective approach for the systematic transfer of beneficial gene editing events into the breeding programs for further phenotypic evaluation of their effects in different genetic backgrounds.
Conclusion
Our study demonstrated that the MGE construct employing endogenous tRNA processing system can be effectively used to induce heritable mutations in the genes controlling multiple agronomic traits in hexaploid wheat. The transgenerational CRISPR-Cas9 activity demonstrated here suggests that effective Cas9-inducible trait transfer can be accomplished by crossing wheat breeding lines with lines expressing CRISPR-Cas9 constructs. Further evaluation of this strategy will be required to assess its value for wheat improvement.
Supplementary Material
Footnotes
Clustered Regularly Interspaced Short Palindromic Repeats.
Acknowledgments
This work is supported by the Agriculture and Food Research Initiative Competitive Grants 2017-67007-25932 from the USDA National Institute of Food and Agriculture and by the Bill and Melinda Gates Foundation grant BMGF: 01511000146. We would like to thank Shichen Wang for assistance with the initial bioinformatical data processing, and Katherine Jordan for the valuable discussions of the presented work. We would like to thank Qi-Jun Chen for providing plasmid pBUN421, and Yinong Yang for providing plasmid pGTR.
Author Disclosure Statement
All the authors declare that no competing financial interests exist.
References
- 1.Wang S, Wong D, Forrest K, et al. . Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 2014;12:787–796. DOI: 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan KW, Wang S, Lun Y, et al. . A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol 2015;16:4–8.. DOI: 10.1186/s13059-015-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krasileva KV, Vasquez-Gross HA, Howell T, et al. . Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci U S A 2017;114:E913–E921. DOI: 10.1073/pnas.1619268114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avni R, Nave M, Barad O, et al. . Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017;357:93–97. DOI: 10.1126/science.aan0032 [DOI] [PubMed] [Google Scholar]
- 5.Uauy C, Wulff BBH, Dubcovsky J. Combining traditional mutagenesis with new high-throughput sequencing and genome editing to reveal hidden variation in polyploid wheat. Annu Rev Genet 2017;51:435–454. DOI: 10.1146/annurev-genet-120116-024533 [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Leal D, Lemmon ZH, Man J, et al. . Engineering quantitative trait variation for crop improvement by genome editing. Cell 2017;171:470–480.e8. DOI: 10.1016/j.cell.2017.08.030 [DOI] [PubMed] [Google Scholar]
- 7.Xing HL, Dong L, Wang ZP, et al. . A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 2014;14:32–7.. DOI: 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science 2013;339:823–826. DOI: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cermak T, Curtin SJ, Gil-Humanes J, et al. . A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017;29:1196–1217. DOI: 10.1105/tpc.16.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Zhang Q, Zhu Q, et al. . A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 2015;8:1274–1284. DOI: 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 12.Nissim L, Perli SD, Fridkin A, et al. . Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 2014;54:698–710. DOI: 10.1016/j.molcel.2014.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Liu B, Weeks DP, et al. . Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 2014;42:10903–10914. DOI: 10.1093/nar/gku806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A 2015;112:3570–3575. DOI: 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, East A, Cheng A, et al. . RNA-programmed genome editing in human cells. Elife 2013;2:e0047–1.. DOI: 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JF, Norville JE, Aach J, et al. . Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 2013;31:688–691. DOI: 10.1038/nbt.2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belhaj K, Chaparro-Garcia A, Kamoun S, et al. . Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 2015;32:76–84. DOI: 10.1016/j.copbio.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 18.Cermak T, Baltes NJ, Cegan R, et al. . High-frequency, precise modification of the tomato genome. Genome Biol 2015;16:23–2.. DOI: 10.1186/s13059-015-0796-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrenson T, Shorinola O, Stacey N, et al. . Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol 2015;16 DOI: 10.1186/s13059-015-0826-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Liang Z, Zong Y, et al. . Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 2016;7:1261–7.. DOI: 10.1038/ncomms12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Cheng X, Shan Q, et al. . Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 2014;32:947–951. DOI: 10.1038/nbt.2969 [DOI] [PubMed] [Google Scholar]
- 22.Upadhyay SK, Kumar J, Alok A, et al. . RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3:2233–2238. DOI: 10.1534/g3.113.008847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang Z, Chen K, Li T, et al. . Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 2017;8:1426–1.. DOI: 10.1038/ncomms14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil-Humanes J, Wang Y, Liang Z, et al. . High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 2017;89:1251–1262. DOI: 10.1111/tpj.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan Q, Wang Y, Li J, et al. . Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 2013;31:686–688. DOI: 10.1038/nbt.2650 [DOI] [PubMed] [Google Scholar]
- 26.Svitashev S, Schwartz C, Lenderts B, et al. . Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 2016;7:1327–4.. DOI: 10.1038/ncomms13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y, Chen L, Du LP, et al. . Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct Integr Genomics 2014;14:341–349. DOI: 10.1007/s10142-014-0380-5 [DOI] [PubMed] [Google Scholar]
- 28.Jaiswal V, Gahlaut V, Mathur S, et al. . Identification of novel SNP in promoter sequence of TaGW2-6A associated with grain weight and other agronomic traits in wheat (Triticum aestivum L.). PLoS One 2015;10:e0129400 DOI: 10.1371/journal.pone.0129400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmonds J, Scott P, Brinton J, et al. . A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor Appl Genet 2016;129:1099–1112. DOI: 10.1007/s00122-016-2686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su Z, Hao C, Wang L, et al. . Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.). Theor Appl Genet 2011;122:211–223. DOI: 10.1007/s00122-010-1437-z [DOI] [PubMed] [Google Scholar]
- 31.Yang Z, Bai Z, Li X, et al. . SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor Appl Genet 2012;125:1057–1068. DOI: 10.1007/s00122-012-1895-6 [DOI] [PubMed] [Google Scholar]
- 32.Nalam VJ, Alam S, Keereetaweep J, et al. . Facilitation of Fusarium graminearum infection by 9-lipoxygenases in Arabidopsis and wheat. Mol Plant Microbe Interact 2015;28:1142–1152. DOI: 10.1094/MPMI-04-15-0096-R [DOI] [PubMed] [Google Scholar]
- 33.Chari R, Mali P, Moosburner M, et al. . Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 2015;12:823–826. DOI: 10.1038/nmeth.3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai Z, Sooksa-nguan T, Vatamaniuk OK. Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol 2009;149:642–652. DOI: 10.1104/pp.108.130260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saintenac C, Zhang W, Salcedo A, et al. . Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 2013;341:783–786. DOI: 10.1126/science.1239022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi W, Zhu T, Tian Z, et al. . High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol 2016;16:5–8.. DOI: 10.1186/s12896-016-0289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.