Abstract
Reactivating post-natal myocardial regeneration potential may be a feasible strategy to regenerate the injured adult heart. Long non-coding RNAs (lncRNAs) have been implicated in regulating cellular differentiation, but whether they can elicit a regenerative response in the post-natal heart remains unknown. In this study, by characterizing the lncRNA transcriptome in human hearts during the fetal-to-adult transition, we found that 3,092 lncRNAs were differentially expressed, and we further identified a novel upregulated fetal lncRNA that we called endogenous cardiac regeneration-associated regulator (ECRAR), which promoted DNA synthesis, mitosis, and cytokinesis in post-natal day 7 and adult rat cardiomyocytes (CMs). Overexpression of ECRAR markedly stimulated myocardial regeneration and induced recovery of cardiac function after myocardial infarction (MI). Knockdown of ECRAR inhibited post-natal day 1 CM proliferation and prevented post-MI recovery. ECRAR was transcriptionally upregulated by E2F transcription factor 1 (E2F1). In addition, ECRAR directly bound to and promoted the phosphorylation of extracellular signal-regulated kinases 1 and 2 (ERK1/2), resulting in downstream targets of cyclin D1 and cyclin E1 activation, which, in turn, activated E2F1. The E2F1-ECRAR-ERK1/2 signaling formed a positive feedback loop to drive cell cycle progression, and, therefore, it promoted CM proliferation. These findings indicated that our newly discovered ECRAR may be a valuable therapeutic target for heart failure.
Keywords: long non-coding RNA, myocardial regeneration, cell cycle, ERK signaling, cardiomyocyte proliferation
Graphical Abstract

Little is known about the role of long non-coding RNAs (lncRNAs) in post-natal myocardial regeneration. Chen et al. compared the lncRNA transcriptome between human fetal and adult hearts, and they identified an lncRNA (ECRAR) that promoted post-natal myocardial regeneration and attenuated aberrant post-infarction remodeling via ERK1/2 signaling.
Introduction
In response to ischemic injury, the adult human heart has a limited capacity for regeneration to compensate for lost myocardial tissue.1 The human heart typically responds to injury through a scarring mechanism rather than through myocardial regeneration, which results in adverse remodeling and subsequent progression to heart failure. Therefore, an understanding of the underlying mechanisms and ability to stimulate the regenerative capacity of the human heart is of the utmost scientific and clinical importance. The adult hearts of lower vertebrates, such as zebrafish and amphibians, have shown robust regenerative capacity.2 In mammals, fetal cardiomyocytes (CMs) can proliferate robustly with heart growth primarily occurring by hyperplasia. Until 7 days after birth, the neonatal rodent heart maintains a significant capacity for cardiac repair and regeneration after injury.3 It was recently reported that human newborns also show an intrinsic capacity to repair myocardial damage and recover cardiac function completely.4 However, the endogenous regeneration capacity of the mammalian heart is largely lost by 7 days after birth, and it remains absent throughout the lifespan.5 It is unclear why this robust regenerative response at birth is lost with age. Exploring the mechanisms that underlie this fetal-to-adult heart transition may reveal new therapeutic strategies to reactivate post-natal myocardial regeneration in the adult heart after injury.
Long non-coding RNAs (lncRNAs) are a novel class of transcripts, larger than 200 nt and with poor protein-coding potential, that can regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels.6 Increasing evidence has indicated that lncRNAs are differentially expressed across various tissues, diseases, and developmental stages and are key regulators in a wide range of biological processes, including cell proliferation, apoptosis, cell cycle control, and cell differentiation.6 lncRNAs are emerging as important players in heart development, heart failure, CM hypertrophy, and atherosclerosis.7 A recent study revealed that lncRNAs have tissue-specific expression, and the authors identified 321 cardiac-expressed lncRNAs and 52 cardiac-enriched lncRNAs in mouse.8 lncRNAs are also dynamically regulated in failing hearts with mechanical circulatory support, and they are regulated in peripheral blood cells of patients with acute myocardial infarction (MI).9 In addition, two studies demonstrated that the lncRNAs Fendrr and Braveheart (Bvht) are required for proper development of the heart and body walls and cardiovascular lineage commitment, respectively.10, 11 Furthermore, bioinformatic analysis of RNA sequencing (RNA-seq) data from both mice and humans revealed that lncRNA expression markedly differs between fetal and adult hearts, and it showed that lncRNA:cis-mRNA gene pairs are involved in heart growth and development.12, 13 Although these studies have implicated specific lncRNAs in heart development or failure, how lncRNA expression profiles may be altered during the human fetal-to-adult heart transition and the precise roles of lncRNAs in regulating post-natal myocardial regeneration have not been well characterized.
In this study, we characterized the human transcriptome expression change in the fetal-to-adult heart transition by analyzing publicly available RNA-seq data of human fetal (13th to 17th week of gestation) and normal adult cardiac tissues. Using an array of in vitro and in vivo approaches, we identified an upregulated fetal lncRNA that we called endogenous cardiac regeneration-associated regulator (ECRAR). We showed that ECRAR fostered rat myocardial regeneration in post-natal day 7 and adult rat hearts and attenuated post-infarction adverse remodeling. We further demonstrated that ECRAR was induced by E2F transcription factor 1 (E2F1) and that the downstream mechanism of post-natal myocardial regeneration triggered by ECRAR was through activating extracellular signal-regulated kinase 1 and 2 (ERK1/2) signaling. It is thus proposed that ECRAR may represent a promising therapeutic target for CM replacement in heart failure.
Results
Differentially Expressed lncRNAs between Fetal and Adult Hearts
The four RNA-seq datasets of fetal and adult human cardiac tissues generated 189 million clear reads, of which over 170 million (≥81.0%) were uniquely aligned to the human genome (hg19) (Figure S1; Table S1). Among the uniquely mapped reads, 87 million (51.0%) reads mapped to intergenic regions, 69 million (40.5%) reads mapped within exons, and 14 million (8.5%) reads mapped to introns (Figure 1A). The chromosome distribution of these mapped reads in fetal heart was similar to that in the adult heart (Figure S2A). In contrast, the proportions of reads mapped to introns and exons were remarkably different between fetal and adult hearts (Figure S2B). The clear reads were first aligned to the hg19 RefSeq. Reads that failed to be mapped were subsequently mapped to the Ensembl gene set, lncRNA database, and the reference genome, respectively. We identified 152,130 (70.9%) transcripts that were annotated to RefSeq genes, 33,073 (15.4%) were annotated to Ensembl genes, and 28,075 (13.1%) were annotated to NONCODE version (v.)4 genes (Figure 1B; Figure S2C). Compared to the proportion of lncRNAs found in the adult heart, lncRNAs accounted for a lower percentage of total genes in the fetal heart (Figure S2C). Among the 3,958 novel transcripts, 3,830 of the novel transcripts with low coding potential were identified as novel lncRNAs (Figure 1C). The novel and known lncRNAs were shorter and less abundant in length than coding genes (mRNA) (Figures 1D and 1E). Conservation analysis revealed that novel and known lncRNA exons were less conserved than coding exons, although introns and promoters were equally conserved (Figure 1F).
Figure 1.
Differentially Expressed Genes in Fetal and Adult Hearts
(A) Pie charts showing read count distributions of exons, introns, and intergenic regions. (B) Pie chart showing composition of RefGene mRNAs (purple), Ensembl mRNAs (blue), known long non-coding RNAs (lncRNAs) (green), and novel lncRNAs (red). (C) Kernel density plot displaying the coding potential of all novel transcripts. (D and E) Transcript length (D) and abundance (E) of mRNAs, known lncRNAs, and novel lncRNAs. (F) PhastCons score distribution of mRNAs, known lncRNAs, and novel lncRNAs. (G) Volcano plot of all coding RNAs. (H) Unsupervised hierarchical clustering of all differentially expressed mRNAs (left) and one representative gene module enriched for cell cycle-related genes (right). (I and J) Gene ontology (GO) enrichment analysis (I) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms (J) (y axis) of differentially expressed coding genes. (K) Volcano plot of highly abundant cardiac lncRNAs. (L and M) GO enrichment analysis of differentially expressed lncRNA:cis-mRNA pairs (L) and lncRNA:trans-mRNA pairs (M).
Globally, the cardiac transcriptome of mRNAs and lncRNAs correlated well within groups in the fetal group or adult heart group (Figure S2D). Volcano plots comparing fold change and probability identified 3,211 significantly increased and 578 significantly decreased mRNAs (Figure 1G). By performing hierarchical clustering of differentially expressed protein-coding RNAs, we found that one module with the most significant differentially expressed genes was related to cell cycle function (Figure 1H; Figure S3A). Gene ontology (GO) classification of upregulated genes in fetal hearts showed enrichment for gene categories that regulate the cell cycle, cell cycle phase, cell cycle process, and mitotic cell cycle, whereas downregulated genes were enriched in other functional categories (Figure 1I; Figures S3B and S3C; Table S2). Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis further revealed that upregulated genes were enriched in proliferation-related pathways, such as the cell cycle (Figure 1J).
We found 3,092 lncRNAs exhibited significant differences (probability >0.8) in expression between fetal and adult hearts, of which 1,343 lncRNAs were upregulated and 1,749 lncRNAs were downregulated in fetal hearts (Figure 1K; Figures S4A and S4B). To explore the potential cis- and trans-regulatory roles of human lncRNAs on protein-coding genes, Pearson correlation coefficients were used to analyze the expression levels of lncRNAs and nearby or distally coding genes. First, to evaluate cis-correlation of expression, we computed interactions between lncRNA-mRNA pairs located within 10 kb upstream and downstream. In total, 54.8% of tested upregulated lncRNA:cis-mRNA gene pairs showed a positive correlation, while 23.1% had a negative correlation (Figure S5A). In contrast, a greater proportion of downregulated lncRNA:cis-mRNA gene pairs were negatively correlated compared to that of those positively correlated (Figure S5B). An example of a cis-acting lncRNA is shown in Figure S5C. GO analysis indicated that both upregulated (r > 0.95) and downregulated (r > 0.95) lncRNA:cis-mRNA pairs were associated with the cell cycle and heart development (Figure 1L).
Next, to investigate the trans-correlation of expression, we examined the interactions of lncRNA-mRNA pairs located either at least 1 Mb apart or present on different chromosomes. We found a greater proportion of upregulated lncRNA:trans-mRNA pairs were positively correlated compared to that of those negatively correlated (Figure S5D), whereas downregulated lncRNA:trans-mRNA pairs showed more frequent negative correlations than positive correlations (Figure S5E). GO classification of upregulated lncRNA:trans-mRNA pairs showed enrichment for heart development, whereas downregulated lncRNA:trans-mRNA pairs showed enrichment for other GO functional categories, such as the glucose metabolic process (Figure 1M). These results indicate that the trans-acting lncRNAs are as prevalent as cis-acting lncRNAs in the human heart during the fetal-to-adult transition.
The mRNAs and lncRNAs encoded by mtDNA accounted for a much greater proportion in the adult heart than in the fetal heart (Figures S6A–S6C), which is a finding that indicates that adult hearts have great energy needs. The mitochondrial lncRNA transcript lengths were shorter than those of nonmitochondrial lncRNAs and mRNAs (Figure S6D). The expression of mitochondrial lncRNA transcripts was more abundant than nonmitochondrial lncRNAs and mRNAs, and mitochondrial lncRNAs were less abundant than mitochondrial mRNAs (Figures S6E–S6G). Hierarchical clustering revealed that both mitochondrial mRNAs and lncRNAs in the fetal heart have lower gene expression than those found in the adult heart (Figures S7A and S7B), whereas GO analysis revealed that mitochondrial genes were enriched in energy metabolism (Figure S7C). These findings indicate that the energy demands of the myocardium change from different development stages.
ECRAR Is Upregulated in the Fetal Heart and Is Potentially Involved in Post-natal Myocardial Regeneration
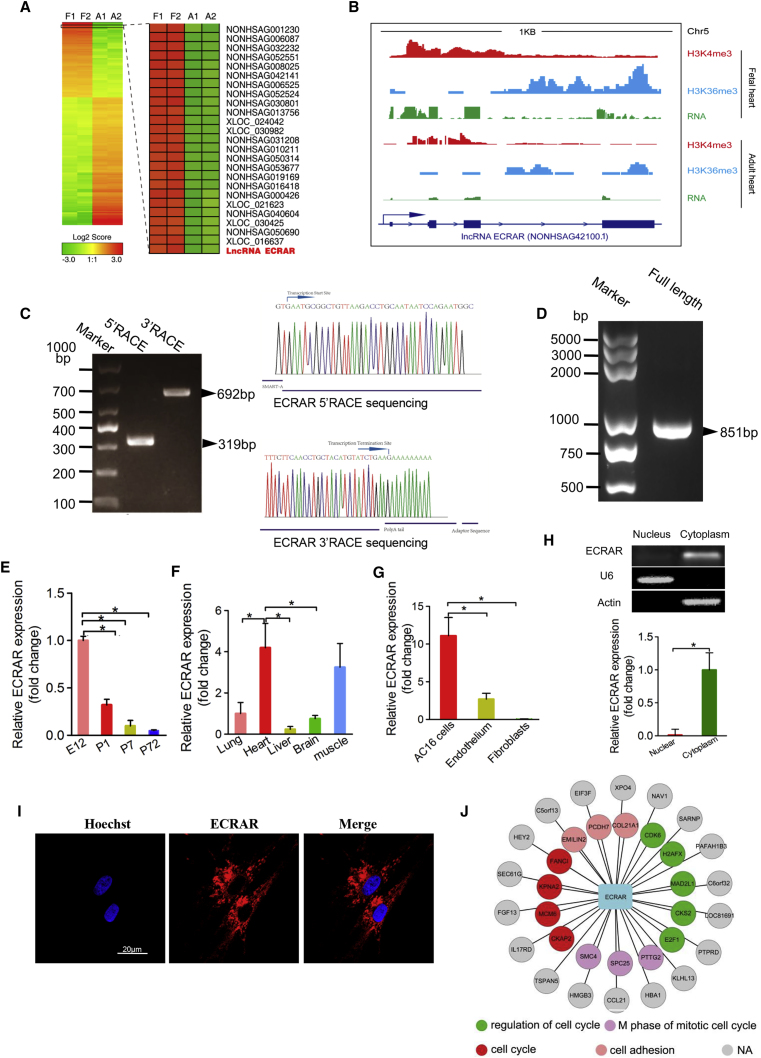
From our transcriptome analysis of the human fetal and adult hearts, we identified an lncRNA that was significantly upregulated in the fetal heart more than 12-fold (Figure 2A), and it matched NONCODE: NONHSAG042100.1 (http://www.noncode.org/) and Ensembl: ENST00000523659.5 (http://asia.ensembl.org/index.html?redirect=no). The gene contains four exons, has a poly (A) tail, and is located on chromosome 5 near pituitary tumor-transforming 1 (PTTG1) (Figure 2B). This transcript was an alternative splicing of PTTG1 gene due to retained intron, and it didn’t contain an open reading frame (ORF), which indicated it could not be translated into protein. Coding potential analysis using the Coding-Non-Coding Index tool (CNCI) (https://github.com/www-bioinfo-org/CNCI) indicated that ECRAR was a non-coding RNA (a value of −0.0414; if the value of CNCI < 0, transcripts were considered to be non-coding), which is similar to that of a well-known lncRNA Xist (value of −0.177). In addition, to evaluate whether there exists some possible coding sequence ability of this transcript, we searched the PDB (http://www.rcsb.org/) and tblastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) databases, and we found no hits by using default parameters.
Figure 2.
ECRAR Is Highly Expressed in Human Fetal Heart
(A) Cluster analysis identified upregulated lncRNAs in fetal heart; red indicates upregulation and green indicates downregulation. The spectrum spans fold changes from 3.0 to −3.0. Fetal tissues are shown on the left and normal adult heart tissues are shown on the right. (B) Read distributions of ECRAR (green) and scoring profiles from chromatin immunoprecipitation (ChIP)-seq of H3K4me3 (red) and H3K36me3 (blue) in both fetal and adult hearts. The respective structure of ECRAR is shown below (dark blue). Each data track shown has the same scale for both fetal and adult hearts. (C) A representative image (right) and sequencing (left) of PCR products from 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE, with the major PCR product indicated by an arrow. (D) Full length of lncRNA ECRAR from 5′ RACE and 3′ RACE. (E) ECRAR expression assayed by qRT-PCR in rat hearts (*p < 0.05 using one-way ANOVA; n = 5 per group). (F) ECRAR expression assayed by qRT-PCR in several different tissues of neonatal rat (*p < 0.05; n = 5 per group). (G) ECRAR expression levels in several cells (*p < 0.05 using one-way ANOVA; n = 5 per group). (H) ECRAR expression in nuclear and cytoplasmic fractions of CMs assayed by qRT-PCR (*p < 0.05 using t test; n = 4). Actin (cytoplasm) and U6 (nucleus) were used as loading controls. Histogram depicts relative ECRAR as fold change versus the respective cytoplasmatic or nuclear loading control. (I) RNA fluorescence in situ hybridization (FISH) assay of ECRAR in CMs. (J) Co-expression network analysis between ECRAR and differentially expressed mRNA. Those gene pairs expected to exhibit high correlations (Pearson correlation coefficients >0.9999) were used to construct the regulatory network by using Cytoscape 3.0. Those mRNA ID lists were submitted online to the DAVID Bioinformatics Resource for Gene Ontology (GO) enrichment.
The enrichment of H3K4me3 and H3K36me3, which are associated with active promoters and active gene bodies, respectively, was observed to be significantly increased in the fetal heart compared to that in the adult heart, which suggests the active chromatin state of ECRAR in the fetal heart (Figure 2B). Conservation analysis revealed that the exons of ECRAR were markedly more conserved than the introns (Figure S8A). By using the BLAST-like alignment tool (BLAT) and Needleman-Wunsch algorithm, we found that the exons were highly conserved across mammalian species, such as chimpanzee, gorilla, pig, rabbit, mouse, and rat, which indicated that this transcript may be a typical non-coding RNA for the mammalian animals (Figure S8B). Rapid amplification of cDNA ends demonstrated that ECRAR was an 851-bp lncRNA (Figures 2C and 2D), which was consistent with the length provided in the Ensembl database (Figure S8C). Using qRT-PCR, we confirmed that ECRAR expression was significantly increased in rat embryonic day (E)12 fetal heart and the expression progressively decreased in rat hearts after birth (Figure 2E). In newborn rat (post-natal day [P]1), we found that ECRAR expression varied among the analyzed tissue types, with expression in the heart and skeletal muscle higher than that found in the brain and liver (Figure 2F). In addition, ECRAR was highly expressed in AC16 cells, while its expression was low in endothelial cells and fibroblasts (Figure 2G).
Findings from the subcellular fractionation assay indicated that ECRAR was mainly located in the cytoplasm (Figure 2H). This subcellular localization was confirmed by confocal microscopy and fluorescence in situ hybridization (FISH) in rat CMs (Figure 2I). To examine the potential functions of ECRAR, we used coexpression analysis, and we found that ECRAR was associated with several important cell cycle-related genes (Figure 2J).The secondary structure analysis by using by RegRNA Server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), with default settings, indicated that ECRAR has a stem-loop structure, which may provide the necessary spatial conformation for interactions (Figure S9A). Further, the identified transcripts based on the secondary structure analysis were used to perform the KEGG pathway analysis, which showed that ECRAR may be involved in the Wnt-signaling pathway (Figure S9B). Overall, the above findings indicate that ECRAR may be involved in the regulation of the cell cycle and, therefore, likely has an important role in CM proliferation.
ECRAR Overexpression Promoted Rat Post-natal CM Proliferation In Vitro
We first investigated whether our newly discovered ECRAR could stimulate proliferation of post-natal CMs in vitro. Rat ventricular CMs (Figures S10A and S10B) were transfected with Ad-ECRAR PBS for 48 hr (the MOI was 10–20 and the CM transfection efficiency was >95%) (Figure S10C). qRT-PCR confirmed that ECRAR was upregulated in the Ad-ECRAR group (Figure S10D).The CM proliferation was assessed by immunostaining of proliferation markers together with the CM-specific marker α-actinin or cTnT. Overexpression of ECRAR enhanced 5-ethynyl-2′-deoxyuridine (EdU) incorporation into CM nuclei, which is a uridine analog incorporated into newly synthesized DNA, suggesting that ECRAR promoted CM proliferation (an increase from 5.7% to 22.7%; Figure 3A). Remarkably, treatment with ECRAR resulted in a significant increase in the number of CMs (Figure 3B).
Figure 3.
ECRAR Promotes Rat Post-natal CM Proliferation In Vitro
(A) Representative images and quantification of post-natal day 7 (P7) CMs positive for 5-ethynyl-2′-deoxyuridine (EdU) and Ki-67 (*p < 0.05 using t test, n = 6 per group). (B) Quantification of total CM numbers in P7 CMs transfected with Ad-ECRAR or control vectors after 7 days, displayed in (A) (*p < 0.05 using t test, n = 5 per group). (C and D) Representative images and quantification of P7 CMs positive for phospho-histone H3 (pH3) (C) and aurora B (D) (*p < 0.05; n = 5–6 per group). (E) Western blot analysis also confirmed that ECRAR induced pH3 and aurora B expression (*p < 0.05 using t test, n = 5 per group). (F) Representative images taken from a 12-hr time-lapse video of cell division in P7 CMs after transfection with Ad-ECRAR or Ad null. (G) Volume analysis of adult CMs in vitro. L, length; W, width; H, height; n = 3 per group, a total of 200 CMs were analyzed. (H) Representative images of fibroblasts positive for EdU. The cardiac fibroblasts (CFs) were label with CD90 and Sca-1.
To further investigate the role of ECRAR on cell mitosis and cytokinesis, transfected cells were assessed by staining for phospho-histone H3 (pH3), a specific marker for mitosis, and for aurora B kinase localization in midbodies, which are transient structures formed during cytokinesis. ECRAR overexpression also resulted in an increased number of pH3-positive CMs (Figure 3C) and aurora B-positive CMs (Figure 3D). Western blot analysis also confirmed that ECRAR induced pH3 and aurora B expression (Figure 3E). To directly visualize newly formed CMs originating from pre-existing CMs, we performed time-lapse imaging of P7 CMs labeled with tetramethylrhodamine ethyl ester (TMRE), a fluorescent mitochondrial dye. We found that the P7 CMs transfected with Ad-ECRAR underwent cell divisions (Video S1; Figure 3F). In contrast, we did not observe any CM division in the P7 CMs transfected with Ad null during the 12-hr time frame (Video S2; Figure 3F).
The effect of ECRAR on cardiac hypertrophy was investigated further. Overexpression of ECRAR did not induce the increased secretion of cardiac hypertrophy-related markers, such as hypertrophy-associated molecular marker atrial natriuretic peptide (ANP), β-myosin heavy chain (β-MHC), α-myosin heavy chain (α-MHC), and natriuretic peptides A-like (BNP) (Figure S11A). Immunostaining of ANP confirmed that overexpression of ECRAR did not induce cardiac hypertrophy (Figure S11B). To investigate whether the ECRAR induced the increase in CM volume, we determined the average volume of CMs, and we found no significant increase in the CM volume (Figure 3G). Wheat germ agglutinin (WGA) staining also showed that the cell size of CMs was not increased after transfection with Ad-ECRAR (Figure S11C). We further investigated the effect of ECRAR on cardiac fibrosis. ECRAR overexpression did not increase rat cardiac fibroblast proliferation. We used vimentin, CD90, and Sca-1 to label the fibroblasts. Immunostaining showed overexpression of ECRAR did not induce cardiac fibroblast proliferation (Figure 3H; Figure S12). These findings indicate that ECRAR induces CM proliferation but does not induce CM hypertrophy or increase fibrosis.
ECRAR Overexpression Promoted Rat Post-natal CM Proliferation In Vivo
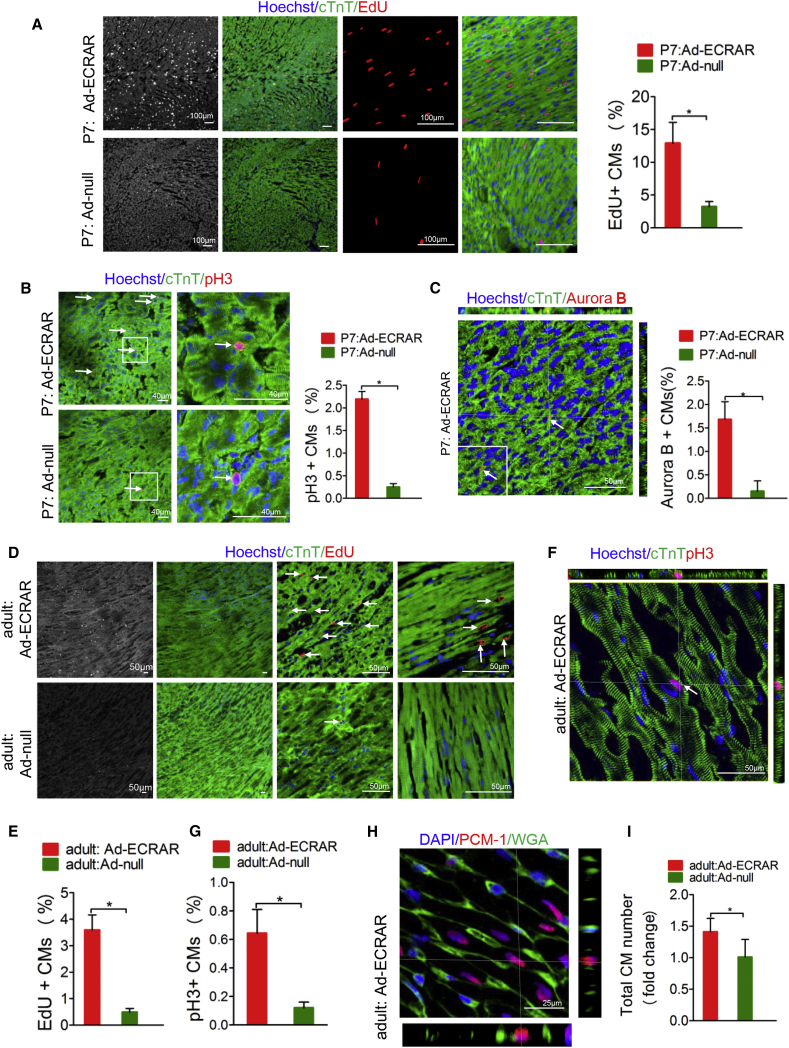
Next, we proceeded to assess the effect of ECRAR on CM proliferation in vivo. Ad-ECRAR or Ad null was directly injected into the hearts of P7 rats. In vivo bioluminescent imaging showed that heart tissues have robust GFP expression in rats that received an intramyocardial injection of Ad-ECRAR after 14 days (Figure S13A). In addition, the distribution of adenovirus (AdV)-mediated ECRAR was restricted to heart tissue (Figure S13A). qRT-PCR confirmed that ECRAR was upregulated in the rats injected with Ad-ECRAR (Figure S13B). Using immunofluorescence analysis, we found a marked increase in the number of EdU-positive CMs (Figure 4A), Ki-67-positive CMs (Figures S14A and S14B), pH3-positive CMs (Figure 4B), and aurora B-positive CMs (Figure 4C) in those rats injected with Ad-ECRAR as compared with those rats injected with Ad null. Moreover, the intracardiac delivery of Ad-ECRAR in adult rats also resulted in an increase in the number of EdU-positive CMs (Figures 4D and 4E) and pH3-positive CMs (Figures 4F and 4G), which suggests that ECRAR induces adult CM proliferation. The P7 rats injected with Ad-ECRAR showed a marked increase in heart weight:body ratio (Figure S14C), while no significant increase was observed in the adult hearts after transfection with the Ad-ECRAR (Figure S14D). Since the identification of CM nuclei in vivo is challenging, we used antibodies against the CM nuclear marker pericentriolar material 1 (PCM-1) to estimate the total numbers of nuclei in the heart (Figure 4H). As previously described,14, 15 we calculated the CM number, and CMs were significantly increased in adult rats after injection with the Ad-ECRAR (Figure 4I; Figure S15).
Figure 4.
ECRAR Promotes Rat Post-natal CM Proliferation In Vivo
(A) Immunostaining and quantification of EdU-positive CMs in P7 rat in vivo (*p < 0.05 using t test; n = 5–6 per group). (B) Immunostaining and quantification of pH3-positive CMs in P7 rat in vivo (*p < 0.05 using t test; n = 5–6 per group). (C) Immunostaining and quantification of aurora B-positive CMs in P7 rat in vivo (*p < 0.05 using t test; n = 5–6 per group). (D and E) Immunostaining (D) and quantification (E) of EdU-positive CMs in adult rat in vivo (*p < 0.05 using t test; n = 5 per group). (F and G) Immunostaining (F) and quantification (G) of pH3-positive CMs in adult rat in vivo (*p < 0.05 using t test; n = 5 per group). (H) The labeling strategy used to identify CM nuclei and the number of nuclei per CM. CM nuclei were labeled with antibodies against PCM-1(red), and the cell borders were labeled with antibodies against WGA (green). (I) Serial section analysis revealed that the number of CMs increased in the adult rat hearts after injection with Ad-ECRAR as compared to injection with Ad null.
ECRAR Overexpression Promoted Post-MI Cardiomyogenesis and Angiogenesis
The P7 and adult male Wistar rats that underwent MI surgery were injected with Ad-ECRAR and Ad null in the peri-infarcted area (Figure S16A). Ischemia was judged from both pallor of the myocardium and ST-segment elevation on the electrocardiogram (Figure S16B). In the peri-infarcted zone, we found that the overexpression of ECRAR enhanced the number of EdU-positive CMs (Figure 5A), pH3-positive CMs (Figure 5B), and aurora B-positive CMs (Figure S16C) in P7 rat hearts at 14 days post-MI. In adult rat hearts, the intracardiac delivery of Ad-ECRAR also resulted in an increase in the number of EdU-positive CMs (Figure 5C) and pH3-positive CMs (Figure 5D). Because CD31 staining can label endothelial cells and alpha-smooth muscle actin (α-SMA) staining can label vascular smooth muscle, we provided both CD31 and α-SMA staining. In the infarcted zone, the capillary and arteriole densities in the Ad-ECRAR group were significantly higher than those in the Ad-null group (Figures 5E–5H). In the peri-infarcted area, a similar trend in capillary density was observed (Figure 5H).
Figure 5.
ECRAR Induces Myocardial Regeneration and Promotes Angiogenesis after MI
(A) Immunostaining and quantification of EdU-positive CMs in P7 rats post-MI in peri-infarct zones (*p < 0.05 using t test; n = 5–6 per group). (B) Immunostaining and quantification of pH3-positive CMs in P7 rats post-MI in peri-infarct zones (*p < 0.05 using t test; n = 5 per group). (C) Immunostaining and quantification of EdU-positive CMs in adult rats post-MI in peri-infarct zones (*p < 0.05 using t test; n = 5–6 per group). (D) Immunostaining and quantification of pH3-positive CMs in adult rats post-MI in peri-infarct zones (*p < 0.05 using t test; n = 5 per group). (E) Representative immunostaining images and quantification of the capillaries by staining with anti-rat CD31 antibody in both the peri-infarct and infarct zones (*p < 0.05 using t test; n = 5 per group). The capillary and arteriole densities were calculated according to the percentages of positively stained areas in relation to those of the whole view field. (F and G) Representative immunostaining images (F) and quantification (G) of α-SMA staining of adult hearts injected with Ad-ECRAR or control vector post-MI (*p < 0.05 using t test; n = 5 per group). (H) Representative immunohistochemical images and quantification of the capillaries by staining with anti-rat CD31 antibody in both the peri-infarct and infarct zones. The capillary and arteriole densities were calculated according to the percentages of positively stained areas in relation to those of the whole view field (*p < 0.05 using t test; n = 6 per group).
ECRAR Overexpression Improved Post-infarction Cardiac Function
We further determined the effect of ECRAR on cardiac function in adult rats post-MI. The assessment of cardiac function by echocardiography at baseline (before MI) and at 1, 14, 30, and 60 days post-MI showed that the left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) were significantly preserved over time in infarcted rats injected with adeno-associated virus (AAV)-ECRAR, whereas they progressively declined over time in the AAV-null group (Figures 6A and 6B). In addition, the anterior wall thickness (AWT) of infarcted hearts injected with AAV-ECRAR markedly improved over time (Figures 6A and 6B). No significant enlargements of left ventricular dimensions (LVEDs) and diastolic left ventricular dimensions (LVEDd) were found in hearts injected with AAV-ECRAR at either 30 or 60 days post-MI (Figures 6A and 6B). We found that the myocardial infarct size, based on triphenyltetrazolium chloride staining, was significantly reduced in rats injected with AAV-ECRAR at 60 days post-MI (Figure 6C). Analysis of Masson’s trichrome staining of heart sections also revealed reduced fibrotic scarring in rats injected with AAV-ECRAR at both 14 and 60 days post-MI compared to that in AAV null-injected hearts (Figure 6D). Consistent with our echocardiography and morphometric observations, ECRAR significantly induced myocardial regeneration via CM proliferation after MI, thereby improving post-infarction cardiac function.
Figure 6.
ECRAR Improved Cardiac Remodeling after MI
(A) Evaluation of cardiac function by echocardiography before and 1, 14, 30, and 60 days after MI. (B) Quantitative analysis of left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), anterior wall thickness (AWT), diastolic left ventricular dimensions (LVEDd), and LV end-systolic diameter (LVESd) measured by echocardiography (AAV-ECRAR versus AAV-null group at the same time, *p < 0.05 using t test; n = 8–9 per group). (C) Representative images and quantification of infarction size based on 2,3,5-triphenyltetrazolium chloride (TTC) staining of heart sections 60 days post-MI. (D) Representative images and quantification of fibrosis area based on Masson’s trichrome staining of heart sections at 14 and 60 days post-MI.
Knockdown of ECRAR Inhibited CM Proliferation and Prevented Post-MI Recovery in Neonatal Hearts
Next, we investigated whether the knockdown of ECRAR affected CM proliferation and post-MI cardiac remodeling. 1-day-old rat ventricular CMs were transfected with short hairpin RNA (sh)-ECRAR and negative control shRNA (sh-NC). qRT-PCR confirmed that ECRAR was downregulated in the sh-ECRAR group (Figure S16D). The knockdown of ECRAR resulted in a significant decrease in the percentage of EdU-positive CMs (Figure 7A), pH3-positive CMs (Figure 7B), and aurora B-positive CMs (Figure 7C). Immunofluorescence analysis showed a marked decrease in the number of EdU-positive CMs (Figure 7D) and pH3-positive CMs (Figure 7E) in those rats injected with sh-ECRAR as compared with those injected with sh-NC. Decreased ECRAR levels inhibited functional post-MI recovery in P1 rat hearts, whereas no effect was observed with the delivery of sh-NC (Figure 7F). We further assessed the effect of ECRAR on human AC16 cells. In vitro administration of sh-ECRAR caused a significant decrease in AC16 CM proliferation, as shown by immunostaining for markers of pH3 and aurora B (Figure 7G).
Figure 7.
ECRAR Knockdown Inhibited CM Proliferation and Prevented Post-MI Recovery in Neonatal Hearts
(A–C) Representative images and quantification of post-natal day 1 (P1) CMs positive for EdU (A), pH3 (B), and aurora B (C) (*p < 0.05 using t test; n = 5–6 per group). (D and E) EdU (D) and pH3 (E) staining of rat hearts transfected with sh-ECRAR or control vectors (*p < 0.05 using t test; n = 5–6 per group). (F) Representative images and quantification of fibrosis area based on Masson’s trichrome staining of P1 rat heart sections at 14 days post-MI (*p < 0.05 using t test; n = 5 per group). (G) Confocal microscopy of aurora B-stained and pH3-stained AC16 cells transfected with sh-ECRAR or sh-NC (*p < 0.05 using t test; n = 4 per group).
ECRAR Is Upregulated through E2F1 Binding at the Promoter Region
We then aimed to identify the transcription factors responsible for ECRAR expression. To promote the binding sites in the promoter region of ECRAR, we used the Jaspar database (http://jaspar.binf.ku.dk/) with default settings, and we predicted that E2F1 might bind to the promoter region of ECRAR (−855 to −866 bp relative to the transcription start site [TSS]) (Figure S17A).Based on our coexpression analysis of ECRAR, we found comparable expression of ECRAR and E2F1 in human fetal and adult hearts (Figure S17B). The AC16 human CMs transfected with E2F1-containing vector increased ECRAR expression as determined by qRT-PCR (Figure S17C). Chromatin immunoprecipitation (ChIP)-qPCR with an anti-E2F1 antibody further confirmed that E2F1 binds to the ECRAR promoter region in CMs (Figures S17D and S17E), which indicates that E2F1 is involved in ECRAR transcription. ECRAR promoter regions with or without an E2F1 motif mutation were cloned into pGL3-basic reporter plasmids (Figure S17F, left). We found that luciferase expression was significantly reduced when the putative E2F1-binding site was mutated (Figure S17F, right). These findings demonstrate that the expression of ECRAR is transcriptionally regulated by E2F1.
ECRAR Promoted CM Proliferation via an ERK1/2-Dependent Mechanism
We performed microarray analysis of gene expression profiles in CMs, and we found that ECRAR overexpression resulted in significant changes in gene expression (1,568 coding genes changed >1.5-fold, all p < 0.05; Figure S18A), with upregulation of many cell proliferation and cell cycle-related genes (Figure 8A; Figure S18B). GO enrichment and KEGG pathway analysis of upregulated genes indicated that the cell cycle pathway was a principal pathway associated with the overexpression of ECRAR (Figures S18C and S18D). These findings suggest ECRAR has a prominent role mediating regulation of the cell cycle during CM proliferation.
Figure 8.
ECRAR Promoted CM Proliferation via an ERK1/2-Dependent Mechanism
(A) Several representative gene modules enriched for cell cycle-related genes from transcriptome microarray analysis in CMs transfected with Ad-ECRAR or control. (B) CMs underwent RNA pull-down analysis, and specific bands were identified of ERK1/2 by mass spectrometry (MS). (C) CMs underwent RNA pull-down analysis and specific bands were assayed by western blot. (D) qRT-PCR detection of the ECRAR retrieved by ERK1/2 antibody compared to IgG in the RNA-binding immunoprecipitation (RIP) assay. (E) Colocalization analysis of ECRAR and ERK1/2. (F) Protein levels of ERK1/2 assayed by western blot (*p < 0.05 using t test; n = 5 per group). (G) Cell fractions were isolated and measured with a western blot detecting ERK1/2. GAPDH (cytoplasm) and PARP (nucleus) were used as loading controls. Histogram depicts relative ERK1/2 levels as fold change versus the respective cytoplasmatic or nuclear loading control. (H) Distribution of cells in G1, S, and G2 phases of the cell cycle (*p < 0.05 using one-way ANOVA; n = 5 per group). (I) Representative images and quantification of post-natal day 7 (P7) CMs positive for pH3 after transfection with Ad-ECRAR (or Ad null) and ERK1/2 inhibitor (*p < 0.05 using one-way ANOVA; n = 5 per group). (J) Representative images and quantification of cyclin D1, cyclin E1, and E2F1 protein levels after transfection with Ad-ECRAR (or Ad null) and ERK1/2 inhibitor (*p < 0.05 versus Ad-ECRAR + control (CN) group using t test; n = 5 per group). (K) Representative images and quantification of E2F1 protein levels after transfection with Ad-ECRAR (or Ad null) and cyclin D1 inhibitor (*p < 0.05 versus Ad-ECRAR + CN group using t test; n = 5 per group). (L) Representative images and quantification of E2F1 protein levels after transfection with Ad-ECRAR (or Ad null) and cyclin E1 inhibitor (*p < 0.05 versus Ad-ECRAR + CN group using t test; n = 5 per group).
We next sought to determine the underlying molecular mechanism of how ECRAR regulates CM proliferation. Because overexpression or knockdown of ECRAR had no effect on the expression of its nearby coding gene PTTG1 (Figures S19A and S19B), we excluded the possibility that ECRAR acts by influencing its nearby gene of PTTG1 in cis. To search for potential ECRAR-interacting proteins, we performed pull-down assays using biotinylated ECRAR followed by mass spectrometry (MS). By screening the MS data, we found three possible related proteins might be involved in the proliferation (Figures S20 and S21). Among them, the mitogen-activated protein kinase 3 (MAPK3), also known as ERK1/2, which has been shown to play a key role in regulating cell proliferation and cell cycle progression (Figure S20; Figure 8B). Prediction of the ECRAR structure indicated that the nucleotide position of 151–202 interacted with ERK1/2 (Figures S22A and S22B). In addition, the binding sites are conserved between human and rats (Figure S22C). Independent immunoblotting further confirmed the identification of ERK1/2 (Figure 8C). We used RNA-binding immunoprecipitation (RIP) to further verify the interaction between ECRAR and ERK1/2 (Figure 8D). RNA FISH and immunofluorescence also showed ECRAR colocalized with ERK1/2 in the cytoplasm (Figure 8E).
The phosphorylation of ERK1/2 is essential for ERK-mediated G1/S transition. During this process, the ERK1/2 translocates into the nucleus and activates the transcription of many genes involved in cell proliferation. We first investigated whether the binding to ECRAR affects ERK1/2 phosphorylation status. As expected, western blot analysis revealed that phosphorylation of ERK1/2 was increased with ECRAR overexpression in P7 CMs (Figure 8F). In addition, overexpression of ECRAR in CMs triggered ERK1/2 translocation into the nucleus (Figure 8G).
The inhibition of ERK1/2 counteracted the decrease in the percentage of G1/G0 phase cells by ECRAR overexpression (Figure 8H). ERK1/2 inhibition also ablated ECRAR-induced CM proliferation (Figure 8I). We further found that activation of ERK1/2 induced by lnc-ECRAR led to the elevated expression of cyclin D1, cyclin E1, and E2F1 proteins, and this elevation could be ablated by using ERK1/2 inhibition (Figure 8J). Overexpression of ECRAR markedly increased the E2F1 expression, whereas both cyclin D1 inhibition and cyclin E1 inhibition could partly counteract the increase (Figures 8K and 8L).
Thus, ECRAR promoted proliferation in post-natal CMs, at least in part, via an ERK1/2-dependent mechanism. The above results, together with the upstream mechanism that ECRAR is transcriptionally upregulated by E2F1, implicate the existence of a positive feedback loop between E2F1 and ECRAR (Figure S23).
Discussion
In this study, we systematically characterized human long non-coding transcriptome changes during the human fetal-to-adult heart transition, and we identified a novel human-derived ECRAR and demonstrated that it promotes post-natal CM proliferation and induces post-MI functional recovery via the ERK1/2-dependent mechanism. Moreover, the E2F1-ECRAR-ERK1/2 pathway forms a positive feedback loop, which provides sustained activation of the ERK1/2 to drive cell cycle reentry (Figure S23). These findings indicated that ECRAR may be a valuable therapeutic target for inducing cardiac regeneration.
Comprehensive analysis of transcriptome changes during the human fetal-to-adult heart transition may reveal novel gene targets that reactivate fetal genes and retain a fetal pattern of regenerative capacity. In this study, we provided a global analysis of mRNA and lncRNA profiles during the human fetal-to-adult heart transition, and we identified over 1,000 differentially expressed lncRNAs. The marked differences in the expression pattern and abundance of these mRNAs and lncRNAs implicated their distinct biological roles in myocardial regeneration, an interpretation supported by the finding that functional annotation of upregulated mRNAs in the fetal heart was enriched in the cell cycle or mitotic cell cycle pathways. In addition, both differentially expressed lncRNA:cis-mRNA gene pairs and differentially expressed lncRNA:trans-mRNA gene pairs were enriched in the cell cycle- or heart development-related functional categories. In contrast to recent findings that only cis-regulation is a major regulatory mechanism of cardiac lncRNAs, our study suggests that trans-acting lncRNAs are as prevalent as cis-acting lncRNAs in the human heart during the fetal-to-adult transition.12, 13 Additionally, we inferred from coexpression network analysis the putative function of differentially expressed lncRNAs, and we found that these lncRNAs may be involved in the role of heart development and the cell cycle. Thus, we proposed that the differentially expressed lncRNAs may be involved in reactivation of the fetal genetic program in the adult heart.
A particularly striking finding in our study was the demonstration that our newly identified ECRAR significantly promoted post-natal CM proliferation, without induced CM hypertrophy or fibroblast proliferation, and attenuated post-MI remodeling. As a field, we have believed for decades that the human adult heart lost its capacity for self-renewal, while new evidence has shown that CMs may renew themselves, albeit at a slow rate (approximately 1% per year at the age of 20 and 0.3% at the age of 75).16 These studies suggest that stimulating endogenous cardiac regeneration may be a viable and promising therapeutic approach. Many researchers have attempted to reactivate adult CM proliferation either by overexpressing cell cycle activators, such as cyclin A2 and cyclin D2, or by removing inhibitors, such as cyclin-dependent kinase (CDK) inhibitors, in the adult heart.17, 18, 19 Other investigators extended the role of microRNAs (miRNAs) in cardiac regeneration and demonstrated that several miRNAs (such as miR-590-3p, miR-199a-3p, and the miR-15 family) could modify cardiac myocyte proliferation and the cell cycle exit via regulating various regulatory pathways.3, 20, 21
In the current study, we not only advanced the current understanding of lncRNA biology in post-natal CM proliferation and post-natal regeneration but also showed a new strategy in that modulation of lncRNA in adult CMs may have a therapeutic benefit. In contrast to CM regeneration, we also found that the overexpression of ECRAR did not cause cardiac hypertrophy or myocardial fibrosis, which indicated that ECRAR might be an ideal gene target for future research. We observed that ECRAR overexpression induced an increase in vascular density. It is likely the paracrine mechanisms augment angiogenesis activity.22 Previous studies have shown that the CM infuences endothelium response via paracrine factors, such as angiopoietins (vascular endothelial growth factor [VEGF]), hepatocyte growth factor (HGF), and fibroblast growth factor (FGF), which efficiently foster angiogenesis.23 Importantly, the transient expression of ECRAR in the adult heart of rats after MI reduced infarct scarring and improved cardiac function. Our study provides important insights into the role of lncRNA ECRAR in cardiac regeneration and cardiac function, and it presents a convincing rationale for the potential therapeutic benefit of ECRAR activation in post-natal CMs.
We found that the phosphorylation of ERK1/2 was induced by Ad-ECRAR along with the process of cytoplasmic-to-nuclear translocation. When ERK1/2 translocates into the nucleus, it can activate transcription of many genes involved in cell proliferation.24 We further demonstrated that ECRAR was induced by transcription factor E2F1 and that the downstream mechanism of post-natal myocardial regeneration triggered by ECRAR was through ERK1/2 signaling. The withdrawal of mammalian CMs from the cell cycle soon after birth is believed to be the primary mechanism for the loss of functional cardiac regenerative capacity.25 Therefore, reactivating CM proliferation through cell cycle reentry is particularly appealing. Previous studies have demonstrated that the genetic manipulation of cell cycle-regulatory factors, such as cyclin D1/D2 and cyclin A2, could induce increased CM-cycling activity and evoke a regenerative response.18 ERK1/2 signaling has been demonstrated to play a central role in cell cycle control.26 Here, we showed that ECRAR could promote CM proliferation by interacting with ERK1/2 and further activating ERK1/2 kinase activity by phosphorylation. ERK1/2 activation plays a fundamental role in the G1/S transition, and it is required for the induction of positive regulators of the cell cycle. We found that the activation of ERK1/2 by ECRAR led to the induction of cell cycle activators of cyclin D1, cyclin E1, and E2F1 proteins. The activation of ERK1/2 has been reported to induce cyclin D1 protein expression through many mechanisms.27 The newly synthesized cyclin D1 interacts with existing CDK4/6 (cyclin-dependent kinases) and forms an active cyclin D1-CDK4/6 complex, which plays a critical role in the G1/S transition.27 In addition, cyclin E is expressed in the later G1 phase and together with Cdk2 to form an active cyclin E1-CDK2 complex, which also promotes the cell cycle entry into the S phase. Moreover, the cyclin D1-CDK4/6 complex and cyclin E1-Cdk2 complex are known to enforce the phosphorylation of the retinoblastoma protein (pRb), resulting in full activation of E2F1.26
The activation of E2F1 further increased the ECRAR transcription in the ECRAR promoter. Sustained activation of ERK1/2 is necessary for successful G1- to S-phase progression. We identified that E2F1-ECRAR-ERK1/2 signaling formed a positive feedback loop, which helps contribute to the irreversibility of the G1/S transition, resulting in cell cycle reentry and induction of CM proliferation.
In conclusion, differentially expressed lncRNAs during the human fetal-to-adult heart transition may be involved in reactivation of the fetal genetic program in the adult heart, and our newly discovered human-derived ECRAR promoted post-natal myocardial regeneration and attenuated aberrant post-infarction remodeling via ERK1/2 signaling. These findings suggest that ECRAR may serve as a novel effective gene target for future therapeutic strategies in heart failure.
Materials and Methods
Animals
All experiments in this study were performed in accordance with Southern Medical University Institutional Guidelines for Animal Research, and the investigation conformed to the NIH’s Guide for the Care and Use of Laboratory Animals. For additional details regarding the animals and methods used in our study, please refer to the Supplemental Materials and Methods.
Transfection of AdV or Adeno-Associated Virus 9 Vectors
In in vitro experiments, the AdV or AAV9-containing GFP vectors for depletion or overexpression of ECRAR were added to cells with an MOI of 10. A scramble vector and PBS were used as controls. After 24 hr, cells were maintained in normal fresh medium for 48 hr before proceeding with experiments and analysis. After transfection, cells were used for ECRAR overexpression experiments.
In P1 and P7 neonatal rats, the AdV or AAV9-containing GFP vectors for depletion or overexpression of ECRAR and empty control vector were injected into the heart ventricles of P1 and P7 rats at 3 sites, at a dose of 3 × 108 viral genome particles per animal, using an insulin syringe with an incorporated 30G needle. In adult rats, the viral genome particles were injected into left ventricles at a dose of 3 × 1011 viral genome particles per animal. Ad-GFP was used to determine the distribution of AdV-mediated ECRAR expression in the myocardium. Rats received an intramyocardial injection of Ad-ECRAR and Ad null as described above. 14 days following injection, the rats were captured to examine GFP fluorescence with a Bruker In-Vivo FX Pro system (Bruker, MA, USA).
Establishment of the MI Model
MI was induced by ligation of the left anterior descending coronary artery as previously described. P1 and P7 rats were anesthetized by cooling on an ice bed for 4 min, whereas adult rats were intraperitoneally anesthetized with 3% pentobarbital sodium (40 mg/kg) following tracheal intubation for artificial ventilation. The left coronary artery (LAD) was ligated with silk suture 2 mm distal from the ascending aorta. Ischemia was judged from both pallor of the myocardium and ST-segment elevation on the electrocardiogram (Figure S16A). Immediately after LAD ligation, Ad-ECRAR (or AAV-ECRAR) or Ad null (or AAV null) was injected into the myocardium bordering the infarct zone (single injection), using an insulin syringe with a 30G needle (Figure S16B). The chest was closed and warmed for several minutes until recovery. EdU was administered intraperitoneally every 2 days for a total period of 10 days.
Computational Methods and Statistical Analysis
All computational procedures were carried out using in-house programs written in R or analyzed in Excel 2007 (Microsoft, Redmond, WA, USA) and Stata 12.0 (Stata, College Station, TX, USA). Quantitative data are expressed as mean ± SD. Differences between two groups were evaluated using unpaired Student’s t tests, and one-way ANOVA with post hoc analysis using Bonferroni tests were used for multiple comparisons with SPSS 16.0 (SPSS, Chicago, IL, USA). For all tests, p < 0.05 was considered significant.
Author Contributions
Y.C. and J.B. conceived the study. Y.C., X.L., and B.L. performed most of the experiments. Y.C. and H.W. performed the bioinformatic analyses. M.L., S.H., Y.S., G.C., X.S., and C.H. helped with various experiments and statistical analyses. Y.L., W.L., and J.B. supervised the study. Y.C. and J.B. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants to J.B. from the National Natural Science Foundation of China (81571698 and 81771857). This study was also supported by grants provided to Y.C. from the National Natural Science Foundation of China (81600319), Guangdong Provincial Science and Technology Plan (No. 2017A020215151), and the Project funded by the China Postdoctoral Science Foundation (2017M610539).
Footnotes
Supplemental Information includes twenty three figures, six tables, Supplemental Materials and Methods, and two videos and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.10.021.
Supplemental Information
References
- 1.Zhang Y., Mignone J., MacLellan W.R. Cardiac Regeneration and Stem Cells. Physiol. Rev. 2015;95:1189–1204. doi: 10.1152/physrev.00021.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao L., Borikova A.L., Ben-Yair R., Guner-Ataman B., MacRae C.A., Lee R.T., Burns C.G., Burns C.E. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA. 2014;111:1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y., Cheng H.W., Qiu Y., Dupee D., Noonan M., Lin Y.D., Fisch S., Unno K., Sereti K.I., Liao R. MicroRNA-34a Plays a Key Role in Cardiac Repair and Regeneration Following Myocardial Infarction. Circ. Res. 2015;117:450–459. doi: 10.1161/CIRCRESAHA.117.305962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haubner B.J., Schneider J., Schweigmann U., Schuetz T., Dichtl W., Velik-Salchner C., Stein J.I., Penninger J.M. Functional Recovery of a Human Neonatal Heart After Severe Myocardial Infarction. Circ. Res. 2016;118:216–221. doi: 10.1161/CIRCRESAHA.115.307017. [DOI] [PubMed] [Google Scholar]
- 5.Kajstura J., Urbanek K., Perl S., Hosoda T., Zheng H., Ogórek B., Ferreira-Martins J., Goichberg P., Rondon-Clavo C., Sanada F. Cardiomyogenesis in the adult human heart. Circ. Res. 2010;107:305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schonrock N., Harvey R.P., Mattick J.S. Long noncoding RNAs in cardiac development and pathophysiology. Circ. Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 8.Matkovich S.J., Edwards J.R., Grossenheider T.C., de Guzman Strong C., Dorn G.W., 2nd Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2014;111:12264–12269. doi: 10.1073/pnas.1410622111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 10.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ounzain S., Micheletti R., Beckmann T., Schroen B., Alexanian M., Pezzuto I., Crippa S., Nemir M., Sarre A., Johnson R. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He C., Hu H., Wilson K.D., Wu H., Feng J., Xia S., Churko J., Qu K., Chang H.Y., Wu J.C. Systematic Characterization of Long Noncoding RNAs Reveals the Contrasting Coordination of Cis- and Trans-Molecular Regulation in Human Fetal and Adult Hearts. Circ Cardiovasc Genet. 2016;9:110–118. doi: 10.1161/CIRCGENETICS.115.001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkass K., Panula J., Westman M., Wu T.D., Guerquin-Kern J.L., Bergmann O. No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell. 2015;163:1026–1036. doi: 10.1016/j.cell.2015.10.035. [DOI] [PubMed] [Google Scholar]
- 15.Brüel A., Nyengaard J.R. Design-based stereological estimation of the total number of cardiac myocytes in histological sections. Basic Res. Cardiol. 2005;100:311–319. doi: 10.1007/s00395-005-0524-9. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S., Zupicich J., Alkass K., Buchholz B.A., Druid H. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng R.K., Asai T., Tang H., Dashoush N.H., Kara R.J., Costa K.D., Naka Y., Wu E.X., Wolgemuth D.J., Chaudhry H.W. Cyclin A2 induces cardiac regeneration after myocardial infarction and prevents heart failure. Circ. Res. 2007;100:1741–1748. doi: 10.1161/CIRCRESAHA.107.153544. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro S.D., Ranjan A.K., Kawase Y., Cheng R.K., Kara R.J., Bhattacharya R., Guzman-Martinez G., Sanz J., Garcia M.J., Chaudhry H.W. Cyclin A2 induces cardiac regeneration after myocardial infarction through cytokinesis of adult cardiomyocytes. Sci. Transl. Med. 2014;6:224ra27. doi: 10.1126/scitranslmed.3007668. [DOI] [PubMed] [Google Scholar]
- 19.Pasumarthi K.B., Nakajima H., Nakajima H.O., Soonpaa M.H., Field L.J. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ. Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 20.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 21.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D., Mammen P.P., Rothermel B.A., Olson E.N., Sadek H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giordano F.J., Gerber H.P., Williams S.P., VanBruggen N., Bunting S., Ruiz-Lozano P., Gu Y., Nath A.K., Huang Y., Hickey R. A cardiac myocyte vascular endothelial growth factor paracrine pathway is required to maintain cardiac function. Proc. Natl. Acad. Sci. USA. 2001;98:5780–5785. doi: 10.1073/pnas.091415198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broughton K.M., Wang B.J., Firouzi F., Khalafalla F., Dimmeler S., Fernandez-Aviles F., Sussman M.A. Mechanisms of Cardiac Repair and Regeneration. Circ. Res. 2018;122:1151–1163. doi: 10.1161/CIRCRESAHA.117.312586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shindo Y., Iwamoto K., Mouri K., Hibino K., Tomita M., Kosako H., Sako Y., Takahashi K. Conversion of graded phosphorylation into switch-like nuclear translocation via autoregulatory mechanisms in ERK signalling. Nat. Commun. 2016;7:10485. doi: 10.1038/ncomms10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meloche S., Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 27.Chambard J.C., Lefloch R., Pouysségur J., Lenormand P. ERK implication in cell cycle regulation. Biochim. Biophys. Acta. 2007;1773:1299–1310. doi: 10.1016/j.bbamcr.2006.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.