Abstract
Chromatin and associated regulatory proteins regulate gene expression in the natural environment of the intact cell nucleus. Specific combinations of DNA-binding transcription factors and recruited coregulatory proteins alter the conformation of chromatin at promoters and enhancers of target genes to stimulate or repress transcription. The dynamic nature of the regulatory proteins active in these processes allows the cell to modulate gene expression very rapidly, an important feature in many physiological processes. Live cell imaging and photobleaching studies of fluorescently-tagged proteins reveal that many transcription factors and other chromatin-associated proteins rapidly move through the nucleoplasm. Transcription factors also transiently interact with specific regulatory sequences in chromatin, suggesting that gene activation does not require the formation of stable long-lived regulatory complexes on the chromatin. In this review we discuss how dynamic interactions allow transcriptional regulatory proteins find their targets within the nucleus, alter target chromatin structure, and modulate physiological gene expression.
Keywords: Chromatin, Nuclear receptor, Dynamics, Living cells, Hit-and-run, Transcription
1. Introduction
A complex chromatin structure compacts the eukaryotic genome within the cell nucleus and plays a key role in regulating gene expression (reviewed in [1]). Local chromatin conformations in the enhancers, promoters, and coding regions of genes are associated with the activation or repression of gene transcription by RNA polymeraseII. A combination of proteins and post-translational modifications interact at specific regions in the genome to determine local chromatin conformation. These DNA–protein conformations must be dynamic, rapidly assuming different functional states in response to extracellular and intracellular signals. This flexible nature of chromatin and associated regulatory proteins is required to generate the time-sensitive control that is commonly observed during physiological modulation of gene expression (reviewed in [2]).
The fundamental unit of chromatin structure is the nucleosome, which consists of an octamer of H2A, H2B, H3 and H4 core histones that is wrapped by approximately two super helical turns of DNA [3]. The nucleosome prevents certain proteins from interacting with DNA in the wrapped regions, providing a mechanism to regulate transcription. Many studies have shown that sequence specific DNA-binding transcription factors recruit coregulatory protein complexes to target chromatin, thus changing chromatin structure and regulating transcription. Nucleosomes can be modified by at least two broad classes of coregulatory proteins. Firstly, core histones can be covalently modified at specific amino acid residues (primarily, but not exclusively, in the N-terminal tail regions) by a number of coregulatory enzymes to change states of acetylation, methylation, ubiquitinylation, etc. [4]. These modifications either affect interactions of the nucleosome with the DNA directly, or serve as marks that recruit other regulatory proteins to the chromatin. Secondly, several ATP-dependent chromatin remodeling complexes can alter local nucleosome– DNA interactions via mechanisms that do not involve covalent modifications [5]. The remodeling events control how transcriptional regulators access target DNA sequences in the chromatin. Higher levels of chromatin organization may also play a role in regulating gene expression [6]. For example, when transcription levels are increased, large regions of surrounding chromatin have been shown to decondense and occupy increased volumes in the nucleus. Additionally, alterations of physical interactions between extremely distant chromosomal regions also regulate transcription [7,8].
The majority of studies that have examined the mechanistic details of chromatin-dependent transcriptional regulation have focused on measuring the steady-state levels of interaction between transcription factors, coregulators, basal transcription machinery, and target chromatin. In many biochemical experiments, extracts are prepared from large numbers of cells or reconstituted from in vitro purified components under conditions designed to stabilize these interactions. Often, these experiments are not designed to measure the dynamic properties of the interactions. This has lead to the assumption that these interactions are stable in vivo, even though this view is inconsistent with the rapid response of physiological gene expression. Recently, a combination of molecular biology and live cell microscopy techniques have allowed observation of transcription factors and chromatin components in real-time within the natural environment of the nucleus. In the living cell, many transcription factors, coregulators and even some structural chromatin components transiently interact with their specific DNA targets and rapidly exchange with the surrounding nucleoplasm [2,9,10]. In this review we explore how an understanding of chromatin dynamics in intact cells reveals fundamental mechanisms of transcriptional control.
2. Measuring chromatin dynamics in intact cells
The characterization and optimization of green fluorescent protein (GFP) has enabled a rapid expansion of our understanding of protein behavior inside living cells [11]. Because GFP is genetically encoded, common molecular cloning techniques can be used to generate vectors that express transcription factors or chromatin components that are fused with the fluorescent tag. These fluorescent fusion proteins often function like their untagged counterparts, but additional experiments are required to confirm this for each fusion protein. After the validated vectors are introduced into cells by transient or stable transfection techniques, the subcellular localization of the fusion proteins is observed by fluorescence microscopy in living or fixed cells. Spectral variants of the fluorescent proteins (FPs) make it possible to simultaneously observe multiple FP-tagged fusion proteins in a single cell [12]. Quantitative digital images are captured by sensitive cameras, or photomultiplier tube systems, that are integrated with the computer controlled fluorescence microscope. In a single image, the relative fluorescence intensity represents the steady-state concentration of fusion protein at each location within the cell.
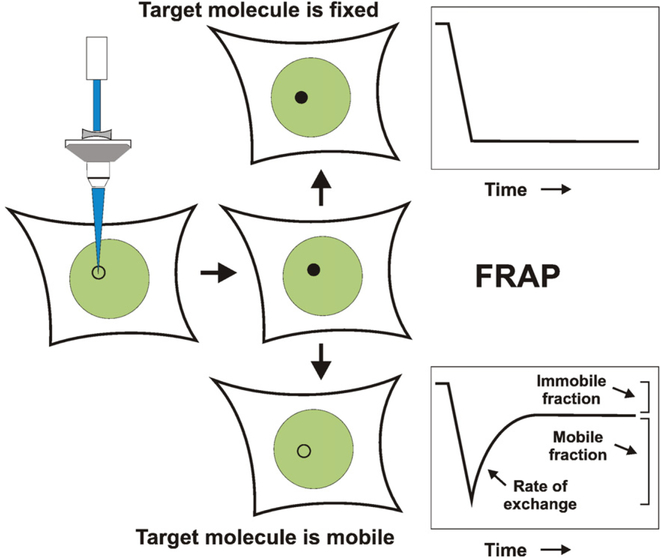
Fluorescence recovery after photobleaching (FRAP) measures protein dynamics within the living cell nucleus [13–16]. In this widely used kinetic microscopy technique, a short set of time-lapse images are captured to obtain initial fluorescence values (Fig. 1). Then a defined region of interest (ROI) within the nucleus is subjected to an extremely short pulse of high intensity laser illumination. This high intensity light photobleaches the FP, reducing the fluorescence in the ROI, but does not destroy the fusion protein [17]. The time-lapse image capture series is then continued, using minimal illumination intensity to prevent photobleaching during prolonged imaging. If the fusion protein is mobile and exchanging with the surrounding nucleoplasm, then the unbleached fusion proteins will move into the ROI and the fluorescence intensity will gradually increase or recover (Fig. 1). If the normalized intensity does not return to the maximal prebleach level, then some fraction of the protein is immobile over the observed time scale. Qualitatively, a faster rate of fluorescent recovery indicates a more mobile protein.
Fig. 1.
FRAP analysis of protein movement in living cells. The diagram shows laser photobleaching of a small subnuclear region, resulting in loss of fluorescence from GFP-labeled transcription factor (GFP-TF) molecules in the region. (Top) If the GFP-TF is immobile, the region will remain less bright than the surrounding nucleoplasm. Bottom) If the GFP-TF exchanges with the surrounding nucleoplasm then the fluorescence in the region will rapidly return over time. (Right panels) Quantification of the fluorescence signals reveals detailed information about protein dynamics. (Bottom, right) The fluorescence intensity will not fully recover to the prebleach level if a fraction of the GFP-TF is immobile.
As we will discuss in later sections, a number of sophisticated analytical methods have been developed to extract quantitative biophysical parameters from FRAP data. However, it should be noted that these analytical techniques continue to evolve. It has recently been noted that different image acquisition and analysis methods can introduce subtle variations in the biophysical parameters that are extracted from the microscopy data [18]. Therefore, care must be taken when comparing precise values from studies using different techniques. Other techniques such as inverse FRAP (iFRAP), fluorescence loss in photobleaching (FLIP) [19], and fluorescence correlation spectroscopy (FCS) [20] also provide useful information about protein dynamics in the living cell nucleus [16,21]. The combination of photoactivatable FPs and time-lapse microscopy have also broadened our understanding of intracellular protein dynamics [22]. When these techniques have been applied to measure nuclear protein dynamics, they have generally yielded results that support the findings derived by FRAP methods.
3. Protein mobility across the nucleoplasm
The nucleoplasmic movement of sequence specific DNA-binding transcription factors and other classes of chromatin-associated proteins provides mechanistic insight into how these proteins rapidly find and affect their specific targets. Members of the nuclear receptor superfamily exhibit several properties that facilitate studies of protein dynamics within the nucleoplasm. The nuclear receptors act as ligand-dependent transcription factors, and the processes underlying this activity have been extensively biochemically characterized. When bound to their specific steroid hormone ligands, the glucocorticoid receptor (GR), progesterone receptor (PR), or estrogen receptor (ER) each dissociates from heat shock protein (hsp) complexes, which prevent the receptor DNA-binding domain from functioning, and interact with target chromatin to control gene expression. Steroid hormone-dependent transcriptional output can be altered within minutes of treatment. Consistent with this rapid onset of function, photobleaching experiments show that ligand bound GR, PR, and ER exchange within the nucleoplasmic volume over a time span of seconds [15,23–26]. In a systematic FRAP study of nine diverse transcription factors in the nucleoplasm, eight exhibit 50% fluorescence recovery in less than 10 s and full recovery within 1 min [27]. Thus, fast nuclear mobility is a characteristic of many classes of transcription factors in addition to the nuclear receptors. The high rate of movement may be required to quickly find the relatively small number of high-affinity response elements among the many non-specific DNA-binding sites present in the nucleus. Interestingly, differences in the recovery rates between the eight highly-mobile factors suggest that not all transcription factors move through the nucleoplasm and interact with chromatin in exactly the same way [27].
In contrast to the rapid full recovery of fluorescence intensity that is often observed for DNA-binding transcription factors, photobleaching studies of GFP-RNA polymerase II (Pol II) in the nucleus reveal a rapid initial recovery, which is followed by a much slower recovery that requires up to 30 min to reach prebleach levels [13,28]. The fast Pol II is hypothesized to represent a diffusing or transiently interacting fraction of molecules. Again, the rapid movement of Pol II may be essential to quickly find transcription initiation sites within the bulk of the chromatin. The slow fraction represents molecules in constant association with chromatin during transcriptional elongation. The time estimated to complete transcription of an average gene is 6–13 min [13,29]. This is consistent with the hypothesis that the recovery time of the slow fraction is due to the release of Pol II that has completed one round of transcription.
The proteins involved in chromatin structure exchange within the nuclear volume at vastly different rates. In the absence of replication, The H3 and H4 core histones are relatively immobile over a time scale of days[30]. However, this study also reveals that a fraction of H2A and H2B exchange with the chromatin over the time scale of hours. These long-lived core histone interactions are hypothesized to be disrupted by processes involved in transcription. For example, histone acetylation is proposed to facilitate the exchange of H2A and H2B by nucleosome chaperones [31]. H2A–H2B dimers may also be displaced from chromatin by Pol II or associated factors during transcription [32], potentially increasing H2A and H2B nuclear mobility. The linker histone H1, which is involved in higher order chromatin folding, is surprisingly mobile over the time scale of minutes both in euchromatin and heterochromatin [33]. The exchange rate is increased or decreased by conditions that respectively simulate or repress transcription [33–35], as well as factors that interact with histones to modify chromatin compaction state, such as the ATP-dependant remodeling enzymes. In general, nuclear protein mobility is conducive to rapid changes in chromatin organization that regulate transcriptional activity.
4. Mechanisms regulating global transcription factor mobility in the nucleus
Quantitative analysis of protein mobility has elucidated some of the mechanisms responsible for global nuclear protein dynamics. Although many transcription factors and other chromatin-associated proteins move very rapidly in the nucleus, these rates are much slower than the rate of native GFP movement. However, GFP movement in the nucleus is consistent with that of similarly sized fluorescent-dextran molecules, indicating that GFP freely diffuses without significant specific interactions [36]. High molecular weight dextrans, which are larger than the transcription factors, also move rapidly in the nucleus[37]. These results suggest that the high mobility of nuclear proteins is generated by a combination of free-diffusion and transient interactions with more immobile components.
Various implementations of reaction–diffusion equation systems have been used to analyze kinetic microscopy data. These computational methods attempt to determine the set of biophysical parameters that most closely describe the observed dynamics of the protein [16,38,39]. Although the dynamic parameters can quantify the fraction of interacting protein at a given concentration, a value that is synonymous with the classic steady-state-binding affinity constant, the set of dynamic parameters also provides additional information. For example, analysis of photobleaching data has been interpreted to indicate two types of transcription factor mobility in the nucleus [16]. Both of these interaction types are transient but differ in their dissociation rates, producing a shorter and longer residency time for the two interacting fractions of a given transcription factor. These two transient interaction states are hypothesized to represent fractions of the transcription factor that are non-specifically and specifically interacting with chromatin. In this approach, Phair et al. have argued that diffusion can be disregarded in calculating the residence times.
Additional studies suggest that diffusion significantly contributes to the nuclear protein mobility [18,39,40]. When diffusion effects are included in the reaction–diffusion analysis, the mobility of many transcription factors and coregulators in the nucleus can be described by a single kinetically distinguishable transient interaction for each protein [18,39,40]. Since there are many more non-specific interaction sites in bulk chromatin compared to the number of high-affinity REs, it is likely that the single interaction type detected by these methods is dominated by non-specific scanning interactions of transcription factors with the global chromatin. Interestingly, wild type p53 and GR mobilities are described by similar kinetic constants when analyzed by the same method [18], suggesting that diverse transcription factors may scan the chromatin via similar mechanisms. Point mutations that block sequence specific DNA-binding do not affect the rapid nuclear mobility of p53 [41]. In contrast, deletion of the GR DNA-binding domain (DBD) or a single GR DBD point mutation causes an increase in mobility [24,42]. Therefore, further mutational analysis of specific versus non-specific chromatin interactions is needed to confirm the hypothesis that a common mechanism allows diverse transcription factors to scan the bulk chromatin.
Rapid scanning by transcription factors may involve three dimensional diffusion between chromatin strands and one dimensional diffusion along the chromatin strand mediated by non-specific interactions [43]. One dimensional movement along the DNA could greatly increase the rate at which transcription factors contact their specific REs [44] and therefore could play a significant role in the regulation of gene expression. Single molecule imaging in living bacteria suggests that the lac repressor protein spends the vast majority of time non-specifically interacting with DNA and diffusing in one dimension along the strand [45]. In addition, p53 rapidly translocates along the DNA strand in vitro via non-specific interactions[46], but it remains to be determined if this occurs on chromatin in the natural environment of the living cell nucleus.
Studies with the ligand-modulated nuclear receptors have also revealed that physiological changes in transcription factor conformation may link global dynamics and regulation of gene expression. Conformation of the steroid receptors is altered when bound by different agonist and antagonist ligands. Strong agonists stimulate transcription by making the receptor DBD accessible and exposing interaction domains that recruit coactivator protein complexes. Antagonists, and some weak agonists, interact with the receptor but are deficient in either one or both of these two stimulatory activities. The global nuclear mobilities of GR, PR, and ER change in the presence of different agonist and antagonist ligands [23,26,47]. In some cases, antagonistic ligands cause the receptors to move more rapidly throughout the nucleus compared to antagonist effects. However, there are some notable exceptions to this trend. For example, the strong antagonist ICI 182,780 immobilizes a large fraction of ER, and the antagonist RU-486 reduces the global mobility of PR [23,26]. Deletion of a portion of the ligand-binding domain (LBD) also increases the mobility of ER and GR, suggesting that regions outside the DBD can modulate receptor movement [24,26,42]. These changes in mobility may be due to altered interactions between the LBD and coregulatory protein complexes. Alternatively, it is possible that changes in the LBD influence the DBD or other domain conformations, which in turn control receptor mobility.
GR and ER nuclear movement also involve activities that are ATP-dependent [25,26,48]. Treatment with MG-132 reduces the global mobility of these receptors, suggesting that proteasome function is one of these energy consuming processes [24–26,42,49]. Reduction of receptor mobility by MG-132 requires a functional receptor DBD and LBD [24,26,42]. When cells are partially permeabilized, GR also becomes immobile in the nucleus [50]. Addition of nuclear extracts or a purified set of chaperone proteins and ATP restores GR mobility. The chaperones exhibit some specificity since they do not restore the mobility of HP1 in permeabilized cells. Thus, chaperones and proteasome complexes may actively disrupt interactions that normally reduce transcription factor movement.
5. Protein dynamics at specific sites of transcription
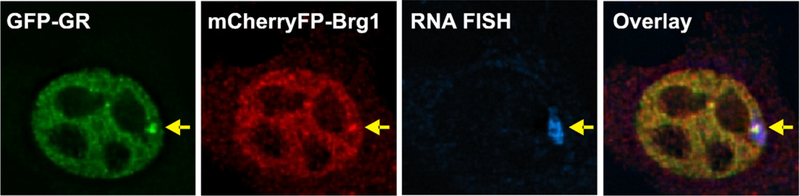
Engineering many repeats of a transcription factor RE at a single genomic site allows the specific transcription factor interactions with the regulatory site to be observed in living cells above the background of global chromatin interactions [51]. The pioneering system for these studies was based on the mouse mammary tumor virus-long terminal repeat (MMTV-LTR) promoter [52]. When bound by hormone, GR protein associates with six REs located in the MMTV-LTR to strongly activate transcription [53–55]. A mammary adenocarcinoma cell line was generated that contains a tandem 200 copy MMTV-LTR reporter gene array integrated at a single genomic locus (referred to as the MMTV array, [15,56,57]). As GFP-GR associates with the 1200 REs in the MMTV array, it concentrates and produces a bright subnuclear domain that is visible by high-resolution fluorescence microscopy (Fig. 2). An important feature of this system is the preservation of the natural promoter structure and regulation. Extensive analysis of the 3617 cell line [56,58] shows that the promoter in the MMTV array manifests chromatin reorganization and transcription activation profiles that are identical to those observed in single copy cells.
Fig. 2.
Direct observation of glucocorticoid receptor binding to response elements. Mammalian cells were engineered to contain multiple tandem copies of an MMTV-LTR reporter construct integrated at a single genomic locus, referred to as the MMTV array [56,57]. These cells harbor 200 copies of a 10 kb promoter–reporter structure in a tandem array with a total length of 2.2×106 bp. The cell derivatives shown here express GFP-GR and Brg1, an ATP-dependent chromatin remodeling protein, labeled with red mCherryFP. Following treatment with 100 nM Dex for 0.5 h, the cells were fixed and processed for RNA FISH to detect the reporter gene and visualized by digital deconvolution microscopy. The images from each fluorescence channel are shown individually (left panels), and merged in the overlay image (far right panel). Arrows show the location of the transcriptionally active MMTV array, where GFP-GR and mCherryFP-Brg1 steady-state concentrations are increased due to specific transient interactions with the MMTV chromatin.
Following photobleaching, the GFP-GR fluorescence fully recovers within 30 s at the MMTV array, with 50% recovery occurring in approximately 5 s [15]. Coregulators that are recruited by GR also exhibit similar kinetics at the MMTV array, arguing against the concept that many components of the regulatory complex are stably bound on regulatory DNA [13]. As expected for a processive enzyme, Pol II requires 13 min to fully recover at this site [13]. Similar slow Pol II kinetics are also observed at another engineered promoter–reporter gene array [59]. Thus, long-term interactions can be detected at the repeated promoter–reporter sequences when the long-term interactions exist. Other steroid receptors, such as PR and AR, also transiently interact with the MMTV array [23,60]. Similar imaging experiments show that ER interacts transiently with regulatory sequences from the prolactin gene, and NF-kB rapidly exchanges with REs in the HIV-LTR [61,62]. Fast transcription factor exchange also occurs on a natural repeat of pol II-regulated genes in yeast [63] and on the clustered pol I-regulated ribosomal genes in the mammalian nucleolus [64], indicating that the dynamic behavior is not an artifact of engineered chromatin. Since each of these divergent transcription factors stimulates expression of the respective repeated reporter gene, transient interactions with REs must modify chromatin structure and activate the transcriptional machinery [9,15,61–63]. These results suggest that many endogenous genes are regulated by transient interactions that do not require the formation of long-lived, stable regulatory complexes on the chromatin.
The Drosophila Heat Shock Factor (HSF), which activates transcription of hsp70 in response to stress, has been reported to maintain significantly longer-lived interactions with target promoters [65]. GFP–HSF interaction with REs can be visualized on polytene amplified chromosome bands in living Drosophila salivary glands. Prior to heat shock, HSF is concentrated on chromosome bands that do not contain the hsp70 genes [65]. Photobleaching results demonstrate that HSF at these chromosome bands rapidly exchanges with the surrounding nucleoplasm. Following heat shock, the HSF redistributes and concentrates at bands containing the hsp70 genes. At this location, HSF binds much more stably, with a half-life greater than 6 min. Since HSF binds with high affinity to REs, it is tempting to speculate that this property alone is responsible for the slow dynamics. However, as a general principle this is disputed by the fact that NF-kB binds its site with very high affinity in vitro, while the interaction is highly transient in vivo [61].
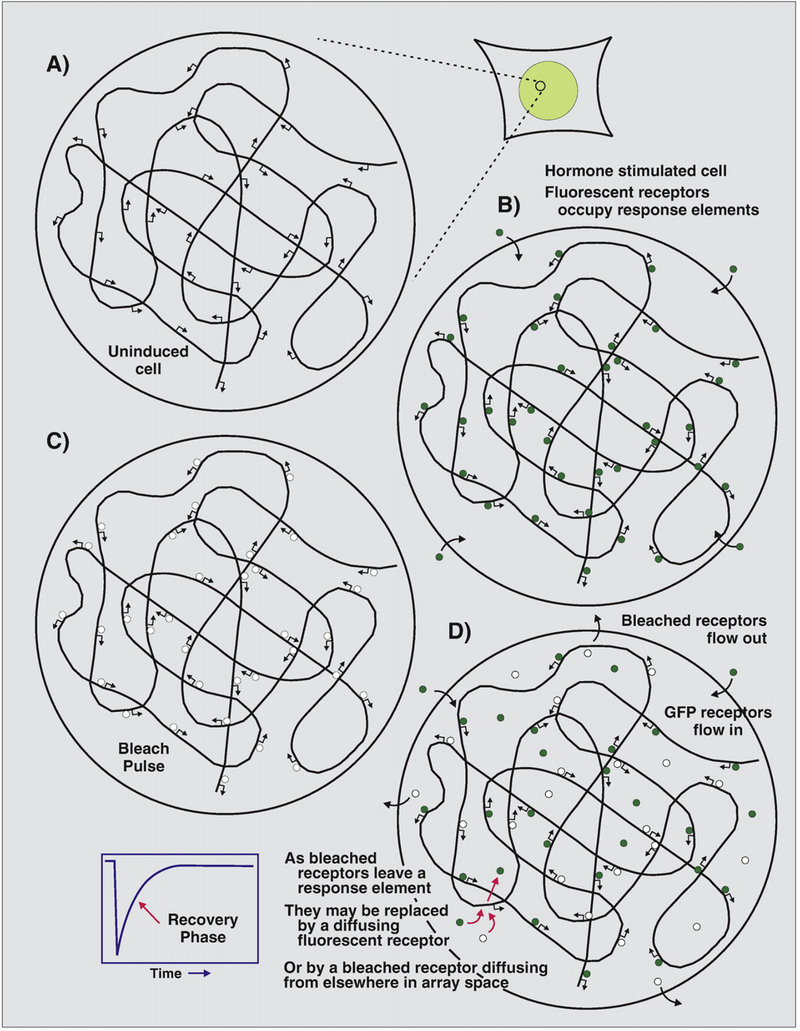
The analysis of protein movement in these amplified gene systems (either natural or engineered), is quite complex. Fig. 3 presents an overview of dynamics in a typical array system, using the glucocorticoid receptor as an example. After activation with ligand, the receptor will bind to a large fraction of available GREs in the array structure (Fig. 3B). Immediately after the bleach pulse (Fig. 3C), all of the receptors in the array space are homogeneously bleached. During the recovery phase (Fig. 3D), however, a complex series of interactions ensue. Receptors, both bleached and unbleached, are rebinding to response elements, and leaving the array space. New receptors, mostly unbleached, enter the space. These considerations apply equally to the Drosophila polytene chromosome bands. These structures occupy a large volume compared to other amplified promoter array systems. As this large domain (containing as many as 1000 gene copies) would harbor a high local concentration of the bleached protein, a significant component of the apparent reduction in protein mobility could be due to rebinding of these molecules (see Fig. 3). Further work is required to determine if other transcription factors bind stably to regulatory chromatin, and if this stable binding occurs outside of specialized polytene chromosomes.
Fig. 3.
Transcription factor movement in an array space during photobleaching. (A) An idealized view of the MMTV array space in 3617 cells is schematically shown. (B) After induction, most of the response elements are occupied with activated glucocorticoid receptor (green filled circles). (C) After the bleach pulse, all receptors in the array space are uniformly bleached (white circles). (D) Many different events are in progress during the recovery phase. New unbleached receptors enter the space and bind to unoccupied response elements. However, some rebinding events will occur with bleached molecules. Furthermore, both bleached and unbleached receptors are leaving the array space.
Studies of the repeated reporter gene arrays have revealed some processes that control transcription factor fast dynamics at their regulatory sites. The movement of transcription factors at these visible subnuclear domains is measurably slower compared to the surrounding nucleoplasm [25,61]. This suggests that additional interactions with specific regulatory sites increase the residency time of the receptor in these domains. However, some common processes regulate mobility both at specific regulatory sites and non-specific nucleoplasmic sites. As for movement in the whole nucleoplasm, GR exchange with the MMTV array is dependent on ATP [25]. This study also demonstrated that chaperone and proteasome activities regulate the exchange of GR at the regulatory sites. The dynamic imaging results support biochemical experiments demonstrating that chaperone complexes disassemble transcription factor complexes at their REs [66]. Additionally, the proteasome machinery is also recruited to the MMTV array, which is consistent with reports in other systems directly linking proteasomal activity to transcriptional regulation [67]. NF-kB exchange with its target sites is also slowed by NF-kB mutations that prevent proteasomal degradation [61]. Considering these results, the active disruption of protein complexes likely reduces the residency times of many transcription factors at their specific regulatory sites in chromatin.
Biochemical studies indicate that nucleosome remodeling complexes also promote transcription factor mobility. This was first detected by in vitro reconstitution experiments in which a region of the MMTV-LTR assembled in chromatin is subjected to the action of purified GR and a Swi/Snf nucleosome remodeling complex [68]. In vivo, GR-dependent recruitment of Swi/Snf complexes and the resulting chromatin remodeling events are essential for full transcriptional activation [69]. As expected, the reconstituted remodeling event is dependent on GR, Swi/Snf complex, and ATP [68]. Surprisingly, the GR is not associated with target sites in chromatin after the reconstituted remodeling reaction. However, the receptor is associated with the target sites in the context of naked DNA, or when the remodeling reaction is blocked by inhibitory conditions, for example omission of ATP. These findings suggest that the remodeling event itself leads to a transient association of GR with the template. This is contrary to the concept that nucleosome remodeling facilitates the stable recruitment of transcription factors to the reorganized chromatin. To overcome the limited time resolution of many biochemical techniques, extremely fast (5 ns) two-photon UV laser crosslinking was used to fix steady-state “snap shots” of the factors on the chromatin at short time intervals during the reconstituted remodeling reaction [70]. Analysis of these fixed complexes shows that the remodeling reaction is transient and periodic. During each cycle, GR loads and recruits Swi/Snf, and is then ejected from the chromatin. The use of a dominant negative Swi/Snf complex and genome wide assays reveals that essentially all GR interactions at endogenous response elements involve chromatin remodeling events [71]. Future photobleaching studies in the presence and absence of this dominant negative Swi/Snf complex will likely shed more light on how transcription factor mobility is controlled in the living cell nucleus.
6. Rapid dynamic flux controls steady-state levels of chromatin interactions
Biochemical time course experiments demonstrate that many promoters are occupied by different transcription factors and coregulators at different times following gene activation. Transcription levels can temporally correlate with these changing interactions, which suggests that they are functionally linked. The biochemically detected alterations in interaction level often evolve over the time course of many minutes or even hours. For example, ER-dependent stimulation of the PS2 promoter causes cyclic promoter interaction and transcriptional profile with a relatively long 60 minute period[72]. Similar behavior is reported from several studies [73,74]. Together, these data have been interpreted to indicate that factors are stably bound at the promoter for 15–20 min, until they are replaced by another set of specific factors. According to this stable-stepwise assembly model, the long residency time is needed to sequentially build a large complex, which in turn modifies the chromatin, and recruits the polymerase. This interpretation is incompatible with the large body of evidence showing that many transcription factors and coregulators only reside very transiently on the target chromatin. In stark contrast, the observation that biochemically measured interaction levels change slowly over time does not directly disagree with the rapid dynamic flux observed in living cells. This is because the biochemical studies measure steady-state interaction levels, which result from a balance of association and disassociation rates that can be very fast or very slow. A change in the balance of these association and disassociation rates moves the system out of true steady-state, causing a time-dependent change in the average level of chromatin association. If the absolute association and dissociation rates for a given factor are very fast but the rates are only slightly out of balance, then the average level of chromatin association will change slowly over longer time periods.
GR-dependent transcription from the MMTV-LTR is transient, with a single pulse of activity which peaks at approximately 30 min followed by attenuation, which is typical of many GR regulated genes [13,75]. In contrast to this single peak of activity, both steady-state levels of NF-kB at a target promoter and transcriptional output repeatedly cycle over a time period of approximately 60 min as measured by biochemical methods [61]. Photobleaching indicates NF-kB resides on the promoter for seconds, suggesting that cyclic changes in binding occur at both extremely short and much longer time scales in the same system [61]. This was confirmed in a yeast study that examined changes in steady-state promoter association by both live cell imaging and biochemical methods [63]. As measured by both techniques, the steady-state levels of the transcription factor Ace1 cycle at CUP1 promoters oscillates with a time period of 30 min. Again, the photobleaching data show that the Ace1 fully exchanges at the regulatory site in less than 2 min. Importantly, single cell analysis of nascent transcripts in this system indicates that transcriptional activation is achieved by the transiently interacting Ace1. Thus, fast exchanging transcription factors at target promoters are functionally relevant and coexist with longer cycles that modulate the steady-state levels of chromatin interaction.
The “Return to Template” model of transcriptional control [2,9,10,76] reconciles the extremely fast dynamic flux and the concurrent slower progression in steady-state levels of transcription factor-chromatin association [2,9]. According to this model, rapid and stochastic interactions allow many regulatory proteins to interact with the promoter in a brief period of time. Many of these stochastic interactions are predicted to be unproductive because several regulatory events must occur on the promoter in the correct order. As predicted, only 1% of Pol II molecules that interact with the target promoter generate a transcript [59]. In agreement with the stochastic/probabilistic view of gene expression, the MMTV-LTR chromatin in single cells exists in many states that are differentiated by widely variable levels of steady-state GR association and transcriptional efficiency [77]. These probabilistic chromatin states transition over time, generating the transcriptional response observed in the average cell population. The rate of GR exchange with the MMTV-LTR changes when the chromatin transitions between conformational states [25]. Thus, changes in steady-state association of transcription factors with chromatin are linked to alterations in fast exchange and probabilistic transitions in the chromatin state. Extremely dynamic transcription factor interactions increase the probability that the correct chromatin transitions will occur, and prevent promoters from becoming blocked for long periods by non-productive complexes. These features of the “Return to Template” model explain many aspects of dynamic transcriptional control in the natural context of the intact cell.
Acknowledgements
This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We would like to acknowledge the assistance of Tatiana Karpova, manager of the LRBGE Fluorescence Imaging Facility.
References
- [1].Li B, Carey M, Workman JL, The role of chromatin during transcription, Cell 128 (2007) 707–719. [DOI] [PubMed] [Google Scholar]
- [2].Hager GL, Elbi C, Johnson TA, Voss TC, Nagaich AK, Schiltz RL, Qiu Y, John S, Chromatin dynamics and the evolution of alternate promoter states, Chromosome Res 14 (2006) 107–116. [DOI] [PubMed] [Google Scholar]
- [3].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature 389 (1997) 251–260. [DOI] [PubMed] [Google Scholar]
- [4].Kouzarides T, Chromatin modifications and their function, Cell 128 (2007) 693–705. [DOI] [PubMed] [Google Scholar]
- [5].Saha A, Wittmeyer J, Cairns BR, Chromatin remodelling: the industrial revolution of DNA around histones, Nat. Rev. Mol. Cell Biol 7 (2006) 437–447. [DOI] [PubMed] [Google Scholar]
- [6].Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T, Large-scale chromatin structure and function, Curr. Opin. Cell Biol 11 (1999) 307–311. [DOI] [PubMed] [Google Scholar]
- [7].Fraser P, Bickmore W, Nuclear organization of the genome and the potential for gene regulation, Nature 447 (2007) 413–417. [DOI] [PubMed] [Google Scholar]
- [8].Kumaran RI, Thakar R, Spector DL, Chromatin dynamics and gene positioning, Cell 132 (2008) 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hager GL, Nagaich AK, Johnson TA, Walker DA, John S, Dynamics of nuclear receptor movement and transcription, Biochim. Biophys. Acta 1677 (2004) 46–51. [DOI] [PubMed] [Google Scholar]
- [10].Hager GL, Elbi CC, Becker M, Protein dynamics in the nuclear compartment, Curr. Opin. Genet. Dev 12 (2002) 137–141. [DOI] [PubMed] [Google Scholar]
- [11].Tsien RY, The green fluorescent protein, Annu. Rev. Biochem 67 (1998) 509–544. [DOI] [PubMed] [Google Scholar]
- [12].Shaner NC, Steinbach PA, Tsien RY, A guide to choosing fluorescent proteins, Nat. Methods 2 (2005) 905–909. [DOI] [PubMed] [Google Scholar]
- [13].Becker M, Baumann CT, John S, Walker D, Vigneron M, McNally JG, Hager GL, Dynamic behavior of transcription factors on a natural promoter in living cells, EMBO Rep 3 (2002) 1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hager GL, Fletcher TM, Xiao N, Baumann CT, Muller WG, McNally JG, Dynamics of gene targeting and chromatin remodeling by nuclear receptors, Steroid Receptor Coactivators and the Remodeling of Chromatin, Biochem. Soc. Trans 28 (2000) 405–410. [PubMed] [Google Scholar]
- [15].McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL, The glucocorticoid receptor: rapid exchange with regulatory sites in living cells, Science 287 (2000) 1262–1265. [DOI] [PubMed] [Google Scholar]
- [16].Phair RD, Misteli T, Kinetic modelling approaches to in vivo imaging, Nat. Rev. Mol. Cell Biol 2 (2001) 898–907. [DOI] [PubMed] [Google Scholar]
- [17].White J, Stelzer E, Photobleaching GFP reveals protein dynamics inside live cells, Trends Cell Biol 9 (1999) 61–65. [DOI] [PubMed] [Google Scholar]
- [18].Mueller F, Wach P, McNally JG, Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching, Biophys. J 94 (2008) 3323–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Koster M, Frahm T, Hauser H, Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies, Curr. Opin. Biotechnol 16 (2005) 28–34. [DOI] [PubMed] [Google Scholar]
- [20].Weiss M, Probing the interior of living cells with fluorescence correlation spectroscopy, Ann. N.Y. Acad. Sci 1130 (2008) 21–27. [DOI] [PubMed] [Google Scholar]
- [21].Hager GL, Studying nuclear receptors with GFP fusions, Methods Enzymol 302 (1999) 73–84. [DOI] [PubMed] [Google Scholar]
- [22].Lippincott-Schwartz J, tan-Bonnet N, Patterson GH, Photobleaching and photoactivation: following protein dynamics in living cells, Nat. Cell Biol (Suppl) (2003) S7–14. [PubMed] [Google Scholar]
- [23].Rayasam GV, Elbi C, Walker DA, Wolford RG, Fletcher TM, Edwards DP, Hager GL, Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro, Mol. Cell. Biol 25 (2005) 2406–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schaaf MJ, Cidlowski JA, Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity, Mol. Cell. Biol 23 (2003) 1922–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG, Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes, Mol. Cell. Biol 24 (2004) 2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, Mancini MA, FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent, Nat. Cell Biol 3 (2001) 15–23. [DOI] [PubMed] [Google Scholar]
- [27].Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager GL,Bustin M, Misteli T, Global nature of dynamic protein–chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins, Mol. Cell. Biol 24 (2004) 6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kimura H, Sugaya K, Cook PR, The transcription cycle of RNA polymerase II in living cells, J. Cell Biol 159 (2002) 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jackson DA, Pombo A, Iborra F, The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells, FASEB J 14 (2000) 242–254. [PubMed] [Google Scholar]
- [30].Kimura H, Cook PR, Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B, J. Cell Biol 153 (2001) 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M, p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone, Genes Dev 14 (2000) 1899–1907. [PMC free article] [PubMed] [Google Scholar]
- [32].Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM, Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription, Mol. Cell 9 (2002) 541–552. [DOI] [PubMed] [Google Scholar]
- [33].Misteli T, Gunjan A, Hock R, Bustin M, Brown DT, Dynamic binding of histone H1 to chromatin in living cells, Nature 408 (2000) 877–881. [DOI] [PubMed] [Google Scholar]
- [34].Contreras A, Hale TK, Stenoien DL, Rosen JM, Mancini MA, Herrera RE, The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation, Mol. Cell Biol 23 (2003) 8626–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dou Y, Bowen J, Liu Y, Gorovsky MA, Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin, J. Cell Biol 158 (2002) 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Braga J, Desterro JM, Carmo-Fonseca M, Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes, Mol. Biol. Cell 15 (2004) 4749–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Seksek O, Biwersi J, Verkman AS, Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus, J. Cell Biol 138 (1997) 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Carrero G, Crawford E, Th’ng J, de Vries G, Hendzel MJ, Quantification of protein–protein and protein–DNA interactions in vivo, using fluorescence recovery after photobleaching, Methods Enzymol 375 (2004) 415–442. [DOI] [PubMed] [Google Scholar]
- [39].Sprague BL, Pego RL, Stavreva DA, McNally JG, Analysis of binding reactions by fluorescence recovery after photobleaching, Biophys. J 86 (2004) 3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Beaudouin J, Mora-Bermudez F, Klee T, Daigle N, Ellenberg J, Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins, Biophys. J 90 (2006) 1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, DiBenedetto E, The DNA binding activity of p53 displays reaction-diffusion kinetics, Biophys. J 91 (2006) 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kino T, Liou SH, Charmandari E, Chrousos GP, Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function, Mol. Med 10 (2004) 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gorski SA, Dundr M, Misteli T, The road much traveled: trafficking in the cell nucleus, Curr. Opin. Cell Biol 18 (2006) 284–290. [DOI] [PubMed] [Google Scholar]
- [44].Mirny L, Cell commuters avoid delays, Nat. Phys 4 (2008) 93–95. [Google Scholar]
- [45].Elf J, Li GW, Xie XS, Probing transcription factor dynamics at the single-molecule level in a living cell, Science 316 (2007) 1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM, Tumor suppressor p53 slides on DNA with low friction and high stability, Biophys. J 95 (2008) L01–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schaaf MJ, Lewis-Tuffin LJ, Cidlowski JA, Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility, Mol. Endocrinol 19 (2005) 1501–1515. [DOI] [PubMed] [Google Scholar]
- [48].Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME, GR and HMGB1 interact only within chromatin and influence each other’s residence time, Mol. Cell 18 (2005) 109–121. [DOI] [PubMed] [Google Scholar]
- [49].Meijsing SH, Elbi C, Luecke HF, Hager GL, Yamamoto KR, The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release, Mol. Cell Biol 27 (2007) 2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL, DeFranco DB, Molecular chaperones function as steroid receptor nuclear mobility factors, Proc. Natl. Acad. Sci. USA 101 (2004) 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Belmont AS, Li G, Sudlow G, Robinett C, Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system, Methods Cell Biol 58 (1999) 203–222. [DOI] [PubMed] [Google Scholar]
- [52].Hager GL, Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus, Prog. Nucleic Acid. Res. Mol. Biol 66 (2001) 279–305. [DOI] [PubMed] [Google Scholar]
- [53].Huang AL, Ostrowski MC, Berard D, Hager GL, Glucocorticoid regulation of the Ha-MuSV p21 gene conferred by sequences from mouse mammary tumor virus, Cell 27 (1981) 245–255. [DOI] [PubMed] [Google Scholar]
- [54].Payvar F, DeFranco DB, Firestone GL, Edgar B, Wrange O, Okret S, Gustafsson JA, Yamamoto KR, Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region, Cell 35 (2 Pt 1) (1983) 381–392. [DOI] [PubMed] [Google Scholar]
- [55].Scheidereit C, Geisse S, Westphal HM, Beato M, The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus, Nature 304 (5928) (1983) 749–752. [DOI] [PubMed] [Google Scholar]
- [56].Kramer P, Fragoso G, Pennie WD, Htun H, Hager GL, Sinden RR, Transcriptional state of the mouse mammary tumor virus promoter can effect topological domain size in vivo, J. Biol. Chem 274 (1999) 28590–28597. [DOI] [PubMed] [Google Scholar]
- [57].Walker D, Htun H, Hager GL, Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods (Companion to Methods in Enzymology) 19 (1999) 386–393. [DOI] [PubMed] [Google Scholar]
- [58].Fragoso G, Pennie WD, John S, Hager GL, The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames, Mol. Cell. Biol 18 (1998) 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Darzacq X, Shav-Tal Y, de T, Brody VY, Shenoy SM, Phair RD, Singer RH, In vivo dynamics of RNA polymerase II transcription, Nat. Struct. Mol. Biol 14 (2007) 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang CY, Hager GL, Saatcioglu F, Ligand-specific dynamics of the androgen receptor at its response element in living cells, Mol. Cell Biol 27 (2007) 1823–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G, A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity, EMBO J 25 (2006) 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE, Mancini MA, Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent, J. Cell Sci 119 (2006) 4101–4116. [DOI] [PubMed] [Google Scholar]
- [63].Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG, Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter, Science 319 (2008) 466–469. [DOI] [PubMed] [Google Scholar]
- [64].Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T, A kinetic framework for a mammalian RNA polymerase in vivo, Science 298 (2002) 1623–1626. [DOI] [PubMed] [Google Scholar]
- [65].Yao J, Munson KM, Webb WW, Lis JT, Dynamics of heat shock factor association with native gene loci in living cells, Nature 442 (2006) 1050–1053. [DOI] [PubMed] [Google Scholar]
- [66].Freeman BC, Yamamoto KR, Disassembly of transcriptional regulatory complexes by molecular chaperones, Science 296 (2002) 2232–2235. [DOI] [PubMed] [Google Scholar]
- [67].Collins GA, Tansey WP, The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev 16 (2006) 197–202. [DOI] [PubMed] [Google Scholar]
- [68].Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS, Hager GL, ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling, Mol. Cell. Biol 22 (2002) 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fryer CJ, Archer TK, Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex, Nature 393 (1998) 88–91. [DOI] [PubMed] [Google Scholar]
- [70].Nagaich AK, Walker DA, Wolford RG, Hager GL, Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling, Mol. Cell 14 (2004) 163–174. [DOI] [PubMed] [Google Scholar]
- [71].John S, Sabo PJ, Johnson TA, Sung MH, Biddie S, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL, Interaction of the glucocorticoid receptor with the global chromatin landscape, Mol. Cell 29 (2008) 611–624. [DOI] [PubMed] [Google Scholar]
- [72].Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M, Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription, Cell 103 (2000) 843–852. [DOI] [PubMed] [Google Scholar]
- [73].Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F, Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter, Cell 115 (2003) 751–763. [DOI] [PubMed] [Google Scholar]
- [74].Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F, Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling, Mol. Cell 11 (2003) 695–707. [DOI] [PubMed] [Google Scholar]
- [75].Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, Adkins NL, Stavreva DA, Wiench M, Georgel PT, Schiltz RL, Hager GL, HDAC1 acetylation is linked to progressive modulation of steroid receptor induced gene transcription, Mol. Cell 22 (2006) 669–679. [DOI] [PubMed] [Google Scholar]
- [76].Nagaich AK, Rayasam GV, Martinez ED, Johnson TA, Elbi C, John S, Hager GL, Subnuclear trafficking and gene targeting by nuclear receptors, Ann. N.Y. Acad. Sci 1024 (2004) 213–220. [DOI] [PubMed] [Google Scholar]
- [77].Voss TC, John S, Hager GL, Single cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligandspecific transcription, Mol. Endocrinol 20 (2006) 2641–2655. [DOI] [PubMed] [Google Scholar]