Abstract
Treatment strategies for dominantly inherited disorders typically involve silencing or ablating the pathogenic allele. CRISPR-Cas nucleases have shown promise in allele-specific knockout approaches when the dominant allele creates unique protospacer adjacent motifs that can lead to allele-restricted targeting. Here, we present a spacer-mediated allele-specific knockout approach that utilizes both SpCas9 variants and truncated single-guide RNAs to achieve efficient discrimination of a single-nucleotide mutation in rhodopsin (Rho)-P23H mice, a model of dominant retinitis pigmentosa. We found that approximately 45% of the mutant P23H allele was edited at the DNA level and that the relative RNA expression of wild-type Rho was about 2.8 times more than that of mutant Rho in treated retinas. Furthermore, the progression of photoreceptor cell degeneration in outer nuclear layer was significantly delayed in treated regions of the Rho-P23H retinas at 5 weeks of age. Our proof-of-concept study therefore outlines a general strategy that could potentially be expanded to examine the therapeutic benefit of allele-specific gene editing approach to treat human P23H patients. Our study also extends allele-specific editing strategies beyond discrimination within the protospacer adjacent motif sites, with potentially broad applicability to other dominant diseases.
Introduction
Conventional gene augmentation therapies have shown great promise in treating recessive diseases, such as inherited retinal degenerations (IRDs) and hemophilia.1,2 Yet, their application to dominant diseases is limited due to the requirement for silencing or ablating the gain-of-function or dominant-negative mutant alleles. While alternative RNA-based suppression and replacement using ribozyme, zinc-finger–based artificial transcription factors or RNAi-based approaches to silence or degrade both mutated and normal RNA transcripts transiently in combination with supplementation of an exogenous wild-type transgene have been actively pursued, the allelic specificity, longevity of the therapy, and regulation of gene expression levels have posed significant challenges for broad application.3–6 Recently developed CRISPR-Cas9* gene editing technologies present an alternative approach to treat dominant diseases by addressing these challenges.
The CRISPR-Cas system has been revolutionary for genome-editing because the Cas9 nuclease can be programmed to generate double-strand DNA breaks at specified genomic sites when targeted by a single-guide RNA (sgRNA),7 which dictates much of the target site specificity.8 The Cas9–sgRNA complex scans the genome for the presence of a protospacer adjacent motif (PAM), the critical first step of DNA target site recognition.8 Recent studies in mice have shown that when the disease-causing allele harbors unique PAM sequences that are not present in its wild-type counterpart, allele-specific genome editing by selective targeting and permanent inactivation of the mutant allele could be achieved while leaving the wild-type allele functionally intact.9–11 However, many dominant alleles do not carry sequences that generate unique PAM sites, making them refractory to this type of allele-specific editing strategy. One potential approach to expand the applicability of allele-specific genome editing is to develop strategies in which the disease-causing allele can be discriminated from the wild-type allele when the mutations are within the spacer region of the Cas9–sgRNA target site. It is a widely recognized challenge that Streptococcus pyogenes Cas9 (SpCas9) is generally ineffective at distinguishing between single nucleotide mismatches in much of its spacer sequence.12–15 However, improvements to SpCas9 targeting specificity by truncating the sgRNA may be leveraged to overcome this hurdle.16 Furthermore, recent studies in human induced pluripotent stem cells (hiPSCs) and rat embryos have provided some encouraging evidence supporting the use of mismatches in the spacer sequence to achieve allele-specific genome editing,17–19 including a most recent report of allele-specific editing against a human Rhodopsin-P23H allele in hiPSCs.20 In the present study, we investigated the feasibility of using this spacer-mediated allele-specific CRISPR-Cas9 gene editing to target an endogenous single-base missense mutation at its native locus in a Rhodopsin-P23H knockin mouse model that genetically and phenotypically recapitulates human autosomal dominant retinitis pigmentosa (adRP).21,22
Retinitis pigmentosa (RP) is the most common IRD, affecting 1:3,500 people.23 Vision loss in IRDs is caused by progressive photoreceptor cell dysfunction and death.23 Mutations in 23 genes have been reported to cause adRP,24 including >180 mutations in the rhodopsin (RHO) gene, which accounts for >25% of adRP cases.25,26 Approximately half of the RHO-associated adRP cases are caused by the P23H mutation.25,26 The mutant P23H rhodopsin protein is thought to misfold and co-aggregate with wild-type rhodopsin, resulting in a gain-of-function or dominant negative effect in rod photoreceptor cells.3,6 In this study, by designing and testing combinations of sgRNAs and Cas9 variants, we have demonstrated effective and specific knockout of the mutant P23H allele in the retina of Rho-P23H heterozygous mice. In treated mice, we observed a significant decrease of the expression of mutant P23H transcript, which in turn led to preservation of the thickness of photoreceptor cell layer in the treated region of the retina. The proof-of-concept presented in this study supports the notion that the spacer-mediated allele-specific CRISPR-Cas9 gene editing approach may be applicable not only to RHO-P23H associated adRP but also to other dominant disorders.
Materials and Methods
Mouse husbandry
Our study conforms to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. All procedures were approved by the Animal Care and Use Committee of the Massachusetts Eye and Ear Infirmary. The Rho-P23H mouse line was purchased from Jackson Laboratories (Bar Harbor, ME; stock no. 017628) and maintained on a 12 h:12 h light/dark cycle. F1 heterozygous progeny were generated by crossing P23H homozygous mice with wild-type mice, and F2 progeny of wild-type, heterozygous, and homozygous mice were generated by intercrossing F1 heterozygous mice.
Plasmid constructs
Plasmids encoding pCAG-SpCas9-VQR-P2A-EGFP, pCAG-SpCas9-VRQR-P2A-EGFP, and pCAG-KKH-SaCas9-P2A-EGFP sequences were generated via isothermal assembly. The Cas9 expression constructs also encode an EGFP gene separated from the nuclease by a P2A sequence to facilitate the identification and sorting of transfected cells. Oligonucleotides to clone sgRNAs were synthesized by Integrated DNA Technologies (Coralville, IA), annealed, and ligated into BsmBI digested BPK2660 (Addgene #70709) or BPK1520 (Addgene #65777) for SaCas9 and SpCas9 sgRNAs, respectively.
Subretinal injection
P0–P2 pups were anesthetized by hypothermia, and plasmid DNAs were injected into the subretinal space using standard methods.27 Briefly, 0.5 μL of DNA solution containing Cas9-encoding plasmid with or without the sgRNA bearing plasmid (absolute amount of DNA is 0.6 μg for Cas9 and 0.3 μg for sgRNA) was injected using a 34G Hamilton syringe, followed by in vivo electroporation using five 90 V, 50 ms square pulses that were delivered with an Electro Square Porator (BTX, Holliston, MA).
Retina dissociation and cell sorting
Eyes were enucleated at P5–P7 days for DNA extraction or at P14 days for RNA isolation. Retinas were isolated in BGJB medium (Thermo Fisher Scientific, Waltham, MA) on ice under a fluorescent dissection microscope to document the transfected region, and then dissociated into single cells by incubation in Solution A containing 1 mg/mL of pronase (Sigma–Aldrich, St. Louis, MO) and 2 mM of EGTA in BGJB medium at 37°C for 20 min. Solution A was gently removed, followed by adding an equal amount of Solution B containing 100 IU/mL of DNase I (New England Biolabs, Cambridge, MA), 0.5% bovine serum albumin, and 2 mM of EGTA in BGJB medium. Cells were collected and re-suspended in 1× phosphate-buffered saline, filtered through a Cell Strainer (BD Biosciences, San Jose, CA), and submitted for fluorescence-activated cell sorting. Enhanced green fluorescent protein (EGFP)-positive and -negative cells were collected using a Cytomation MoFlo Cell Sorter (Cytomation, Fort Collins, CO).
Genomic DNA extraction and polymerase chain reaction
Genomic DNA was extracted from sorted cells using DNA Extraction solution (Epicenter, Madison, WI), and 100 ng of genomic DNA was used as polymerase chain reaction (PCR) template to amplify the sequence flanking the sgRNAs' on-target and predicted top 10 off-target sequences. Primer sets are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/crispr).The PCR amplicons were then subjected to next-generation sequencing (NGS) analysis.
RNA extraction and reverse transcription PCR
Total RNA from sorted EGFP positive and negative cells at P14 was extracted using the RNeasy Micro Kit (Qiagen, Hilden, Germany). Total RNA was reverse transcribed with the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, Rockford, IL). cDNA was PCR amplified using the condition described above and subjected to NGS analysis.
Targeted deep sequencing
The on- and off-target activities of Cas9–sgRNA pairs used in this study were evaluated by targeted deep sequencing. Briefly, PCR or reverse transcription (RT)-PCR amplicons of the target site or predicted off-target sites from treated and untreated cells were analyzed by NGS using the primers listed in Supplementary Table S1. Paired-end sequencing of the resulting TruSeq-compatible paired-end Illumina libraries was performed on the Illumina MiSeq platform. Sequencing data were analyzed using the CRISPResso software followed by a custom program that corrects nonsense single nucleotide polymorphisms (SNPs) due to NGS errors and consolidates indel counts.28 Allele frequencies from treated retinas and untreated heterozygous control retinas were analyzed via calculating the percentage of paired reads for each of the wild-type, P23H, or edited alleles.
Immunostaining and microscopy
Eyes from Cas9–sgRNA treated mice were isolated and processed for retinal immunostaining experiments 5 weeks post injection, as described previously.29 Retinal cryosections were cut and evaluated using an anti-rhodopsin antibody at 1:1,000 dilution (MAB5356; EMD Millipore, Billerica, MA), followed by Alexa 555-conjugated secondary antibodies. Images were taken using the Eclipse Ti fluorescence microscope. Outer nuclear layer (ONL) thickness of the EGFP-positive region and the corresponding region in the control retinas were measured.
Statistical analysis
All data are presented as mean ± standard deviation. A two-tailed unpaired t-test was used for data analysis. Statistical significance was considered at p < 0.05.
Results
Design and allele-specificity screening of sgRNAs for targeting the Rho-P23H allele in mouse retina
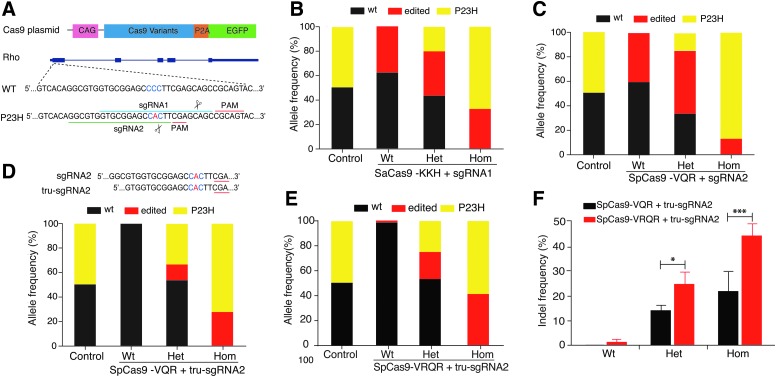
The endogenous sequence surrounding the mouse Rho-P23H mutation does not contain any canonical NGG PAM sites for wild-type SpCas9. Therefore, for an alternative approach, we designed two sgRNAs (sgRNA1 and sgRNA2) against sites with non-canonical NNCAGT and NGA PAMs that are accessible when using engineered SaCas9-KKH and SpCas9-VQR nucleases, respectively (Fig. 1A).30,31 For these two target sites, the P23H mutation is located either 12 or 4 base pairs (bp) upstream of the PAM in the sgRNA1 or sgRNA2 target sites, respectively. To determine the cleavage efficiency and allele-specificity of the sgRNAs along with their compatible Cas9 variants, DNA plasmids were transfected into the retinas of wild-type, heterozygous and homozygous P23H mice at age P0–P2 via subretinal injection and in vivo electroporation, followed by NGS analysis of the genomic DNA at 5–7 days post injection. The transfection efficiency was approximately 1–3% on average, measured by the percentage of EGFP-positive cells compared to all retinal cells. Targeted deep sequencing analysis of PCR products amplified from the Cas9–gRNA transfected cells revealed robust cutting efficiency; however, neither nuclease–sgRNA pair was able to distinguish the mutant P23H allele from the wild-type Rho allele (Fig. 1B and C). Cutting efficiencies of 37.8% and 40% were observed for the SaCas9-KKH/sgRNA1 and SpCas9-VQR/sgRNA2 pairs, respectively, in the injected wild-type mice, even though both nucleases preferentially targeted the mutant allele in heterozygous mice (Fig. 1B and C and Supplementary Table S2). As expected, the frequencies of wild-type and P23H alleles in the control heterozygous retinas were approximately 50% each when evaluated by NGS analysis (Fig. 1B–E).
FIG. 1.
P23H allele-specific single-guide RNA (sgRNA) screening in vivo. (A) Schematic overview of CRISPR-Cas9 constructs and their target sites in the Rho-P23H allele. Cas9 construct (top panel). Protospacer adjacent motif (PAM) sequence is underlined. (B–D) Allele frequencies of wild-type (wt), P23H, and edited alleles in injected wt, P23H heterozygous (het) and homozygous (hom) mice, and uninjected het mice (control). Allele frequencies were determined by the percentage of paired reads detected by next-generation sequencing (NGS). (B) sgRNA1 with SaCas9-KKH; (C) sgRNA2 with SpCas9-VQR; (D) tru-sgRNA2 + SpCas9-VQR; (E) tru-sgRNA2 + SpCas9-VRQR. (F) Comparison of targeting efficiency of SpCas9-VQR versus SpCas9-VRQR combined with tru-sgRNA2 in wt, het, and hom mice determined by NGS. *p < 0.05; ***p < 0.001 (n = 3).
Given that both conventional full-length sgRNA1 and sgRNA2 were not capable of allele-specific editing, we therefore examined whether a truncated 17 nt version of sgRNA2(tru-sgRNA2) bearing a shortened 5′ end could improve allele discrimination. This approach has been shown to increase specificity in a previous study.16 When paired with tru-sgRNA2, SpCas9-VQR exclusively edited the P23H mutant allele with a cleavage efficiency of 28% in the homozygous mutant retinas and no detectable cleavage in the wild-type controls (Fig. 1D and Supplementary Table S2).
Improvement of cleavage efficiency of the P23H allele in vivo
To potentially improve on-target P23H allele editing efficiency, we next tested tru-sgRNA2 with SpCas9-VRQR, an improved version of SpCas9-VQR that has enhanced activity against NGA PAM sites (Fig. 1E).32 On-target editing of the P23H allele with SpCas9-VRQR and tru-sgRNA2 (44.8 ± 4.8%) was significantly improved relative to that of SpCas9-VQR (22.1 ± 8.1%; p < 0.001) in homozygous mice, with a very low level of nuclease-induced editing of the wild-type allele (1.3 ± 0.3%) in wild-type mice (Fig. 1E and F). These data demonstrate that tru-sgRNA2 paired with SpCas9-VRQR cleaves the P23H allele with greater efficiency. Thus, this nuclease/guide pair was used in subsequent experiments.
Indel profiles analysis of the edited P23H allele
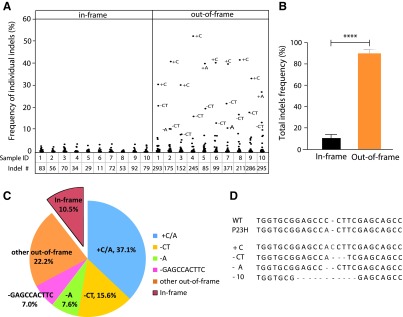
To assess whether the insertion or deletion mutations (indels) created by the activity of SpCas9-VRQR and tru-sgRNA2 could lead to the knockout of mutant P23H allele, we analyzed the indel profiles of the edited P23H alleles across 10 independent groups of mice that were treated on different days. We observed that 89.5 ± 3.3% of the indels at the targeted P23H mutation site were frameshifts (Fig. 2B and C and Supplementary Fig. S1). These frameshift indels are expected to create a premature stop codon at protein position 81 aa when the indel was 3n + 1 bp insertion or –(3n + 2) bp deletion or position 142 aa when the indel was 3n + 2 bp insertion or –(3n + 1) bp deletion, respectively. The frameshift indels are predicted to result in nonsense-mediated decay of the edited P23H transcripts.33 Moreover, consistent with a previous report, the indel pattern observed was nonrandom,34 as the top two most frequent indels across 10 different experimental replicates were a 1 bp insertion (C or A) and 2 bp deletion (–CT), accounting for 50.2 ± 4.7% of all indels (Fig. 2A, C, and D).
FIG. 2.
Indel profile of tru-sgRNA2 with SpCas9-VRQR. (A) Distribution and frequency of individual in-frame (left) and out-of-frame (right) indels from P23H het mice injected with SpCas9-VRQR and tru-sgRNA2 in 10 independent groups of mice. The total number of indel patterns for each sample is listed at bottom. (B) Average percentage of in-frame versus out-of-frame indels. ***p < 0.001 (n = 10). (C) Average percentage of top five and other out-of-frame and in-frame indels. (D) Representative sequences of top five indel patterns.
Decreased expression level of P23H mutant mRNA in CRISPR-Cas9-treated cells
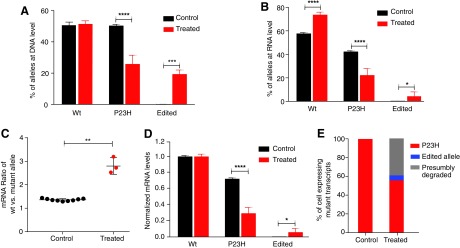
We next determined whether the indels introduced by SpCas9-VRQR/tru-sgRNA2 at the P23H DNA level could influence the expression level of mutant P23H mRNA via non-sense mediated decay. We performed targeted deep sequencing of RT-PCR products to compare the amount of mRNA transcripts of wild-type, P23H, and edited alleles in heterozygous retinas 14 days after transfection, at which time the expression of rhodopsin is easily detectable. At the DNA level, the percentage of reads for wild-type, P23H, and edited alleles were 51.2 ± 2.4%, 25.7 ± 5.0%, and 22.8 ± 2.9%, respectively, in the treated cells (Fig. 3A). A comparison at the relative mRNA levels demonstrated that the percentage of wild-type transcript was significantly higher in treated cells versus untreated cells (75.5 ± 2.3% vs. 57.6 ± 0.9%; Fig. 3B), and the percentage of P23H transcript was significantly lower in the treated cells versus the control samples (22.1 ± 5.8% vs. 42.1 ± 0.8%; Fig. 3B). Notably, the relative percentage of edited allele was reduced from 22.8 ± 2.9% at DNA level to only 4.8 ± 4.2% at mRNA level in the treated cells (Fig. 3B). Sequence analysis also revealed that the majority of the edited alleles detected at mRNA levels were in-frame indels (data not shown). Thus, the ratio of wild-type mRNA versus mutant mRNA was significantly increased in the treated cells (2.8 ± 0.35) compared to that in the control cells (1.37 ± 0.05; Fig. 3C). When mRNA levels were normalized to the wild-type allele, the ratio of wild-type versus mutant mRNA was 1:0.35 in the treated cells and 1:0.73 in the untreated cells (Fig. 3D), indicating a 38% decrease of the mutant mRNA in the treated cells (Fig. 3E). This result is consistent with our finding at the DNA level that 89.5% of edited alleles harbor an out-of-frame indel. As the majority of the aberrant transcripts of mutant alleles with out-of-frame indels are predicted to be removed via nonsense mediated decay, this suggests that of all cells transfected with SpCas9-VRQR/tru-sgRNA2, presumably 38% of them express only wild-type rhodopsin mRNA.
FIG. 3.
Expression of P23H allele after gene editing with tru-sgRNA2 and SpCas9-VRQR in P23H heterozygous mice. Relative DNA level (A) and RNA level (B) of wt, P23H, and edited alleles in the treated retinas (n = 3) with tru-sgRNA2 and SpCas9-VRQR versus untreated retinas (n = 9) in P23H het mice 14 days post injection. (C) The ratio of wt to mutant (P23H + edited) allele at mRNA levels in the treated (n = 3) and untreated retinas (n = 9). (D) Normalized mRNA levels of wt, P23H, and edited alleles in the treated (n = 3) and untreated retinas (n = 9). (E) Percentage of cells presumably expressing wt/P23H, wt/edited P23H, and wt only in the treated and untreated retinas. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Photoreceptor cell preservation in treated Rho-P23H mouse retinas
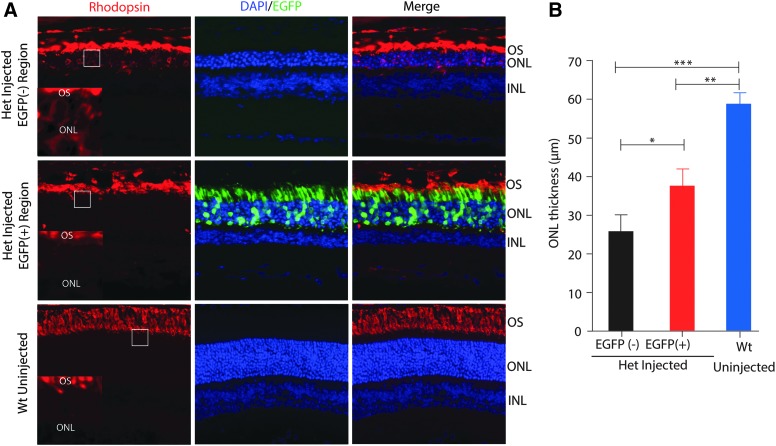
In order to gauge whether ablation of the mutant P23H allele could result in prevention or delay of photoreceptor degeneration, we evaluated the retinal structure in the regions transfected with SpCas9-VRQR/tru-sgRNA2 plasmids in Rho-P23H heterozygous mice at 5 weeks of age, at which time the retinas of P23H mice demonstrate significant degeneration histologically and functionally.21,22 Immunofluorescence analysis showed that within the transfected region of retina, approximately 30% of photoreceptor cells were treated with Cas9 components, as counted by the EFGP-positive cells over all cells stained with DAPI in ONL (Fig. 4A). Measurements of ONL thickness revealed that there were significantly more photoreceptor cells in the treated EGFP positive region (five to six rows of cells, 37.3 ± 3.7 μm) than in the adjacent untreated EGFP-negative area (three to four rows of cells, 25.9 ± 3.9 μm; p < 0.05; Fig. 4A and B). Also, we observed that the rhodopsin signal in the ONL was less intense in the GFP-positive region than that in the GFP-negative region (Fig. 4A, left panels, insets). These results suggest that CRISPR-Cas9-mediated disruption of the P23H mutant RHO allele resulted in photoreceptor cell preservation in the treated retinas.
FIG. 4.
Photoreceptor cell preservation after gene editing with tru-sgRNA2 and SpCas9-VRQR in P23H heterozygous mice. (A) Retinal images from the treated region (enhanced green fluorescent protein [EGFP] positive) and adjacent untreated region (EGFP negative) in the P23H het mice at 5 weeks post injection. Wild-type uninjected retina was used as control (bottom). Rhodopsin expression was detected by a rhodopsin antibody (red), the EGFP signal indicates transfection (green), and nuclei were counterstained with Hoechst (blue). OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer. Insets at the bottom left corner of each rhodopsin panel were high magnification images of the regions outlined in white. (B) Comparison of ONL thickness of the treated (EGFP positive) area to its adjacent untreated (EGFP negative) central area and to uninjected wild-type retinas (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001 (n = 3).
Evaluation of off-target activity
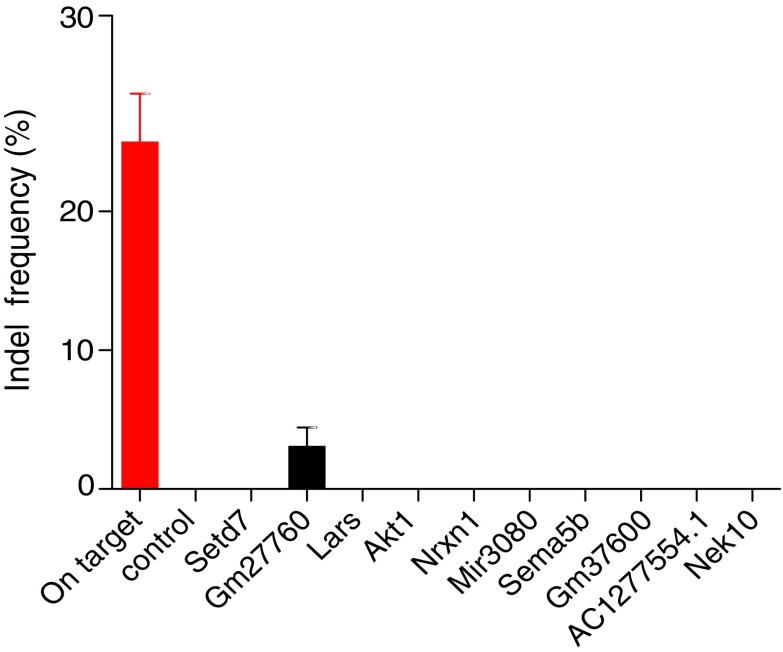
To evaluate whether tru-sgRNA2 together with SpCas9-VRQR would cause unwanted off-target cleavage in the mouse genome, we utilized targeted deep sequencing to assess off-target cutting at the top 10 potential tru-sgRNA2 binding sites out of 2,074 identified using CCTop (listed in Supplementary Table S3).35 NGS analysis showed no detectable off-target activity for 9/10 candidate sites (Fig. 5), each of which harbors between two and four mismatches compared to the on-target P23H sequence (Supplementary Table S3). Low off-target activity (3.12 ± 2.28%) was detected at an off-target site which harbors a 1 bp mismatch at position −11 from the PAM, in the region of a predicted miRNA, Gm27760 (Fig. 5).
FIG. 5.
Off-target evaluation of tru-sgRNA2 with SpCas9-VRQR in vivo. Off-target analysis was conducted at the in silico predicted potential top 10 off-target sites. Injected retinas were collected and analyzed by PCR followed by targeted deep sequencing 7 days post injection. Targeting activities on the P23H site (on-target) and 10 off-target sites were determined by NGS reads of polymerase chain reaction amplicons. Data are represented as mean ± standard deviation (n = 3). Control: uninjected mice.
Discussion
Allele-specific CRISPR-Cas9 genome editing provides a promising means to treat dominantly inherited diseases. Here, we demonstrated that an in vivo allele-specific genome editing approach based on sequence differences in the spacer region of the sgRNA can selectively inactivate a single-base missense dominant allele at its native locus in the RHO-P23H knock-in mouse model of adRP. By allowing for targeting of mutations that occur outside of PAM sequences, this approach significantly broadens the potential application of allele-specific CRISPR-Cas9 genome editing for the treatment of dominant genetic diseases.
We accomplished allele-specific targeting using a truncated sgRNA, along with an SpCas9 variant designed to target an alternative PAM sequence. Although the targeting specificity of CRISPR-Cas9 is largely determined by the 10–12 bp PAM-proximal seed sequences of sgRNAs,14,36–38 it is notable that single mismatches within the seed sequence, even at the intended break point, as in the case of sgRNA2, may not provide sufficient allele-specific targeting. This finding indicates that this spacer-mediated allele-specific knockout strategy works in a site-dependent manner. Thus, empirical evaluation of each individual allele-specific knockout approach is required.
It is particularly encouraging that, despite the inability of the 20 nt sgRNA2 to exhibit allele specificity, a truncated 17 nt version of sgRNA2 paired with SpCas9-VQR provided exclusive allele specificity in this study. It is also of interest that in comparison to SpCas9-VQR, the SpCas9-VRQR nuclease provided an approximately twofold increase in on-target activity on the mutant allele, albeit with an increase in the targeting of the wild-type allele from 0% to 1.3 ± 0.3%. Notably, this low-level disruption of the wild-type rhodopsin allele did not abrogate the observed therapeutic benefit, as evidenced by the significant increase of the relative levels of wild-type versus mutant Rho mRNA in the treated cells (Fig. 3). This finding is consistent with previous observation that an increased ratio of normal versus mutant protein may be therapeutically beneficial for dominant diseases caused via a dominant negative mechanism.39 Additional studies will be needed to seek the appropriate balance between cutting efficiency of the mutant allele and the level of modification of the normal allele.
It is worth mentioning that the allele-specific CRISPR-Cas9 gene knockout approach can be designed not only to target the mutation alone, but also can be broadened to common or rare SNPs that are in cis with the mutation. For example, the common SNPs that represent a given haplotype can all be used for allele-specific knockout of mutant alleles caused by different mutations,10 which could make the allele-specific therapeutic strategy more generic and cost-effective. Moreover, as the expansion of the CRISPR-Cas toolbox continues, it may be possible to achieve highly efficient and allele-specific knockout of most, if not all, human dominant alleles via a similar allele-specific editing approach.
The potential off-target effects at unintended sites raises safety concerns for translating CRISPR-Cas9-based genome editing therapies to humans. In this proof-of-concept study, we performed off-target analysis for 10 in silico predicted sites in the mouse genome and found a low-level off-target activity in a predicted miRNA Gm27760. No information regarding the function of this predicted miRNA is available to date. Since the tru-sgRNA2 cannot be translated to human use due to sequence mismatches between species, we elected not to study the effect of off-target activity at the Gm27760 site further. Rather, we anticipate more comprehensive studies of potential off-target effects associated with the development of a human specific P23H Cas9–sgRNA system, which we believe is warranted based on the results of this and other studies.20,40 Furthermore, multiple means with the potential to improve targeting specificity, such as truncated gRNAs,16 Cas9 nickases,41,42 and the more recently engineered Cas9s (eSpCas9,43 SpCas9-HF1,32 and HypaCas915) can be explored to minimize off-target activity.
Our study demonstrates that the allele-specific knockout of the mutant P23H allele in mouse photoreceptor cells results in a significant reduction of the mutant Rho-P23H mRNA in the transfected cells, which in turn leads to local preservation of photoreceptor cells in the treated regions of Rho-P23H retinas. Two recent studies have reported successful CRISPR-Cas9-mediated cleavage of a human RHO-P23H allele in mouse and pig transgenic models with similar phenotypic rescue observed in the hRHO-P23H transgenic mice.20,40 Although encouraging, the disruption of the human RHO-P23H transgene was achieved by exploiting the sequence differences between the human transgene and the endogenous allele in the host animals.20,40 Thus, further investigation is required to validate the allele-specificity of the human RHO Cas9/sgRNA systems thoroughly, including the SaCas9/sgRNA used in previous studies20,40 and the corresponding SpCas9-VQR/tru-sgRNA2 system used in this study, in a heterozygous P23H human cell line or a humanized knock-in animal model harboring both a human P23H and a normal hRHO gene. It should also be noted that the therapeutic efficacy in our study was restricted by the delivery approach using plasmid DNA and in vivo electroporation. We are hopeful that other more relevant and effective in vivo delivery approaches, such as the use of dual adeno-associated virus vectors encoding the Cas9 and sgRNAs separately, will result in treatment of more photoreceptor cells. This dual-vector approach has been successfully applied to the delivery of Cas9–sgRNA-mediated genome editing components into the mouse retina.44–47
Conclusion
Our proof-of-concept study demonstrates that an allele-specific CRISPR-Cas9 editing approach reduces photoreceptor cell death in Rho-P23H knock-in mice by selectively targeting the endogenous mutant P23H allele. Our study also provides evidence that spacer-mediated allele-specific genome editing approaches are feasible for the treatment of dominant genetic diseases.
Supplementary Material
Footnotes
Clustered Regularly Interspaced Short Palindromic Repeats.
Acknowledgments
We are grateful to all the members in the Ocular Genomics Institute for their inputs and technical assistance in this study. Special thanks to Drs. Jason Comander and Xiaowu Gai for their critical inputs in the NGS data analyses. This study is supported by grants from the Fighting Blindness Foundation (Q.L.); Research to Prevent Blindness Foundation (Q.L.); the National Eye Institute RO1 EY012910 (E.A.P., Q.L.) and P30 EY014104 (MEEI core support); Banting (Natural Sciences and Engineering Research Council of Canada) (B.P.K.); a Charles A. King Trust Postdoctoral Fellowship (B.P.K.); NIH R35 GM118158 (B.P.K., M.S.P., and J.K.J.); and a Desmond and Ann Heathwood MGH Research Scholar Award (J.K.J.).
Author Disclosure Statement
B.P.K. is a consultant for Avectas. J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Monitor Biotechnologies, Pairwise Plants, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. No competing financial interests exist for the remaining authors.
References
- 1.Nienhuis AW, Nathwani AC, Davidoff AM. Gene therapy for hemophilia. Mol Ther 2017;25:1163–1167. DOI: 10.1016/j.ymthe.2017.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell S, Bennett J, Wellman JA, et al. . Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, Phase 3 trial. Lancet 2017;390:849–860. DOI: 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaVail MM, Yasumura D, Matthes MT, et al. . Ribozyme rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc Natl Acad Sci U S A 2000;97:11488–11493. DOI: 10.1073/pnas.210319397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang L, Zhang H, Dizhoor AM, et al. . Long-term RNA interference gene therapy in a dominant retinitis pigmentosa mouse model. Proc Natl Acad Sci U S A 2011;108:18476–18481. DOI: 10.1073/pnas.1112758108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botta S, Marrocco E, de Prisco N, et al. . Rhodopsin targeted transcriptional silencing by DNA-binding. Elife 2016;5:e1224–2.. DOI: 10.7554/eLife.12242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mussolino C, Sanges D, Marrocco E, et al. . Zinc-finger-based transcriptional repression of rhodopsin in a model of dominant retinitis pigmentosa. EMBO Mol Med 2011;3:118–128. DOI: 10.1002/emmm.201000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science 2013;339:823–826. DOI: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262–1278. DOI: 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malkki H. Huntington disease: selective deactivation of Huntington disease mutant allele by CRISPR-Cas9 gene editing. Nat Rev Neurol 2016;12:614–615. DOI: 10.1038/nrneurol.2016.151 [DOI] [PubMed] [Google Scholar]
- 10.Shin JW, Kim KH, Chao MJ, et al. . Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 2016;25:4566–4576. DOI: 10.1093/hmg/ddw286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtney DG, Moore JE, Atkinson SD, et al. . CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther 2016;23:108–112. DOI: 10.1038/gt.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Foden JA, Khayter C, et al. . High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–826. DOI: 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PD, Scott DA, Weinstein JA, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832. DOI: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattanayak V, Lin S, Guilinger JP, et al. . High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013;31:839–843. DOI: 10.1038/nbt.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JS, Dagdas YS, Kleinstiver BP, et al. . Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017;550:407–410. DOI: 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Sander JD, Reyon D, et al. . Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 2014;32:279–284. DOI: 10.1038/nbt.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith C, Abalde-Atristain L, He C, et al. . Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther 2015;23:570–577. DOI: 10.1038/mt.2014.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto Y, Makiyama T, Harita T, et al. . Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum Mol Genet 2017;26:1670–1677. DOI: 10.1093/hmg/ddx073 [DOI] [PubMed] [Google Scholar]
- 19.Yoshimi K, Kaneko T, Voigt B, et al. . Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat Commun 2014;5:424–0.. DOI: 10.1038/ncomms5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burnight ER, Gupta M, Wiley LA, et al. . Using CRISPR-Cas9 to generate gene-corrected autologous iPSCs for the treatment of inherited retinal degeneration. Mol Ther 2017;25:1999–2013. DOI: 10.1016/j.ymthe.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakami S, Maeda T, Bereta G, et al. . Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem 2011;286:10551–10567. DOI: 10.1074/jbc.M110.209759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakami S, Kolesnikov AV, Kefalov VJ, et al. . P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Hum Mol Genet 2014;23:1723–1741. DOI: 10.1093/hmg/ddt561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006;368:1795–1809. DOI: 10.1016/S0140-6736(06)69740-7 [DOI] [PubMed] [Google Scholar]
- 24.RetNet. www.sph.uth.tmc.edu/Retnet/ (last accessed February5, 2018)
- 25.Sung CH, Davenport CM, Hennessey JC, et al. . Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A 1991;88:6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dryja TP, McGee TL, Hahn LB, et al. . Mutations within the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. N Engl J Med 1990;323:1302–1307. DOI: 10.1056/NEJM199011083231903 [DOI] [PubMed] [Google Scholar]
- 27.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A 2004;101:16–22. DOI: 10.1073/pnas.2235688100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinello L, Canver MC, Hoban MD, et al. . Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol 2016;34:695–697. DOI: 10.1038/nbt.3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Q, Lyubarsky A, Skalet JH, et al. . RP1 is required for the correct stacking of outer segment discs. Invest Ophthalmol Vis Sci 2003;44:4171–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinstiver BP, Prew MS, Tsai SQ, et al. . Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481–485. DOI: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleinstiver BP, Prew MS, Tsai SQ, et al. . Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol 2015;33:1293–1298. DOI: 10.1038/nbt.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinstiver BP, Pattanayak V, Prew MS, et al. . High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490–495. DOI: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 2004;16:293–299. DOI: 10.1016/j.ceb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 34.van Overbeek M, Capurso D, Carter MM, et al. . DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol Cell 2016;63:633–646. DOI: 10.1016/j.molcel.2016.06.037 [DOI] [PubMed] [Google Scholar]
- 35.Stemmer M, Thumberger T, Del Sol Keyer M, et al. . CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 2015;10:e012463–3.. DOI: 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Bikard D, Cox D, et al. . RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013;31:233–239. DOI: 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M, Chylinski K, Fonfara I, et al. . A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821. DOI: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao H, James T, Jr., Schwein A, et al. . AAV delivery of wild-type rhodopsin preserves retinal function in a mouse model of autosomal dominant retinitis pigmentosa. Hum Gene Ther 2011;22:567–575. DOI: 10.1089/hum.2010.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latella MC, Di Salvo MT, Cocchiarella F, et al. . In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids 2016;5:e38–9.. DOI: 10.1038/mtna.2016.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mali P, Aach J, Stranges PB, et al. . CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013;31:833–838. DOI: 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ran FA, Hsu PD, Lin CY, et al. . Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154:1380–1389. DOI: 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slaymaker IM, Gao L, Zetsche B, et al. . Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84–88. DOI: 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung SS, Chrysostomou V, Li F, et al. . AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest Ophthalmol Vis Sci 2016;57:3470–3476. DOI: 10.1167/iovs.16-19316 [DOI] [PubMed] [Google Scholar]
- 45.Ruan GX, Barry E, Yu D, et al. . CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber congenital amaurosis 10. Mol Ther 2017;25:331–341. DOI: 10.1016/j.ymthe.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu W, Mookherjee S, Chaitankar V, et al. . Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun 2017;8:1471–6.. DOI: 10.1038/ncomms14716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung SS, Li F, Wang JH, et al. . Methods for in vivo CRISPR/Cas editing of the adult murine retina. Methods Mol Biol 2018;1715:113–133. DOI: 10.1007/978-1-4939-7522-8_9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.