Abstract
In the past decade, an exciting realization has been that diverse liver diseases, ranging from non-alcoholic steatohepatitis, alcoholic steatohepatitis, and cirrhosis, to hepatocellular carcinoma, are not unrelated but fall along a spectrum. Recent work on the biology of the gut-liver communication axis has assisted in understanding the basic biology of both alcoholic and nonalcoholic fatty liver disease. Of immense importance is the massive advancement in understanding of the role of the microbiome, driven by high-throughput DNA sequencing and improved computational techniques that allow the complexity of the microbiome to be interrogated, together with improved experimental designs. Here, we review the gut-liver communications of these various forms of liver disease, explore the molecular, genetic and microbiome relationships, discuss prospects for exploiting the microbiome to determine the stage of liver disease, and to predict the effects of pharmaceutical, dietary, and other interventions at a population and individual level. We conclude that although much remains to be done in understanding the relationship between the microbiome and liver disease, rapid progress towards clinical applications is being made, especially in study designs that complement human intervention studies with mechanistic work in mice that have been humanized in multiple respects, including the genetic, immunological and microbiome characteristics of individual patients. These “avatar mice” may be especially useful for guiding new microbiome-based or microbiome-informed therapies.
Introduction
The crosstalk between the gut and liver is increasingly recognized, strengthened by the parallel rise in liver diseases and gastrointestinal (GI) and immune disorders.1,2 The most common type of liver disease, nonalcoholic fatty liver disease (NAFLD), alone affects more than 65 million Americans with a cost burden of $103 billion annually within the US itself.3 To manage the socio-economic burden of GI-associated liver diseases by developing new therapeutic modalities, we must elucidate specific molecular events that facilitate interaction between the gut and the liver. As we begin to appreciate these links, animal models4–6 as well as well-designed, clinical studies7–9 are already revealing key components of these interactions.
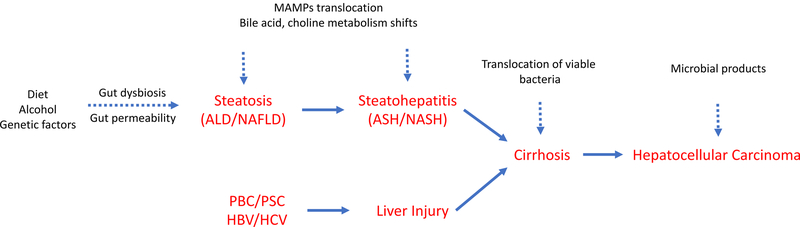
The present understanding of the etiology of the spectrum of liver diseases (Figure 1) is underpinned by proinflammatory changes in the host. Intestinal dysbiosis and increased intestinal permeability leads to translocation of microbes and microbial products including cell-wall components (endotoxins from gram-negative bacteria, β-glucan from fungi) and DNA, together referred to as microbial- (or pathogen-) associated molecular patterns (MAMPs/PAMPs). These patterns are recognized by immune receptors on liver cells such as Kupffer cells and hepatic stellate cells and lamina propria (an immune cell-rich tissue beneath the intestinal epithelium) which initiate and maintain inflammatory cascades that ultimately lead to liver damage in the form of fibrosis.10–13 This damage can progress from cirrhosis (severe fibrosis) to hepatocellular carcinoma (HCC), the most predominant form (more than 80%) of primary liver cancers.14 Previously demonstrated associations between intestinal health and several different types of neoplasia suggest a potential role of the microbiome in HCC.15,16 Additionally, the liver and microbiome engage in co-metabolism of xenobiotics including carcinogens, which can independently predispose the host to HCC.17,18
Figure 1: Physiological manifestations of liver injury along a spectrum of progression.
Risk factors such as alcohol abuse, unbalanced diet, infection (HBV/HCV) or immune dysfunction (PBC/PSC) can independently lead to liver injury. Alcohol-abuse patients and obese individuals often develop steatosis (fatty liver), which is characterized by increased intestinal permeability and dysbiosis. Subsequently, bile acid and choline homeostasis is disturbed along with increased translocation of MAMPs across the gut-barrier, leading to steatohepatitis, the progressive form of liver damage. Both, steatosis-dependent and steatosis-independent liver damage can progress to cirrhosis (end-stage liver damage), which is marked by translocation of viable bacteria to the liver and severe inflammation. As liver function is progressively compromised, tumor-promoting metabolites and xenobiotics accumulate. These could activate oncogenic pathways causing hepatocellular carcinoma, the most predominant form of primary liver cancers.
(MAMPs: Microbial-associated molecular patterns; ALD: Alcoholic liver disease; NAFLD: Nonalcoholic fatty liver disease; ASH: Alcoholic steatohepatitis; NASH: Nonalcoholic steatohepatitis; HBV: Hepatitis B virus; HCV: Hepatitis C virus; PSC: Primary sclerosing cholangitis; PBC: Primary biliary cholangitis)
The missing links in the complex interaction network between host and microbes are being discovered piece-by-piece using various experimental designs (detailed later). These findings encourage microbiome-oriented therapeutic modalities to treat liver-associated as well as other metabolic diseases. Here, we review the current understanding of the etiology of liver diseases and discuss the open research questions (Box 3) to motivate focused research in this area with special attention to the role of the microbiome.
Box 3: Open research questions.
Mounting evidence implicates the gut microbiome in the development and progression of different forms of liver disease. However, several questions remain open and must be answered to advance the field.
Is there a set of microbes (beneficial or harmful) that can read out the current extent, or predict the future extent, of disease progression in patients with ALD and NAFLD?
Can microbiome research using a consistent set of methodologies, including multi-omics profiling, provide a consistent mechanistic picture that unifies our understanding of the relationships among forms of liver disease?
Can fecal microbiota transplant, or collections of probiotic strains isolated from human feces, be expanded as a therapeutic modality for liver disease?
Does introducing a humanized microbiome into an HCC avatar mouse improve its fidelity in terms of responding to therapeutic options like an individual patient?
How do the liver and gut communicate?
The gut and liver communicate via tight bidirectional links through the biliary tract, portal vein and systemic circulation (Figure 2). The liver communicates with the intestine by releasing bile acids and many bioactive mediators into the biliary tract and the systemic circulation. In the intestine, host and microbes metabolize endogenous (bile acids, amino acids) as well as exogenous substrates (from diet and environmental exposure), the products of which translocate to the liver through the portal vein and influence liver functions.19 Some crucial links between the gut and liver are discussed below.
Figure 2: Bidirectional communication between gut and liver.
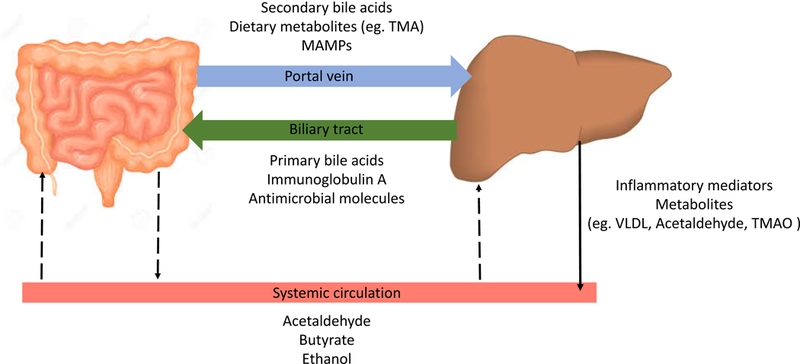
The liver transports bile salts and antimicrobial molecules (IgA, angiogenin 1) to the intestinal lumen through the biliary tract. This maintains gut eubiosis by controlling unrestricted bacterial overgrowth. Bile salts also act as important signaling molecules via nuclear receptors (such as FXR, TGR5) to modulate hepatic bile acid synthesis, glucose metabolism, lipid metabolism and energy utilization from diet. On the other hand, gut-products such as host and/or microbial metabolites and MAMPs translocate to the liver via the portal vein and influence liver functions. Additionally, systemic circulation extends the gut-liver axis by transporting liver metabolites from dietary, endogenous or xenobiotic substances (eg. FFAs, choline metabolites, ethanol metabolites) to the intestine through the capillary system. Owing to this medium of transport and ease of diffusion of systemic mediators across blood capillaries, these could affect the intestinal barrier both, positively (eg. butyrate) or negatively (eg. acetaldehyde)
(TMA: Trimethylamine; TMAO: Trimethylamine N-oxide; MAMPs: Pathogen-associated molecular patterns; VLDL: Very low-density lipoprotein; FXR: Farnesoid X receptor; TGR5: Takeda G-protein coupled receptor 5; FFA: Free fatty acid)
Enterohepatic circulation of bile acids
Bile acids (BAs) are amphipathic molecules synthesized from cholesterol in the pericentral hepatocytes. These are conjugated to glycine or taurine and released in the biliary tract. On reaching the small intestine through the duodenum, BAs, together with other biliary components, facilitate emulsification and absorption of dietary fats, cholesterol, and fat-soluble vitamins. About 95% of the BAs are actively reabsorbed in the terminal ileum and transported back to the liver 20,21. The remaining five percent are deconjugated, dehydrogenated and dehydroxylated by the intestinal microbiota to form secondary bile acids, which reach the liver via passive absorption into the portal circulation. The liver recycles BAs and secretes them back to the biliary tract completing the “enterohepatic circulation” i.e. a system of exchange between the gut and the liver.
A carrier-mediated process transports hydrophilic primary bile acids across cell membranes for uptake into intestinal epithelial cells. Regulatory effects of BAs have been best studied with respect to farnesoid X receptor (FXR) and Takeda G-protein-coupled receptor 5 (TGR5). BAs bind to FXR in the enterocytes and induce transcription of an enterokine, fibroblast growth factor 19 or FGF19 (FGF15 in mouse). FGF-19 reaches the liver through the portal vein and down-regulates de novo bile acid synthesis by inhibiting CYP7A1 in hepatocytes, forming a feedback system for modulating BA production.22 FXR activation is known to affect glucose and lipid metabolism.23,24 Additionally, BAs bind to TGR5 on the plasma membrane and act on tissues beyond enterohepatic circulation. This binding mediates host energy expenditure,25,26 glucose homeostasis27 and anti-inflammatory immune responses.28,29
BAs and the gut microbiota closely interact and modulate each other. BAs exert direct control on the intestinal microbiota. By binding to FXR, they induce production of antimicrobial peptides such as angogenin1 and RNAse family member 4, which are directly involved in inhibiting gut microbial overgrowth and, subsequently, gut barrier dysfunction.30,31 Intestinal dysbiosis shifts the balance between primary and secondary bile acids and their subsequent enterohepatic cycling, the metabolic effects of which are not comprehensively understood. However, because of differences in the affinity of these two classes of BAs for the FXR, these shifts have been associated with increased hepatic bile acid synthesis and metabolic stress.32–35 An imbalance in BAs and gut bacteria elicits a cascade of host immune responses relevant to the progression of liver diseases.
Intestinal permeability
The central components of the intestinal barrier are enterocytes that are tightly bound to adjacent cells by apical junctional proteins that include claudins, occludins, junctional adhesion molecules (JAMs) and E-cadherins.36 This barrier restricts movement of microbes and molecules from the gut lumen, while allowing permselective, active transport of nutrients across the tight junctions. The intestinal barrier is further strengthened by several additional lines of defense:
-
(1)
Mucins (heavily glycosylated protein aggregates) form a physical barrier between luminal bacteria and the underlying epithelial layer36
-
(2)
Antibacterial lectins, such as regenerating islet-derived protein III-gamma (REG3G), which are produced by intestinal paneth cells to target bacteria associated with mucosal lining37,38
-
(3)
Immunoglobulins, specifically sIgA, produced by plasma cells and transported into the lumen through the intestinal epithelial cells that neutralize microbial pathogens by blockading epithelial receptors39
-
(4)
Commensal bacteria are closely associated with the gut mucosa, and reinforce barrier integrity by stimulating cell-mediated immunity via toll-like receptor mediated signaling37,40 or by producing metabolites that directly strengthen tight junctions (short chain fatty acids)41–43 and inhibit other microbes44–46
Breakdown of one or more of these barrier components compromises gut-barrier integrity. The major drivers of increased permeability include gut inflammation and dysbiosis47,48 which have been linked to consumption of high-fat, Western diet,49–51 chronic alcohol consumption,52–54 prolonged antibiotic usage,55 and immune-mediated inflammatory diseases such as IBD.56 An important association between the gut microbiota, inflammation and gut-barrier integrity is provided by Akkermansia muciniphila, a gram-negative anaerobe that colonizes the intestinal mucus layer. Reduced abundance of A. muciniphila has been associated with thinning of mucus layer and increased inflammation promoting both, alcoholic and nonalcoholic liver damage.57,58 When the gut barrier is compromised, microbes and microbe-derived molecules can translocate to the liver through the portal system, causing inflammation and hepatic injury. Some translocated intestinal products may also directly interact with host factors and contribute to exacerbation of liver disease.59–64
Systemic circulation
Bacteria and MAMPs
Intestinal permeability is characterized by compromised tight junctions between enterocytes, and is consistently seen across the spectrum of liver diseases.65,66 Liver damage is associated with small intestinal bacterial overgrowth (SIBO) and microbial dysbiosis of the lower gastrointestinal tract.67 Together, these lead to increased translocation of MAMPs into the portal circulation. On reaching the liver, MAMPs induce localized inflammation through pattern recognition receptors (PRRs) on Kupffer cells68 and hepatic stellate cells.69,70 Endotoxin-mediated activation of Toll-like Receptor-4 (TLR4) 69,68 along with TLR9 (activated by methylated DNA70 and TLR2 (activated by gram-positive bacteria)71 are the primary drivers of immune response in liver disease. TLR signaling in Kupffer cells activates downstream proinflammatory cascade, leading to MyD88 mediated activation of NF-kB.13 Additionally, TLR4 signaling also promotes fibrosis by down regulating Bambi, a decoy receptor for TGF-β. 13 These lead to expression of inflammatory cytokines, oxidative and endoplasmic reticulum (ER) stress, and subsequent liver damage.72
Choline metabolites
Choline is a macronutrient that is important for liver function, brain development, nerve function, muscle movement, and maintaining a healthy metabolism.73 (Rodents fed a choline-deficient diet have been used to model human nonalcoholic steatohepatitis.74–76) Choline is processed into phosphatidylcholine (lecithin) by the host, which assists in excretion of very-low density lipoproteins (VLDL) particles from the liver. This prevents hepatic accumulation of triglycerides (liver steatosis). Additionally, choline can also be converted to trimethylamine (TMA) by intestinal bacteria. TMA can translocate to the liver through the portal circulation where it is converted to trimethylamine N-oxide (TMAO).77
The significance of methylamines is increasingly being recognized with respect to liver, cardiometabolic and more recently, mental disorders77,78. Increased systemic circulation of TMAO is concomitant with reduced levels of host-produced phosphatidylcholine, an imbalance characteristic of intestinal dysbiosis. This has been linked with liver damage due to increased triglyceride accumulation (hepatic steatosis)9,77,79–81 and consequently, non-alcoholic fatty liver disease9 and liver tumorigenesis.82.
Free fatty acids
Free fatty acids include short-chain fatty acids (SCFA) and saturated long-chain fatty acids (LCFA). Butyrate and propionate (products of bacterial fermentation) are the dominant short chain fatty acids in the large intestine. Butyrate is an energy source for the enterocytes and facilitates maintenance of intestinal barrier.41–43 Alcohol-induced liver injury is suggestively marked by reduced butyrate and propionate83,84 and increased acetate (possibly produced by ethanol metabolism in the lumen, but predominantly derived from ethanol metabolism in the liver). Increased acetaldehyde can weaken gut barrier59,85 and induce hepatic stress 86,87 on translocation of intestinal antigens to the liver. Butyrate supplementation in the form of a glycerol ester, tributyrin, reduced ethanol-induced intestinal permeability and subsequent liver injury in mice on a short-term alcohol diet. However, how tributyrin mechanistically protects intestinal barrier remains to be established.
Luminal species of LCFAs include pentadecanoic acid (C15:0), palmitic acid (C16:0), heptadecanoic acid (C17:0), and stearic acid (C18:0). In mice fed alcohol chronically, C15:0 and C17:0, which are only produced by bacterial fermentation,88 are significantly reduced when compared to control on isocaloric diet.83,89. There is also an overall reduction in total saturated LCFAs highlighting the importance of bacterial contribution in LCFA homeostasis. Of the remnant LCFA species, C16:0 and C18:0 were in highest concentration which suggests lower microbiome involvement (which is disrupted by alcohol consumption) in their production.83 Lactobacillus spp. are known metabolizers of saturated LCFAs and a reduction in their concentration is concomitant with decreased luminal Lactobacilli.83 To our knowledge, restoring Lactobacillus spp. by LCFA supplementation has not been experimentally demonstrated. However, dietary supplementation of Lactobacillus rhamnosus has been shown to increase luminal LCFAs,89 suggesting that Lactobacillus-induced increase in intestinal FFAs contribute to its probiotic effects.90–96
Ethanol and acetaldehyde
The mucosa of the GI tract absorbs ethanol by simple diffusion. Within the GI tract, the majority of ethanol from food and beverages is absorbed by the stomach (~ 20%) and small intestine (~ 70%).97,98 Although, microbial fermentation contributes to luminal ethanol concentration, the biggest share of alcohol in the large intestine comes from the systemic circulation.59
Gut microbiota and enterocytes express alcohol-metabolizing enzymes such as alcohol dehydrogenase, aldehyde dehydrogenase co-metabolizing ethanol into acetaldehyde and, to a lesser-studied extent, acetate59,85,86The liver also responds to circulating levels of ethanol by upregulating its ethanol metabolism pathway.86,87 The importance of microbes for xenobiotics metabolism was underscored by a study that demonstrated an increase in hepatic expression of ethanol metabolizing genes in germ-free mice, and subsequent liver damage.86,87
Non-alcoholic and alcoholic liver diseases (Figure 3; Table 1) are characterized by increased luminal and circulating levels of ethanol and its metabolites, acetaldehyde and acetate.64,99 These metabolites have independently been associated to liver damage.61–63 Acetaldehyde has been implicated in weakening the intestinal tight junctions compromising the gut barrier and allowing translocation of microbial products.100–105 It has also been associated with downregulating the expression of antimicrobial peptides (AMPs) in the intestine,106,107 and eliciting inflammatory and adaptive host immune responses.108–110 Additionally, ALD is marked by reduced intestinal butyrate83,111,112 (an energy source for enterocytes) which is linked to weakening of intestinal tight junctions and hence, permeability.84,113–115
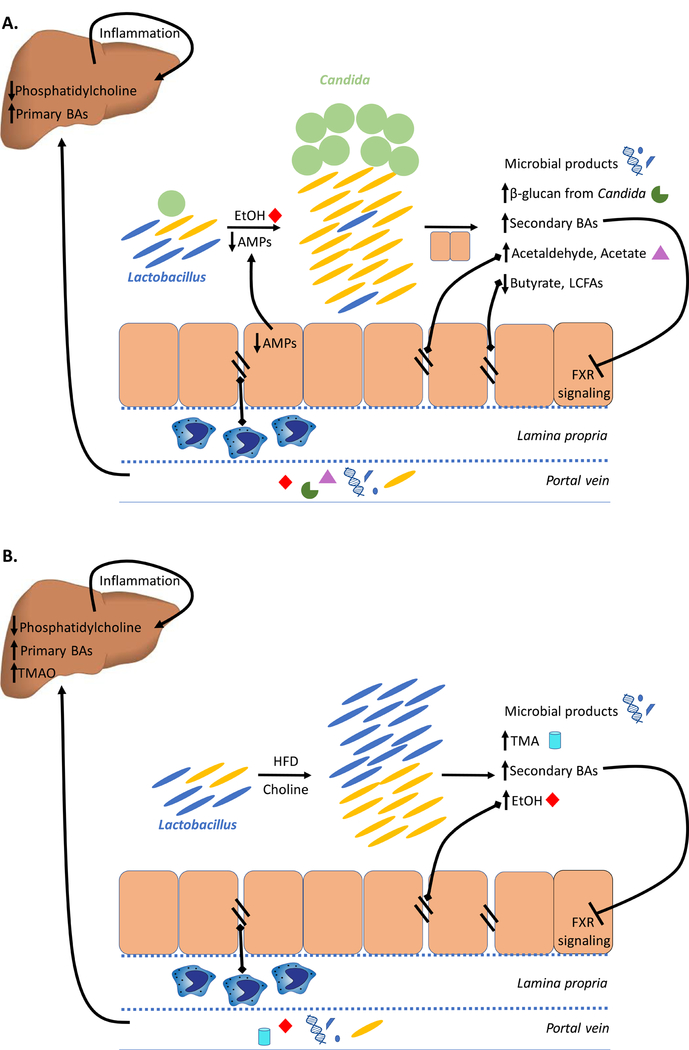
Figure 3. Interplay between the liver and gut microbiome in (A) Alcoholic liver disease (ALD) and (B) Nonalcoholic fatty liver disease (NAFLD).
Intestinal dysbiosis and bacterial overgrowth is observed in both, ALD and NAFLD. Bacterial overgrowth causes an increase in secondary BAs which disrupts FXR-mediated modulation of BA levels, leading to an overall increase in hepatic BA synthesis. A reduction in hepatic phosphatidylcholine is also seen in both ALD and NAFLD, which causes triglyceride accumulation in the liver (fatty liver). While ALD-associated dysbiosis is characterized by reduction in Lactobacillus and Candida overgrowth, NAFLD patients have higher abundance of Lactobacillus (effects on fungal population remain to be investigated). Both, in ALD and NAFLD, increased ethanol and its metabolite acetaldehyde in the intestinal lumen mediates weakening of intestinal tight junctions. Consequently, increased translocation of MAMPs (seen in ALD and NAFLD) and gut metabolites such as acetaldehyde, acetate (seen in ALD) and TMA (seen in NAFLD) elicits intestinal and hepatic inflammatory responses, leading to progressive liver damage.
(AMP: Antimicrobial peptides; BA: Bile acids; EtOH: Ethanol; FXR: Farnesoid X receptor; HFD: High-fat diet; LCFA: Long-chain fatty acids; TMA: Trimethylamine; TMAO: Trimethylamine N-oxide)
Table1.
Comparison of alcoholic and nonalcoholic liver disease
| Alcoholic Liver Disease (ALD) | Nonalcoholic Liver Disease (NAFLD) | |
|---|---|---|
| SIBO | Observed106,214,215 | Observed65 |
| Gut microbiota | ↑ Enterobactericeae (humans)133, 132 ↓ Lactobacillus215, 133 (humans and mice), Bacteroidetes (humans)132,134, Akkermansia muciniphila (humans and mice)57 Gut microbiota protects against alcohol -induced liver injury86 Reduced fungal diversity; Candida overgrowth8 |
↑ Enterobactericeae (humans)216,64, Lactobacillus (humans)216, 217, Bacteroides (humans and mice)158, 125, Ruminococcus (humans)125 ↓ Prevotella (humans)125, 216, Akkermansia muciniphila (mice)58 Gut microbiota mediates HFD-induced liver steatosis218, 219 (Fungal dysbiosis not demonstrated) |
| Reversibility of gut dysbiosis |
Partial reversibility on abstinence113, 94 | (Reversibility not demonstrated) |
| Inflammation |
↑ Intestinal TNF-α (mice)103 ↑ Systemic inflammatory markers (humans)47, 220 |
↑ Intestinal TNF-α, IFNγ, IL-6 (humans and mice)221, 216 ↑ Systemic inflammatory markers (humans)222 |
| Transferability via microbiome | FMT from alcoholic hepatitis patients caused severe liver inflammation and injury in mice198 FMT from ALD resistant to ALD susceptible mice prevented liver injury in recipient131 |
Cohousing inflammasome deficient, NASH mice with WT mice exacerbated liver steatosis WT cage mates48 FMT from NAFLD-susceptible mice promoted liver injury in recipient223 |
| Translocation | ↑ PAMPs translocation (endotoxins224, 47, 225, 226, β-glucan8, viral/bacterial DNA227, 224 (humans and mice)) |
↑ PAMPs translocation(endotoxins225, 228, viral/bacterial DNA229 (humans and mice)) |
| Bile acids | ↑ Total plasma bile acids (humans)230 ↑ Hepatic bile acid synthesis (humans and mice)141, 142 |
↑ Total serum bile acids (humans)231 ↑ Hepatic bile acid synthesis (humans)35 ↑ Total fecal bile acids, primary to secondary bile acid ratio (humans)35 |
| Choline | ↓ Phosphatidylcholine in plasma and liver (rats)232, 233 (Changes in trimethylamine not demonstrated) |
↓ Phosphatidylcholine in plasma (mice)234 ↑ Intestinal trimethylamine (mice)234 |
| Free-fatty acids | ↓ Bacterial fatty-acid biosynthesis (mice)83 LCFA and SCFA supplementation reduced ethanol-induced liver injury (mice)83,115 |
↑ Free-fatty acids in the liver235 |
| Ethanol | ↑ Blood ethanol, luminal acetaldehyde130 ↑ Systemic acetate234, 83 |
↑ Blood ethanol64, 236, 237 |
Links between the microbiome and specific liver diseases
Nonalcoholic fatty liver disease (NAFLD)
NAFLD refers to a spectrum of liver disease that can be broadly classified into two categories: nonalcoholic fatty liver (NAFL), the non-progressive form of NAFLD, and nonalcoholic steatohepatitis (NASH), the progressive form of NAFLD.116 NASH is generally linked to type 2 diabetes, cardiovascular risk factors and obesity117, 118, although incidences have also been reported in lean individuals, emphasizing that genetic and environmental factors also contribute to disease development. 119,120,121,122
Several studies have stressed on the role of the gut microbiota in NAFLD, but, causality is yet to be established123. Patients with NAFLD have a higher prevalence of small intestinal bacterial overgrowth (SIBO)65,124 and microbial dysbiosis. Using 16S amplicon sequencing, Boursier et al.125 found that the bacterial genera, Bacteroides and Ruminococcus were significantly increased, and Prevotella was reduced in NASH patients with stage 2 fibrosis or higher. Loomba et al. (2017)7 utilized whole genome metagenomics to characterize the gut microbiota in NAFLD patients with and without advanced fibrosis (stages 3 and 4) and showed an increased abundance of Escherichia coli, and Bacteroides vulgatus in advanced fibrosis patients. An enrichment of Escherichia (genera) was also seen in pediatric NASH patients compared to obese controls.64 Consistent with preclinical studies, these studies indicate an association between gram-negative bacteria and progression of liver fibrosis.126
Genetically modified mouse models have been used to study NAFLD-associated gut dysbiosis and permeability for mechanistic insights in liver disease progression. Rahman et al. (2016)127 used JAM-A (junctional adhesion molecule-A protein) knockout mice to demonstrate that deficiency in this tight junction protein is linked to increased intestinal permeability and liver inflammation. This inflammation could be alleviated by administering antibiotics, underscoring the importance of microbial translocation in promoting immune response in the liver. Another group used muc-2 knockout mice and found that there was a compensatory increase in intestinal levels of Reg3b and Reg3g genes leading to an overall protective response against NAFLD.107
The contribution of liver-damaging inflammation in response to translocation of microbes and MAMPs was elucidated by Henao-Mejia and colleagues (2012)48. Using NLRP3- and NLRP6- (inflammasome-) deficient mice models, they demonstrated an increase in influx of TLR4 and TLR9 in portal circulation, which enhanced the expression of hepatic tumor-necrosis factor (TNF)-α driving NASH progression. Furthermore, cohousing inflammasome-deficient mice with wild-type controls exacerbated hepatic steatosis and obesity in healthy cage mates, suggesting transferability of disease via the microbiome.
Increasing links between NAFLD and the gut microbiome at both the observational and mechanistic levels have gut microbiota an attractive source of biomarkers for early diagnosis of NAFLD. In a comparison between obese children with and without NASH, Zhu and colleagues64 observed significantly elevated gut microbial production of ethanol in NASH patients. NAFLD patients also show increased systemic TMAO9 and hepatic bile-acid synthesis35, and decreased production of phosphatidylcholine.128 Recently, Loomba et al. further observed differences in carbon and amino acid metabolism in gut microbiome of patients with NAFLD-associated advanced fibrosis. This proof of concept study provides preliminary evidence to support the utility of a microbiome-derived metagenomics signature to detect advanced fibrosis and as well as candidacy for anti-fibrotic treatment trials in NAFLD.
Alcoholic liver disease (ALD)
The manifestation of ALD in chronic alcohol abuse patients is a consequence of multifactorial interactions involving genetics, immune system, gut microbiome and environmental factors.100,129,130,131 Like NAFLD, non-progressive form of ALD is characterized by accumulation of fat inside liver (fatty liver or steatosis), while it’s progressive form is marked by inflammation and liver injury (alcoholic steatohepatitis or ASH).
Our understanding of the compositional and mechanistic contributions of the gut microbiota in ALD is improving with the increasing number of studies investigating this link. As in NAFLD, SIBO has been demonstrated as an important hallmark of alcohol-associated liver disease in humans35 and mice.106,131 Intestinal dysbiosis in alcohol-abuse patients is characterized by significant enrichment of Enterobactericaea (family) and reduction in Bacteroidetes and Lactobacillus (genera). 132,106,133,134 It has also been demonstrated that alcohol-induced dysbiosis is only partially reversible by alcohol-withdrawal or probiotic treatment.94,113 Interestingly, alcohol-dependent patients also displayed reduced fungal diversity and Candida overgrowth, presenting the first evidence of the role of gut mycobiome in pathogenesis of liver diseases.8
Genetically-modified murine models have advanced our mechanistic understanding of the contribution of various components of the gut-barrier in the etiology and progression of ALD. Using Reg3b(−/−) or Reg3g(−/−) mice, it was found that REG3 lectins protected against alcoholic steatohepatitis by reducing mucosa-associated microbiota, thereby preventing translocation of viable bacteria.135 Muc-2 deficient mice were protected against alcohol-induced inflammation (similar to HFD-induced inflammation in NAFLD model) due to a compensatory increase in Reg3g and Reg3b lectins.107 Furthermore, IgA knockout in mice led to increased levels of IgM and a net protective effect against ASH progression.136
In response to ethanol-induced gut-barrier dysfunction and translocation, TLRs and other pathogen recognition receptors activate hepatic Kupffer cells and macrophages, as was demonstrated in male Wistar rats.137 This initiates inflammatory cascades releasing TNF-alpha, IL-1, IL-10, IL-12, and TGF-beta. 138,139,140 Using TLR-4 chimeric mice, it was shown that endotoxin-induced release of TGF-beta is mediated by MyD88-NF-kappaB-dependent pathway providing explanatory mechanism of inflammation-induced liver damage.68 Furthermore, increased translocation of fungal β-glucan also induced liver inflammation via CLEC7A receptor on hepatic Kupffer cells such that treatment of mice with antifungal agents reduced intestinal fungal overgrowth, decreased β-glucan translocation, and ameliorated ethanol-induced liver disease.8
Concomitant with immunological responses to barrier dysfunction, ALD is also marked by system-wide changes in many bioactive compounds. Alcohol consumption leads to an increase in hepatic bile acid synthesis humans and mice. 141,142 This could be explained by dysbiosis-associated disruption in FXR activation in the enterocytes as FXR deficient mice were more likely to develop ethanol-induced steatohepatitis143, and treatment with an FXR agonist (WAY-362450) had protective effects against liver damage.144 Alcohol-associated dysbiosis in mice was further linked to reduced LCFA biosynthesis such that LCFA supplementation restored eubiosis. In fact, a significant correlation between Lactobacillus spp. and bacterial LCFA (C15:0 and C17:0) was found in ALD patients but not in healthy controls.83 Butyrate (SCFA) production was also negatively altered following ethanol exposure and administration of butyrate in the form of tributyrin mitigated alcohol-induced liver injury in mice.84
With increasing evidence of mechanistic links between the gut microbiota and liver disease progression, fecal microbiota transplant (FMT) is being explored as a therapeutic option for ALD. However, larger, carefully designed trials across multiple ethnic groups are needed before FMT can be considered safe in routine clinical practice for managing ALD.
Cirrhosis
Cirrhosis (or end-stage liver disease) is an extreme manifestation of chronic liver injury characterized by loss of liver cells, thick fibrous scar, and regenerating nodules. This topic has been extensively reviewed recently so we only provide a brief discussion here.145 NAFLD, ALD, primary biliary cholangitis (PBC; Box 1), primary sclerosing cholangitis (PSC; Box 2) or hepatitis can each progress to cirrhosis and constitute its subtypes. Currently, NASH is the second leading cause of adult cirrhosis in the USA.146 Depending upon the etiology of cirrhosis, there is a variable risk of developing HCC.
Box 1: Primary Biliary Cholangitis (PBC) .
Primary biliary cholangitis (PBC) is characterized by inflammation-mediated damage to the small bile ducts inside the liver gradually progressing to liver fibrosis and cirrhosis. Previously considered as a typical autoimmune disorder, the modified etiological understanding of PBC considers proinflammatory changes in the gut-microbiota, intestinal bile acid disruptions, and gut-barrier dysfunction.201–204 Consequently, MAMPs ascend within the biliary duct, perpetuating infection. An immune attack against the biliary epithelial cells is mediated by antibodies that recognize E2 subunit of pyruvate dehydrogenase complex (PDC-E2) due to cross-reactivity with conserved proteins in Escherichia coli,201 Lactobacillus delbrueckii,205 and Novosphingobium aromaticivorans.206 In fact, genetically susceptible mouse strains developed liver lesions mimicking PBC when infected with Novosphingobium aromaticivoransm, which further grounds the implications of microbiome associations in this disease.207 Ursodeoxycholic acid, a tertiary bile acid produced by Ruminococcus has been approved for PBC-treatment.208 Thus, microbiome-based treatment modalities hold promise for managing PBC and should be studied further.
Box 2: Primary Sclerosing Cholangitis (PSC).
Primary sclerosing cholangitis (PSC), is also an immune-mediated disease of the bile ducts. However, unlike PBC, PSC can affect bile ducts, both inside and outside of the liver. Gut dysbiosis-mediated bile dysregulation, intestinal permeability and translocation of proinflammatory molecules in the portal vein characterizes PSC.201,209,210 The immune reaction in PSC is mediated by autoantibodies including p-ANCAs (perinuclear antineutrophil cytoplasmic antibody) that recognize the ubiquitously expressed bacterial antigen, FtsZ.211 Furthermore, increase in microbe-associated TLR expression and T helper type 17 (Th17) cells has been reported in PSC which strongly suggests microbiome involvement in disease pathogenesis.212,213 PSC is closely associated with inflammatory bowel disease (IBD), in particular ulcerative colitis and shares some of its characteristic features (such as increased Th17 cells). Thus, a common disease mechanism may be at play, and novel treatment avenues by targeting microbe-associated immune pathways can be explored.
Alterations in the gut microbiome including dysbiosis and SIBO have been associated with cirrhosis and its complications.147–149 Treatment for portal systemic encephalopathy and decompensated cirrhosis includes treatment with nonsystemic antibiotics to reduce intestinal microbiota overgrowth.150–152 Gut microbiome alterations were observed in alcohol- and hepatitis-associated cirrhotic patients in a Chinese cohort,153 which observed an invasion of the lower intestinal tract by oral bacteria. Concordant with these findings, Chen and colleagues (2016) also found an overrepresentation of genera including Veillonella, Megasphaera, Dialister, Atopobium, and Prevotella in the duodenum of cirrhosis patients. The genera, Neisseria and Gemella were discriminative between hepatitis-B-virus- and PBC-related cirrhosis.154 Recently, Bajaj and colleagues observed significant fungal dysbiosis in cirrhosis patients and showed that Bacteroidetes to Ascomycota ratio could independently predict hospitalization in these patients.155
All experimental models of liver fibrosis result in gut microbial dysbiosis and increased intestinal permeability and treatment of GI tract with nonabsorbable antibiotics decreases liver fibrosis. Mice with genetic ablations of the receptors for bacterial product ligands, TLR2, TLR4, TLR9, and NLP3, are protected from experimental liver fibrosis.156 The current treatment philosophy involves decreasing the bacterial product ligands or blocking their receptors, which results in decreased inflammatory and fibrogenic signaling in the liver, although no antifibrotic drug is currently available for routine clinical practice.
Hepatocellular carcinoma (HCC)
The etiology of non-viral HCC follows a “multiple-hit” pathway, whereby liver steatosis followed by oxidative stress, endoplasmic reticulum (ER) stress together with intestinal dysbiosis and inflammation contribute to the final manifestation of cancer.
The gut microbiota changes in composition dramatically in hosts suffering from HCC. Clostridium species have been found to be enriched in obesity-induced mouse models of HCC157,158, but clinical studies with HCC patients detect an overgrowth of intestinal Escherichia coli.159 Murine models as well as human studies have reported a migration of Helicobacter species to HCC tumor tissues.160–163 Notably, members of this genus are known to promote tumor-development by activating NF-kB and WNT signaling and suppressing anti-tumor immunity, and might play a potential role in HCC development.160,164
To get insights into the molecular events explaining the progression of liver disease to HCC, various murine models (diet-based, toxin plus diet-based and genetic plus diet-based models) have been explored. However, most of these have proven suboptimal because they either do not develop all intermediate pathological & metabolic stages, or they manifest HCC incompletely (Febbraio and Karin, submitted). We have highlighted some frequently-used rodent models, their usage and caveats in Table 2 to aide future research.
Table 2.
Experimental Mouse models for liver disease
| Model | Description | Liver pathology | Microbiome Features |
| Diet | |||
| High Fat diet | Diet using higher saturated fat, or supplemented with cholesterol compared to chow |
Induces fatty liver and hepatic steatosis. Associated with metabolic syndrome phenotype.238 |
Common model for inducing dysbiosis; associated with changes in the microbiome |
| Choline Deficient Diet |
A high fat diet with choline and methionine omitted. |
Induces fatty liver, steatosis and inflammation and fibrosis. The model does not contribute to metabolic syndrome.5 |
Small study suggests diet- induced changes239 |
| Ethanol supplemented liquid diet |
A model of chronic alcohol abuse administered as an isocaloric diet where ethanol or maltose and dextrose are supplemented. Diet can be administered orally (Lieber-DeCarli240) or intragastrically (Tsukamoto-French241) |
Oral supplementation leads to inflammation and fatty liver, representing a good model for early ALD Intragastric administration leads to severe steatosis and mild fibrosis4 |
Diet affects the abundance of several taxa and is associated with changes in the microbiome57 |
| Genetic Manipulations | |||
| Knock out model | A mouse line where both copies of a gene have been removed |
This depends on the gene. For example, FxR−/− mice have more fatty liver accumulation on a high fat diet.31 Muc2−/− are protected from diet- induced liver injury.107 GSTA4−/−, PPAR-α−/− double knockout mice have increased inflammation and fibrosis compared to either single mutant or WT242 |
The microbiome of lineage- derived mice is distinct from wild type mice. This is likely to be an effect of microbiome drift within the colonies, rather than a direct effect of the genotype.243 |
| Littermate controls | Mice from a heterozygous cross that lead to Wild Type and knockout littermates. |
Much of the mouse microbiome is acquired through vertical transmission; littermates are better microbial controls.244 |
|
| Cre-Lox localized mutation |
A genetic cross that allows for tissue-specific knockout of a gene |
This is gene dependent on the gene. A Cre/Lox model of liver specific E-cadherin knockout shows pathology like primary sclerosing cholangitis, and increases susceptibility to cancer.245 The loss of TLR 5 in hepatocytes leads to increased inflammation and fibrosis in a high fat diet induced model of NASH246 |
Microbiome considerations depend on the how the controls are selected. |
| Avatar Mice | Mice transplanted with solid state tumors from cancer patients. |
Human hepatocellular carcinoma can be transplanted into the mouse.247 |
There is no specific effect on the microbiome. |
| Microbiome | |||
| Antibiotic treatment |
Treatment with a broad- spectrum antibiotic |
No direct effect on liver disease; Antibiotics can moderate the effect of other interventions. |
Antibiotics can have off target effects and significantly alter the microbial community in addition to decreasing the bacterial load194 |
| Probiotic manipulation |
Microbial supplementation to modify the microbiome |
No direct effect on liver disease; probiotics can modulate the effect of other treatments: Lactobacillus to ameliorate alcohol-induced liver injury)106 |
Can lead to the over- abundance of a specific organism or correct defects in the community. However, not all probiotics colonize. |
| Germ-Free Mice | Raised without any bacterial community |
Germ free mice have immune defects.248 These mice are also more susceptible to alcohol- induced liver injury.86 |
Useful to demonstrate the importance of bacterial communities for a phenotype. |
| Monoculture gnotobiotic mice |
Germ free mice that have been colonized with a single bacterium or defined bacterial community |
No direct effect; depends on the community transplanted and challenge |
Can test whether the defined community can modulate the phenotype |
| Mouse transplant | Bacterial communities from mice transplanted into germ free mice |
No direct effect; depends on the community transplanted and challenge |
Demonstrates whether mouse phenotype is transferable or can be modulated through the microbial community. |
| Humanized mice | Germ free mice which have been gavage with the microbiome from a human donor |
No direct effect; depends on the community transplanted and challenge |
Demonstrates whether human phenotype is transferable or can be through the microbial community. |
Accumulating evidence suggests that HCC-associated dysbiosis is accompanied by gut-barrier dysfunction, bacterial translocation, systemic circulation of their tumor-promoting metabolites and activation of proinflammatory and oncogenic signaling pathways.165 The intestinal poly-immunoglobulin receptor (PIgR) regulates the transport of IgA into the intestinal lumen and maintains microbial homeostasis.166 A recent study showed that PIgR−/− mice modelling NASH-induced HCC had increased systemic and liver IgA, and a concomitant increase in hepatic tumorigenesis due to localized inhibition of liver cytotoxic T cells that prevent HCC development. Further, the application of broad spectrum antibiotics has been shown to attenuate liver inflammation and HCC-development in mice157,167 highlighting the role of the intestinal microbiome in liver tumorigenesis. In another mouse model where HCC was induced by diethylnitrosamine (a carcinogen), activation of TLR4 due to LPS translocation upregulated the hepatic mitogen EREG in HSCs and activated NF-kB, resulting in enhanced tumor cell proliferation.167 Additionally, deoxycholic acid (DCA), a gut bacterial metabolite was shown to upregulate proinflammatory genes, such as IL6 and TNFa to provoke a senescence-associated secretory phenotype in HSCs.157,168–170
In addition to its role in HCC development, the gut microbiome also modulates pro-tumorigenic adaptive immune response via Th17 cells, which produce the proinflammatory cytokine IL-17A.171–173 The therapeutic efficacy of the anticancer drug cyclophosphamide depended on the interplay between Th17 signaling and gut microbiome such that germ-free tumor bearing mice or mice given non-absorbable antibiotics had reduced Th17 response and a subsequent resistance to therapeutic effects of cyclophosphamide was seen.174
Increased understanding of the role of the gut microbiota has motivated successful microbiome-based therapeutic modalities for HCC, such as treating with synthetic bile acids to reduce HCC risk in NAFLD patients,175 non-selective beta-blockers in the intestinal mucosa to prevent bacterial translocation and liver inflammation176 and administering probiotics in rodents modeling HCC to slow tumor growth and reduce tumor size.
Experimental design of microbiome studies
Given the intense recent interest in links between the microbiome and liver disease, we provide a brief overview of experimental models useful for researchers entering this field.
Much of our knowledge of the human microbiome comes from association studies that use either a cross-sectional or case-control design. Well-designed case-control studies are critical to demonstrate there may be a relationship between microbes and a disease of interest. However, these studies cannot establish causality, and are often subject to confounding variables. Most studies are conducted at a single time point in a population with the disease, and no long term follow up is performed. Consequently, these studies can only identify microbes that differentiate individuals with the disease and the control population. While these may have been causative agents, it is nearly impossible to separate this from secondary effects associated with the condition. For example, medication plays a major role in shaping the microbiome; a study of Type II diabetics found that treatment with Metformin had a larger effect on the microbiome than the disease.177 Similarly, we hypothesize the physiology of the disease may also contribute to changes in community structure.
Association studies are also often confounded by the selection of poor controls. The microbiome is dynamic,178,179 and cumulative exposures over an individual’s life, shaped by their diet,180 lifestyle,181 medical history177,182,177,182, genetics,183 and other factors184 create a unique community. Therefore, if cases and controls are not correctly selected, association studies may detect differences due to confounding factors. Matching cases and controls based on age and sex is often not sufficient. In cases where this is not possible, it is critically important to collect information about potential confounding factors.
Comparisons across current cross sectional studies are also challenging due to large effects due to technical parameters, including sample collection, storage, primer selection, and analysis techniques.184 Differences across studies increase the challenge of meta-analysis and increase the challenge of identifying causative clades.185 Some of these problems can be ameliorated by using consistent methodology.184,185
Twin studies provide a potential antidote to some of the problems with association studies. Twin pairs are naturally controlled for age and some early life exposures.186 Monozygotic twin pairs also share the same genetic background, further limiting potential confounders.186 Twin studies can be leveraged in two ways. First, identifying differences between discordant and concordant twin pair represent more powerful association studies, due to the partial internal control. Although these are particularly useful in young children, the approach can also be used with adults.187 Second, twin studies are critical to examine genetic control of the microbiome. A recent study of the UK Twins cohort suggested strong association of the microbiome and genes, including those associated with dietary preference and serum lipids.188
As the cost of microbiome analysis decreases, longitudinal studies are becoming more common. Understanding temporal fluctuation in the microbiome, and the role of microbes in contributing to disease etiology will rely on studies over time. Recent work suggests that community instability may, in and of itself, be a characteristic of an unhealthy ecosystem.189,190 Prospective studies, such as a recent study to looking at death from hepatocellular carcinoma in individuals with nonalcoholic fatty liver disease help identify the role of exposures and etiological factors in contributing to disease outcomes.191 Incorporating microbiome samples into these long term studies will help look at the role of microbial communities - either at a single time point or the community dynamics - as a contributing factor to complex conditions.192
Model animals also play an important role in shaping our understanding of the microbiome in disease (Table 2). Although rodent microbial communities are distinct from the human microbiome, there are some shared physiological and microbial-shared traits.193 Both rodent and human communities are dominated by the same set of bacterial phyla, although a smaller percentage of genera are shared. As such, experimental findings implicating individual organisms or genera in rodents should be taken with caution until they are validated in humans. Instead, rodent models can show phenotypic consequences of microbiome manipulation. This makes mice a useful model system to investigate causality, explore interactions, and test early interventions.
Both antibiotics and probiotics have been used to study the effect of changing the conventional murine microbiome on a phenotypic outcome. Antibiotics decrease the total bacterial load, as well as causing major perturbations in the microbial communities.194 In some cases, such as in liver disease models, this can demonstrate the role of bacterial products like lipopolysaccharide (LPS) in modulating inflammation.127 In other cases, like a recent addiction model, it can be used to demonstrate the importance of an intact microbiome in regulating behavior.195 Probiotics can also be used to look at the effect of a specific bacteria or bacterial cocktail within a controlled environment. A study of alcoholic fatty liver disease demonstrated an attenuation of the microbiome-mediated inflammation when a probiotic was used.106
Gnotobiotic, or germ free mice, can be used in multiple contexts. Comparisons of specific pathogen free laboratory mice and germ free mice can be used to examine the role of the microbiome in modulating an expressed or induced phenotype.196,197 More importantly, gnotobiotic mice can be humanized with a donor’s stool. This creates a system in which an individual’s microbiota can be tested, either for its ability to modulate a disease phenotype or as a target for intervention.187,196 For instance, in a recent small study, mice received their microbiome from either a donor with severe alcoholic hepatitis or no liver disease. Following alcohol treatment, the mice with the microbiome from the patient with alcoholic hepatitis showed greater liver damage than mice that received stool from the healthy donor.198
Well-designed mouse models that combine our current understanding of liver disease with humanized microbiomes offer some of the greatest potential for preclinical interventions. Avatar, or sometime called Patient-derived Xerograph (PDX) mice, are widely used in the cancer community to test the efficacy of chemotherapeutics for individual tumors, including HCC.199,200 This model better re-capitulates the complexity of a tumor than cell culture. Avatar mice can be further personalized by introducing an human immune system into an immuno-compromised mouse, along with the tumor.199 Generating this model in germ-free mice with a humanized microbiome as well as immune system expands our capacity to understand the role of the microbiome in modulating cancer. For example, this model could be used to study whether the microbiome of a patient with alcoholic liver disease leads to more tumor growth than the microbiome from a healthy control.
The use of well-designed experiments in both mice and humans promise to expand our understanding of the role of the microbiome in the development and progression of liver disease.
Conclusions
An accumulating body of research suggests that the disparate observations in liver disease-related studies could be unified and explained by the microbiome. It is now widely accepted that liver damage is a result of an extensive interplay between gut microbiota via specialized molecules such as TMA, acetaldehyde, LPS and host-immune system via Kupffer cells-mediated liver inflammation. However, a comprehensive understanding of the exchange between the microbiome and the liver still evades us. Animal models, particularly rodents, have been instrumental in elucidating many important mechanistic pathways in disease etiology. The introduction of the microbiome into these models will provide a more complete view of the cancer ecosystem. Because microbiome research is sensitive to technical variability that often masks underlying biological signal, there is a need for consistency in technical platforms and standardized protocols, so that findings from different laboratories (and model organisms) can be replicated and validated. Additionally, it is also critical to use an animal model that mimics human disease as closely as possible in all its physiological and metabolic manifestations.
We are slowly advancing from observation-based studies in human patients as recent research establishes grounds for microbiome-based therapeutic modalities such as fecal microbiota transplant (FMT) and probiotic interventions. However, effectively translating and applying findings accrued through animal models to humans requires well-designed, large-scale clinical trials spanning multiple disease etiologies and patient ethnicities. As the role of microbiota in liver disease development, prognosis and treatment is increasingly recognized, we emphasize on the need for focused, microbiome-aware efforts to efficiently tackle the socio-economic burden of this spectrum of liver diseases.
Key points.
The liver and intestine communicate extensively through the biliary tract, portal vein and systemic mediators. Liver products primarily influence the gut-microbiome composition and gut barrier integrity, whereas intestinal factors regulate bile acid synthesis, glucose and lipid metabolism in the liver.
Diverse liver diseases (ALD/ASH, NAFLD/NASH, PBC, PSC) are not unrelated, but converge along a common path of progression. Proinflammatory changes in the liver and intestine mediate development of fibrosis, cirrhosis and ultimately, hepatocellular carcinoma (HCC).
Alcoholic and nonalcoholic liver diseases share key characteristics such as intestinal dysbiosis, gut permeability and shifts in levels of bile acids, ethanol and choline metabolites.
Precise contributions of the microbiome to liver diseases may differ based on etiology. Improvements in experimental design and development of animal models is rapidly elucidating causal mechanisms.
Recent advances in understanding the gut-liver axis encourage research into microbiome-based, diagnostic, prognostic, and therapeutic modalities to improve management of liver diseases.
Acknowledgements
MK is supported by R01 AI043477 and R01 CA118165. RL is supported in part by the grant R01-DK106419-03. Research reported in this publication was supported in part by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number P42ES010337. BS is supported by NIH grants R01 AA020703, U01 AA021856, U01AA24726, and by Award Number I01BX002213 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development. JD is supported by the Robert Wood Johnson Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author biographies
Anupriya Tripathi is a Ph.D. student in Division of Biological Sciences at University of Calfornia San Diego (labs of Prof. Rob Knight and Prof. Pieter Dorrestein). She is interested in links between the gut microbiome, metabolome and cardio-metabolic diseases.
Justine Debelius is a postdoctoral research associate in the Knight Lab at the University of California San Diego. She is interested in links between the microbiome and the immune system.
David Brenner is Vice Chancellor for Health Sciences and Dean of the School of Medicine at the University of California San Diego. He is a physician-scientist who explores liver fibrosis in humans and animal models.
Michael Karin is a Professor of Pharmacology at the University of California San Diego and a Member of the National Institute of Medicine. He is an immunologist who has revolutionized our understanding of the links between the immune system and cancer.
Rohit Loomba is a Professor of Gastroenterology at the University of California San Diego, where he directs the NAFLD Research Center, and a board-certified hepatologist. He is a transplant hepatologist who is primarily interested in NAFLD and NASH.
Bernd Schnabl is an Associate Professor of Gastroenterology at the University of California San Diego, and is a staff physician and attending at the VA San Diego Medical Center in La Jolla and the UCSD Medical Center. His main research interests are in genetic and molecular mechanisms of chronic liver disease progression, and the gut-liver axis.
Rob Knight is a Professor of Pediatrics and Computer Science & Engineering at the University of California San Diego, where he directs the Center for Microbiome Innovation. His main research interest is in methods to characterize the microbiome in high throughput.
Footnotes
Competing interests
The authors declare no competing interests.
References
- •.Yang A-M et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest 127, 2829–2841 (2017). (first study to implicate the mycobiome in alcoholic liver disease) [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Leclercq S et al. Role of intestinal permeability and inflammation in the biological and behavioral control of alcohol-dependent subjects. Brain. Behav. Immun 26, 911–918 (2012). (role of inflammation in ALD and reversibility on abstinence in humans) [DOI] [PubMed] [Google Scholar]
- •.Henao-Mejia J et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–85 (2012). (role of inflammation in NAFLD and transferability of symptoms by co-housing mice) [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Zhu L et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). (elevated ethanol production by gut microbiota in pediatric NASH patients) [DOI] [PubMed] [Google Scholar]
- •.Cresci GA et al. Prophylactic tributyrin treatment mitigates chronic-binge alcohol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol (2017). (example of microbiome-based therapeutics for liver disease management) [DOI] [PMC free article] [PubMed]
- •.Ferrere G et al. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J. Hepatol 66, 806–815 (2017). (FMT could prevent alcohol-induced liver damage) [DOI] [PubMed] [Google Scholar]
- •.Bajaj JS et al. Fungal dysbiosis in cirrhosis. Gut gutjnl-2016–313170 (2017). (role of mycobiome in cirrhosis) [DOI] [PubMed]
- •.Yoshimoto S et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). (Deoxycholic acid, a gut microbiota-derived bile acid promotes HCC) [DOI] [PubMed] [Google Scholar]
- •.Dapito DH et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell 21, 504–516 (2012). (role of gut microbiota in HCC development) [DOI] [PMC free article] [PubMed] [Google Scholar]
- •.Zaneveld JR, McMinds R & Vega Thurber, R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol 2, 17121 (2017). (explains how instability as a community characteristic may be important to understanding inflammatory disease) [DOI] [PubMed] [Google Scholar]
- •.Etienne-Mesmin L, Vijay-Kumar M, Gewirtz AT & Chassaing B Hepatocyte Toll-Like Receptor 5 Promotes Bacterial Clearance and Protects Mice Against High-Fat Diet-Induced Liver Disease. Cell. Mol. Gastroenterol. Hepatol 2, 584–604 (2016). (well-designed mouse study showing the importance of both inflammatory and tolerizing bacteria in regulating liver inflammation) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.Schnabl B & Brenner DA Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146, 1513–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartmann P, Seebauer CT & Schnabl B Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol. Clin. Exp. Res 39, 763–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi ZM et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64, 1577–1586 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Bertola A, Mathews S, Ki SH, Wang H & Gao B Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc 8, 627–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelz S, Stock P, Brückner S & Christ B A methionine-choline-deficient diet elicits NASH in the immunodeficient mouse featuring a model for hepatic cell transplantation. Exp. Cell Res 318, 276–87 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Itagaki H, Shimizu K, Morikawa S, Ogawa K & Ezaki T Morphological and functional characterization of non-alcoholic fatty liver disease induced by a methionine-choline-deficient diet in C57BL/6 mice. Int. J. Clin. Exp. Pathol 6, 2683–96 (2013). [PMC free article] [PubMed] [Google Scholar]
- 7.Loomba R et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 25, 1054–1062. e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang A-M et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest 127, 2829–2841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y-M et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci. Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csak T et al. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology 54, 133–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uesugi T, Froh M, Arteel GE, Bradford BU & Thurman RG Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology 34, 101–8 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Anand G, Zarrinpar A & Loomba R Targeting Dysbiosis for the Treatment of Liver Disease. Semin. Liver Dis 36, 37–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seki E & Schnabl B Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J. Physiol 590, 447–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jemal A et al. Global cancer statistics. CA. Cancer J. Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Schwabe RF & Jobin C The microbiome and cancer. Nat. Rev. Cancer 13, 800–812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett WS Cancer and the microbiota. Science (80-. ) 348, 80–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koppel N, Maini Rekdal V. & Balskus EP Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolba R, Kraus T, Liedtke C, Schwarz M & Weiskirchen R Diethylnitrosamine (DEN)-induced carcinogenic liver injury in mice. Lab. Anim 49, 59–69 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Stärkel P & Schnabl B Bidirectional Communication between Liver and Gut during Alcoholic Liver Disease. Semin. Liver Dis 36, 331–339 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Chiang JYL Bile acid metabolism and signaling. Compr. Physiol 3, 1191–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahlström A, Sayin SI, Marschall HU & Bäckhed F Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metabolism 24, (2016). [DOI] [PubMed] [Google Scholar]
- 22.Zarrinpar A & Loomba R Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther 36, 909–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Copple BL & Li T Pharmacology of bile acid receptors: Evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol. Res 104, 9–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinal CJ et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–44 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Pols TWH, Noriega LG, Nomura M, Auwerx J & Schoonjans K The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol 54, 1263–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broeders EPM et al. The Bile Acid Chenodeoxycholic Acid Increases Human Brown Adipose Tissue Activity. Cell Metab 22, 418–26 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Thomas C et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 10, 167–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perino A & Schoonjans K TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends Pharmacol. Sci 36, 847–57 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Schaap FG, Trauner M & Jansen PLM Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol 11, 55–67 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Inagaki T et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U. S. A 103, 3920–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parséus A et al. Microbiota-induced obesity requires farnesoid X receptor. Gut 66, 429–437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arab JP, Karpen SJ, Dawson PA, Arrese M & Trauner M Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology 65, 350–362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang C et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest 125, 386–402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridlon JM, Kang DJ, Hylemon PB & Bajaj JS Bile acids and the gut microbiome. Curr. Opin. Gastroenterol 30, 332–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouzaki M et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One 11, e0151829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner JR Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol 9, 799–809 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Abreu MT Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol 10, 131–144 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Gallo RL & Hooper LV Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol 12, 503–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantis NJ, Rol N & Corthésy B Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 4, 603–611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S & Medzhitov R Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–41 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Yaku K et al. The enhancement of phase 2 enzyme activities by sodium butyrate in normal intestinal epithelial cells is associated with Nrf2 and p53. Mol Cell Biochem 370, 7–14 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Wächtershäuser A & Stein J Rationale for the luminal provision of butyrate in intestinal diseases. Eur. J. Nutr 39, 164–71 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Ziegler K, Kerimi A, Poquet L & Williamson G Butyric acid increases transepithelial transport of ferulic acid through upregulation of the monocarboxylate transporters SLC16A1 (MCT1) and SLC16A3 (MCT4). Arch. Biochem. Biophys 599, 3–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lobos O, Barrera A & Padilla C Microorganisms of the Intestinal Microbiota of Oncorhynchus Mykiss Produce Antagonistic Substances Against Bacteria Contaminating Food and Causing Disease in Humans. Ital. J. food Saf 6, 6240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh CJ, Guinane CM, O’ Toole PW & Cotter PD A Profile Hidden Markov Model to investigate the distribution and frequency of LanB-encoding lantibiotic modification genes in the human oral and gut microbiome. PeerJ 5, e3254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graham CE, Cruz MR, Garsin DA & Lorenz MC Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. U. S. A 114, 4507–4512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leclercq S et al. Role of intestinal permeability and inflammation in the biological and behavioral control of alcohol-dependent subjects. Brain. Behav. Immun 26, 911–918 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Henao-Mejia J et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Medina M et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63, 116–24 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Serino M et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 61, 543–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pendyala S, Walker JM & Holt PR A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology 142, 1100–1101. e2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y et al. Effects of alcohol on intestinal epithelial barrier permeability and expression of tight junction-associated proteins 9, 2352–6 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Fukui H, Brauner B, Bode JC & Bode C Plasma endotoxin concentrations in patients with alcoholic and non-alcoholic liver disease: reevaluation with an improved chromogenic assay. J. Hepatol 12, 162–9 (1991). [DOI] [PubMed] [Google Scholar]
- 54.Schäfer C, Parlesak A, Schütt C, Bode JC & Bode C Concentrations of lipopolysaccharide-binding protein, bactericidal/permeability-increasing protein, soluble CD14 and plasma lipids in relation to endotoxaemia in patients with alcoholic liver disease. Alcohol Alcohol 37, 81–6 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Tulstrup MV-L et al. Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. PLoS One 10, e0144854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forbes JD, Van Domselaar G & Bernstein CN The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol 7, 1081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grander C et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut gutjnl-2016–313432 (2017). doi: 10.1136/gutjnl-2016-313432 [DOI] [PubMed]
- 58.Everard A et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Nati. Acad. Sci. USA 110, 9066–9071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elamin EE, Masclee AA, Dekker J & Jonkers DM Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr. Rev 71, (2013). [DOI] [PubMed] [Google Scholar]
- 60.Filliol A et al. RIPK1 protects hepatocytes from Kupffer cells-mediated TNF-induced apoptosis in mouse models of PAMP-induced hepatitis. J. Hepatol 66, 1205–1213 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Ni YH, Huo LJ & Li TT [Effect of interleukin-22 on proliferation and activation of hepatic stellate cells induced by acetaldehyde and related mechanism]. Zhonghua Gan Zang Bing Za Zhi 25, 9–14 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Wu X, Wang Y, Wang S, Xu R & Lv X Purinergic P2X7 receptor mediates acetaldehyde-induced hepatic stellate cells activation via PKC-dependent GSK3β pathway 43, 164–171 (2017). [DOI] [PubMed] [Google Scholar]
- 63.López-Lázaro M A local mechanism by which alcohol consumption causes cancer. Oral Oncol 62, 149–152 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Zhu L et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Miele L et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 49, 1877–1887 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Pascual S et al. Intestinal permeability is increased in patients with advanced cirrhosis. Hepatogastroenterology 50, 1482–6 (2003). [PubMed] [Google Scholar]
- 67.Philips CA et al. Healthy Donor Fecal Microbiota Transplantation in Steroid-Ineligible Severe Alcoholic Hepatitis: A Pilot Study. Clin. Gastroenterol. Hepatol 15, 600–602 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Seki E et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med 13, 1324–1332 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Isayama F et al. LPS signaling enhances hepatic fibrogenesis caused by experimental cholestasis in mice. Am. J. Physiol. Gastrointest. Liver Physiol 290, G1318–28 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Gäbele E et al. Role of TLR9 in hepatic stellate cells and experimental liver fibrosis. Biochem. Biophys. Res. Commun 376, 271–276 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Hartmann P, Haimerl M, Mazagova M, Brenner DA & Schnabl B Toll-Like Receptor 2–Mediated Intestinal Injury and Enteric Tumor Necrosis Factor Receptor I Contribute to Liver Fibrosis in Mice. Gastroenterology 143, 1330–1340. e1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lebeaupin C et al. ER stress induces NLRP3 inflammasome activation and hepatocyte death. Cell Death Dis 6, e1879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeisel SH & da Costa K-A Choline: an essential nutrient for public health. Nutr. Rev 67, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han J et al. Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obesity 25, 1069–1076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muraki Y, Makita Y, Yamasaki M, Amano Y & Matsuo T Elevation of liver endoplasmic reticulum stress in a modified choline-deficient l -amino acid-defined diet-fed non-alcoholic steatohepatitis mouse model. Biochem. Biophys. Res. Commun 486, 632–638 (2017). [DOI] [PubMed] [Google Scholar]
- 76.RUTENBURG AM et al. The role of intestinal bacteria in the development of dietary cirrhosis in rats. J. Exp. Med 106, 1–14 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Velasquez M, Ramezani A, Manal A & Raj D Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins (Basel) 8, 326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Del Rio D et al. The Gut Microbial Metabolite Trimethylamine-N-Oxide Is Present in Human Cerebrospinal Fluid. Nutrients 9, 1053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spencer MD et al. Association Between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver With Choline Deficiency. Gastroenterology 140, 976–986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gogiashvili M et al. Metabolic profiling of ob/ob mouse fatty liver using HR-MAS 1H-NMR combined with gene expression analysis reveals alterations in betaine metabolism and the transsulfuration pathway. Anal. Bioanal. Chem 409, 1591–1606 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Sherriff JL, OSullivan TA, Properzi C, Oddo J-L & Adams LA Choline, Its Potential Role in Nonalcoholic Fatty Liver Disease, and the Case for Human and Bacterial Genes. Adv. Nutr. An Int. Rev. J 7, 5–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen J et al. Liver tumorigenicity of trimethylarsine oxide in male Fischer 344 rats--association with oxidative DNA damage and enhanced cell proliferation. Carcinogenesis 24, 1827–1835 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Chen P et al. Supplementation of Saturated Long-Chain Fatty Acids Maintains Intestinal Eubiosis and Reduces Ethanol-induced Liver Injury in Mice. Gastroenterology 148, 203–214. e16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cresci GA et al. Prophylactic tributyrin treatment mitigates chronic-binge alcohol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol (2017). doi: 10.1111/jgh.13731 [DOI] [PMC free article] [PubMed]
- 85.Hamarneh SR et al. Intestinal Alkaline Phosphatase Attenuates Alcohol-Induced Hepatosteatosis in Mice. Dig. Dis. Sci 62, 2021–2034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen P et al. Microbiota Protects Mice Against Acute Alcohol-Induced Liver Injury. Alcohol. Clin. Exp. Res 39, 2313–2323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ansari R, Husain K & Rizvi S Role of Transcription Factors in Steatohepatitis and Hypertension after Ethanol: The Epicenter of Metabolism. Biomolecules 6, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolk A, Furuheim M & Vessby B Fatty acid composition of adipose tissue and serum lipids are valid biological markers of dairy fat intake in men. J. Nutr 131, 828–33 (2001). [DOI] [PubMed] [Google Scholar]
- 89.Shi X et al. Hepatic and Fecal Metabolomic Analysis of the Effects of Lactobacillus rhamnosus GG on Alcoholic Fatty Liver Disease in Mice. J. Proteome Res 14, 1174–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim D-H et al. Dual function of Lactobacillus kefiri DH5 in preventing high-fat-diet-induced obesity: direct reduction of cholesterol and upregulation of PPARα in adipose tissue. Mol. Nutr. Food Res 1700252 (2017). doi: 10.1002/mnfr.201700252 [DOI] [PubMed] [Google Scholar]
- 91.Nanji AA, Khettry U & Sadrzadeh SM Lactobacillus feeding reduces endotoxemia and severity of experimental alcoholic liver (disease). Proc. Soc. Exp. Biol. Med 205, 243–7 (1994). [DOI] [PubMed] [Google Scholar]
- 92.Forsyth CB et al. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol 43, 163–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Loguercio C et al. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. J. Clin. Gastroenterol 39, 540–3 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Kirpich IA et al. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol 42, 675–682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stadlbauer V et al. Effect of probiotic treatment on deranged neutrophil function and cytokine responses in patients with compensated alcoholic cirrhosis. J. Hepatol 48, 945–951 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Chen R-C et al. Lactobacillus rhamnosus GG supernatant promotes intestinal barrier function, balances T reg and T H 17 cells and ameliorates hepatic injury in a mouse model of chronic-binge alcohol feeding. Toxicol. Lett 241, 103–110 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Levitt MD et al. Use of measurements of ethanol absorption from stomach and intestine to assess human ethanol metabolism. Am. J. Physiol 273, G951–7 (1997). [DOI] [PubMed] [Google Scholar]
- 98.Norberg A, Jones AW, Hahn RG & Gabrielsson JL Role of Variability in Explaining Ethanol Pharmacokinetics. Clin. Pharmacokinet 42, 1–31 (2003). [DOI] [PubMed] [Google Scholar]
- 99.Setshedi M, Wands JR & de la Monte SM Acetaldehyde Adducts in Alcoholic Liver Disease. Oxid. Med. Cell. Longev 3, 178–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rao RK in Methods in molecular biology (Clifton, N.J.) 447, 171–183 (2008). [DOI] [PubMed] [Google Scholar]
- 101.Mir H et al. Occludin deficiency promotes ethanol-induced disruption of colonic epithelial junctions, gut barrier dysfunction and liver damage in mice. Biochim. Biophys. Acta - Gen. Subj 1860, 765–774 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chaudhry KK et al. Glutamine supplementation attenuates ethanol-induced disruption of apical junctional complexes in colonic epithelium and ameliorates gut barrier dysfunction and fatty liver in mice. J. Nutr. Biochem 27, 16–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen P, Stärkel P, Turner JR, Ho SB & Schnabl B Dysbiosis-induced intestinal inflammation activates tumor necrosis factor receptor I and mediates alcoholic liver disease in mice. Hepatology 61, 883–894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Forsyth CB, Voigt RM, Burgess HJ, Swanson GR & Keshavarzian A Circadian rhythms, alcohol and gut interactions. Alcohol 49, 389–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yan AW & Schnabl B Bacterial translocation and changes in the intestinal microbiome associated with alcoholic liver disease. World J. Hepatol 4, 110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan AW et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 53, 96–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hartmann P et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology 58, 108–119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park B, Lee H-R & Lee Y-J Alcoholic liver disease: focus on prodromal gut health. J. Dig. Dis 17, 493–500 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Wang H, Lafdil F, Kong X & Gao B Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int. J. Biol. Sci 7, 536–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mottaran E et al. Lipid peroxidation contributes to immune reactions associated with alcoholic liver disease. Free Radic. Biol. Med 32, 38–45 (2002). [DOI] [PubMed] [Google Scholar]
- 111.Xie G et al. Chronic Ethanol Consumption Alters Mammalian Gastrointestinal Content Metabolites. J. Proteome Res 12, 3297–3306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Couch RD et al. Alcohol Induced Alterations to the Human Fecal VOC Metabolome. PLoS One 10, e0119362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leclercq S et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci 111, E4485–E4493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arroyo V et al. Acute-on-chronic liver failure in cirrhosis. Nat. Rev. Dis. Prim 2, 16041 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Cresci GA, Bush K & Nagy LE Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcohol. Clin. Exp. Res 38, 1489–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Spengler EK & Loomba R Recommendations for Diagnosis, Referral for Liver Biopsy, and Treatment of Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Mayo Clin. Proc 90, 1233–1246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Loomba R, Abraham M & Unalp A Association between diabetes, family history of diabetes and risk of nonalcoholic steatohepatitis and fibrosis. Hepatology 56, 943–951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doycheva I et al. Non-invasive screening of diabetics in primary care for NAFLD and advanced fibrosis by MRI and MRE. Aliment. Pharmacol. Ther 43, 83–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loomba R et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology 149, 1784–1793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cui J et al. Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology 64, 1547–1558 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Caussy C et al. Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J. Clin. Invest 127, 2697–2704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao B & Bataller R Alcoholic Liver Disease: Pathogenesis and New Therapeutic Targets. Gastroenterology 141, 1572–1585 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wieland A, Frank DN, Harnke B & Bambha K Systematic review: microbial dysbiosis and nonalcoholic fatty liver disease. Aliment. Pharmacol. Ther 42, 1051–1063 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Kapil S et al. Small intestinal bacterial overgrowth and toll-like receptor signaling in patients with non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol 31, 213–21 (2016). [DOI] [PubMed] [Google Scholar]
- 125.Boursier J et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63, 764–775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bajaj JS et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 62, 1260–1271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rahman K et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterology 151, 733–746. e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arendt BM et al. Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl. Physiol. Nutr. Metab 38, 334–340 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Rao RK, Seth A & Sheth P Recent Advances in Alcoholic Liver Disease I. Role of intestinal permeability and endotoxemia in alcoholic liver disease. AJP Gastrointest. Liver Physiol 286, G881–G884 (2004). [DOI] [PubMed] [Google Scholar]
- 130.Ferrier L et al. Impairment of the Intestinal Barrier by Ethanol Involves Enteric Microflora and Mast Cell Activation in Rodents. Am. J. Pathol 168, 1148–1154 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ferrere G et al. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J. Hepatol 66, 806–815 (2017). [DOI] [PubMed] [Google Scholar]
- 132.Mutlu EA et al. Colonic microbiome is altered in alcoholism. AJP Gastrointest. Liver Physiol 302, G966–G978 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tuomisto S et al. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol 14, 40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen Y et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54, 562–72 (2011). [DOI] [PubMed] [Google Scholar]
- 135.Wang L et al. Intestinal REG3 Lectins Protect against Alcoholic Steatohepatitis by Reducing Mucosa-Associated Microbiota and Preventing Bacterial Translocation. Cell Host Microbe 19, 227–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Inamine T et al. Genetic Loss of Immunoglobulin A Does Not Influence Development of Alcoholic Steatohepatitis in Mice. Alcohol. Clin. Exp. Res 40, 2604–2613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Adachi Y, Bradford BU, Gao W, Bojes HK & Thurman RG Inactivation of Kupffer cells prevents early alcohol-induced liver injury. Hepatology 20, 453–60 (1994). [PubMed] [Google Scholar]
- 138.Seo W & Jeong W Il. Hepatic non-parenchymal cells: Master regulators of alcoholic liver disease? World J. Gastroenterol 22, 1348–1356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ju C & Mandrekar P Macrophages and Alcohol-Related Liver Inflammation. Alcohol Res 37, 251–62 (2015). [PMC free article] [PubMed] [Google Scholar]
- 140.Tilg H, Moschen AR & Szabo G Interleukin-1 and inflammasomes in alcoholic liver disease/acute alcoholic hepatitis and nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology 64, 955–965 (2016). [DOI] [PubMed] [Google Scholar]
- 141.Axelson M, Mörk B & Sjövall J Ethanol has an acute effect on bile acid biosynthesis in man. FEBS Lett 281, 155–9 (1991). [DOI] [PubMed] [Google Scholar]
- 142.Xie G et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J 27, 3583–3593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu W-B et al. Excessive bile acid activated NF-kappa B and promoted the development of alcoholic steatohepatitis in farnesoid X receptor deficient mice. Biochimie 115, 86–92 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Wu W-B et al. Agonist of farnesoid X receptor protects against bile acid induced damage and oxidative stress in mouse placenta – A study on maternal cholestasis model. Placenta 36, 545–551 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Bhat M et al. Implication of the intestinal microbiome in complications of cirrhosis. World J. Hepatol 8, 1128–1136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Charlton MR et al. Frequency and Outcomes of Liver Transplantation for Nonalcoholic Steatohepatitis in the United States. Gastroenterology 141, 1249–1253 (2011). [DOI] [PubMed] [Google Scholar]
- 147.Bajaj JS et al. Gut Microbiota Alterations can predict Hospitalizations in Cirrhosis Independent of Diabetes Mellitus. Sci. Rep 5, 18559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jun DW et al. Association Between Small Intestinal Bacterial Overgrowth and Peripheral Bacterial DNA in Cirrhotic Patients. Dig. Dis. Sci 55, 1465–1471 (2010). [DOI] [PubMed] [Google Scholar]
- 149.Yao J, Chang L, Yuan L & Duan Z Nutrition status and small intestinal bacterial overgrowth in patients with virus-related cirrhosis. Asia Pac. J. Clin. Nutr 25, 283–91 (2016). [DOI] [PubMed] [Google Scholar]
- 150.Mas A et al. Comparison of rifaximin and lactitol in the treatment of acute hepatic encephalopathy: results of a randomized, double-blind, double-dummy, controlled clinical trial. J. Hepatol 38, 51–8 (2003). [DOI] [PubMed] [Google Scholar]
- 151.Bajaj JS et al. Rifaximin Improves Driving Simulator Performance in a Randomized Trial of Patients With Minimal Hepatic Encephalopathy. Gastroenterology 140, 478–487. e1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vlachogiannakos J et al. Long-term administration of rifaximin improves the prognosis of patients with decompensated alcoholic cirrhosis. J. Gastroenterol. Hepatol 28, 450–455 (2013). [DOI] [PubMed] [Google Scholar]
- 153.Qin N et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014). [DOI] [PubMed] [Google Scholar]
- 154.Chen Y et al. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci. Rep 6, 34055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bajaj JS et al. Fungal dysbiosis in cirrhosis. Gut gutjnl-2016–313170 (2017). doi: 10.1136/gutjnl-2016-313170 [DOI] [PubMed]
- 156.Fouts DE, Torralba M, Nelson KE, Brenner DA & Schnabl B Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J. Hepatol 56, 1283–1292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yoshimoto S et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- 158.Xie G et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 7, 19355–19366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grąt M et al. Relevance of Pre-Transplant α-fetoprotein Dynamics in Liver Transplantation for Hepatocellular Cancer. Ann. Transplant 21, 115–24 (2016). [DOI] [PubMed] [Google Scholar]
- 160.Fox JG et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 59, 88–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rogers AB Distance burning. Gut Microbes 2, 52–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang Y et al. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J. Clin. Pathol 57, 1273–1277 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krüttgen A et al. Study on the association of helicobacter species with viral hepatitis-induced hepatocellular carcinoma. Gut Microbes 3, 228–233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ward JM et al. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst 86, 1222–7 (1994). [DOI] [PubMed] [Google Scholar]
- 165.Mima K et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett 402, 9–15 (2017). [DOI] [PubMed] [Google Scholar]
- 166.Brandtzaeg P Secretory IgA: Designed for Anti-Microbial Defense. Front. Immunol 4, 222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dapito DH et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell 21, 504–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xie G et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int. J. Cancer 139, 1764–1775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ruhland MK et al. Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat. Commun 7, 11762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Demaria M et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gomes AL et al. Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 30, 161–175 (2016). [DOI] [PubMed] [Google Scholar]
- 172.Li J et al. Interleukin 17A Promotes Hepatocellular Carcinoma Metastasis via NF-kB Induced Matrix Metalloproteinases 2 and 9 Expression. PLoS One 6, e21816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hammerich L, Heymann F & Tacke F Role of IL-17 and Th17 Cells in Liver Diseases. Clin. Dev. Immunol 2011, 1–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Viaud S et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science (80-. ) 342, 971–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pár A & Pár G Nem alkoholos zsírmáj és hepatocellularis carcinoma – 2016. Orv. Hetil 157, 987–994 (2016). [DOI] [PubMed] [Google Scholar]
- 176.Thiele M, Wiest R, Gluud LL, Albillos A & Krag A Can non-selective beta-blockers prevent hepatocellular carcinoma in patients with cirrhosis? Med. Hypotheses 81, 871–874 (2013). [DOI] [PubMed] [Google Scholar]
- 177.Forslund K et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Caporaso JG et al. Moving pictures of the human microbiome. Genome Biol 12, R50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Halfvarson J et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol 2, 17004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wu GD et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science (80-. ) 334, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Noguera-Julian M et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine 5, 135–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jackson MA et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut 65, 749–756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Goodrich JK et al. Human Genetics Shape the Gut Microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Debelius J et al. Tiny microbes, enormous impacts: what matters in gut microbiome studies? Genome Biol 17, 217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lozupone CA et al. Meta-analyses of studies of the human microbiota. Genome Res 23, 1704–1714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.van Dongen J, Slagboom PE, Draisma HHM, Martin NG & Boomsma DI The continuing value of twin studies in the omics era. Nat. Rev. Genet 13, 640–653 (2012). [DOI] [PubMed] [Google Scholar]
- 187.Smith MI et al. Gut microbes of Malawian twin pairs discordant for kwashiorkor. Science (80-. ) 339, 548–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Goodrich JK et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 19, 731–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Weingarden A et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3, 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zaneveld JR, McMinds R & Vega Thurber R Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol 2, 17121 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Piscaglia F et al. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: A multicenter prospective study. Hepatology 63, 827–838 (2016). [DOI] [PubMed] [Google Scholar]
- 192.Kostic AD et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe 17, 260–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nguyen TLA, Vieira-Silva S, Liston A & Raes J How informative is the mouse for human gut microbiota research? Dis. Model. Mech 8, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Manichanh C et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 20, 1411–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kiraly DD et al. Alterations of the Host Microbiome Affect Behavioral Responses to Cocaine. Sci. Rep 6, 35455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sampson TR et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 167, 1469–1480. e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Markle JGM et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–8 (2013). [DOI] [PubMed] [Google Scholar]
- 198.Llopis M et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65, 830–839 (2016). [DOI] [PubMed] [Google Scholar]
- 199.Byrne AT et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 17, 254–268 (2017). [DOI] [PubMed] [Google Scholar]
- 200.Hidalgo M et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 4, 998–1013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mattner J Impact of Microbes on the Pathogenesis of Primary Biliary Cirrhosis (PBC) and Primary Sclerosing Cholangitis (PSC). Int. J. Mol. Sci 17, 1864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Verdier J, Luedde T & Sellge G Biliary Mucosal Barrier and Microbiome. Viszeralmedizin 31, 156–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Miyake Y & Yamamoto K Role of gut microbiota in liver diseases. Hepatol. Res 43, 139–146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Pflughoeft KJ & Versalovic J Human Microbiome in Health and Disease. Annu. Rev. Pathol. Mech. Dis 7, 99–122 (2012). [DOI] [PubMed] [Google Scholar]
- 205.Bogdanos D-P et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology 42, 458–465 (2005). [DOI] [PubMed] [Google Scholar]
- 206.PADGETT K et al. Phylogenetic and immunological definition of four lipoylated proteins from, implications for primary biliary cirrhosis. J. Autoimmun 24, 209–219 (2005). [DOI] [PubMed] [Google Scholar]
- 207.Mohammed JP et al. Identification of Cd101 as a Susceptibility Gene for Novosphingobium aromaticivorans-Induced Liver Autoimmunity. J. Immunol 187, 337–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Lee J-Y et al. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res 54, 3062–3069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Olsson R et al. Bile duct bacterial isolates in primary sclerosing cholangitis: a study of explanted livers. J. Hepatol 28, 426–32 (1998). [DOI] [PubMed] [Google Scholar]
- 210.Pollheimer MJ, Halilbasic E, Fickert P & Trauner M Pathogenesis of primary sclerosing cholangitis. Best Pract. Res. Clin. Gastroenterol 25, 727–739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Toyoki Y et al. Semiquantitative evaluation of hepatic fibrosis by measuring tissue hydroxyproline. Hepatogastroenterology 45, 2261–4 (1998). [PubMed] [Google Scholar]
- 212.Karrar A et al. Biliary Epithelial Cell Antibodies Link Adaptive and Innate Immune Responses in Primary Sclerosing Cholangitis. Gastroenterology 132, 1504–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 213.Katt J et al. Increased T helper type 17 response to pathogen stimulation in patients with primary sclerosing cholangitis. Hepatology 58, 1084–1093 (2013). [DOI] [PubMed] [Google Scholar]
- 214.Bode JC, Bode C, Heidelbach R, Dürr HK & Martini GA Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology 31, 30–4 (1984). [PubMed] [Google Scholar]
- 215.Bull-Otterson L et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One 8, e53028 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jiang W et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep 5, 8096 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Raman M et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol 11, 868–75–3 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Bäckhed F et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A 101, 15718–23 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bäckhed F, Manchester JK, Semenkovich CF & Gordon JI Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U. S. A 104, 979–84 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Achur RN, Freeman WM & Vrana KE Circulating cytokines as biomarkers of alcohol abuse and alcoholism. J. Neuroimmune Pharmacol 5, 83–91 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Luck H et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab 21, 527–42 (2015). [DOI] [PubMed] [Google Scholar]
- 222.Luther J et al. Hepatic Injury in Nonalcoholic Steatohepatitis Contributes to Altered Intestinal Permeability. Cell. Mol. Gastroenterol. Hepatol 1, 222–232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Le Roy T et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62, 1787–94 (2013). [DOI] [PubMed] [Google Scholar]
- 224.Bala S, Marcos M, Gattu A, Catalano D & Szabo G Acute binge drinking increases serum endotoxin and bacterial DNA levels in healthy individuals. PLoS One 9, e96864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bode C, Kugler V & Bode JC Endotoxemia in patients with alcoholic and non-alcoholic cirrhosis and in subjects with no evidence of chronic liver disease following acute alcohol excess. J. Hepatol 4, 8–14 (1987). [DOI] [PubMed] [Google Scholar]
- 226.Parlesak A, Schäfer C, Schütz T, Bode JC & Bode C Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J. Hepatol 32, 742–7 (2000). [DOI] [PubMed] [Google Scholar]
- 227.Roh YS, Zhang B, Loomba R & Seki E TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am. J. Physiol. Gastrointest. Liver Physiol 309, G30–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jin R et al. Fructose induced endotoxemia in pediatric nonalcoholic Fatty liver disease. Int. J. Hepatol 2014, 560620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mridha AR et al. TLR9 is up-regulated in human and murine NASH: pivotal role in inflammatory recruitment and cell survival. Clin. Sci. (Lond) 131, 2145–2159 (2017). [DOI] [PubMed] [Google Scholar]
- 230.Alm R, Carlson J & Eriksson S Fasting serum bile acids in liver disease. A comparison with histological features. Scand. J. Gastroenterol 17, 213–8 (1982). [DOI] [PubMed] [Google Scholar]
- 231.Ferslew BC et al. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Dig. Dis. Sci 60, 3318–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Fernando H, Bhopale KK, Kondraganti S, Kaphalia BS & Shakeel Ansari G. A. Lipidomic changes in rat liver after long-term exposure to ethanol. Toxicol. Appl. Pharmacol 255, 127–37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fernando H et al. 1H and 31P NMR lipidome of ethanol-induced fatty liver. Alcohol. Clin. Exp. Res 34, 1937–47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dumas M-E et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U. S. A 103, 12511–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu J, Han L, Zhu L & Yu Y Free fatty acids, not triglycerides, are associated with non-alcoholic liver injury progression in high fat diet induced obese rats. Lipids Health Dis 15, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Volynets V et al. Nutrition, intestinal permeability, and blood ethanol levels are altered in patients with nonalcoholic fatty liver disease (NAFLD). Dig. Dis. Sci 57, 1932–41 (2012). [DOI] [PubMed] [Google Scholar]
- 237.Engstler AJ et al. Insulin resistance alters hepatic ethanol metabolism: studies in mice and children with non-alcoholic fatty liver disease. Gut 65, 1564–71 (2016). [DOI] [PubMed] [Google Scholar]
- 238.Nakamura A & Terauchi Y Lessons from Mouse Models of High-Fat Diet-Induced NAFLD. Int. J. Mol. Sci 14, 21240–21257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ishioka M, Miura K, Minami S, Shimura Y & Ohnishi H Altered Gut Microbiota Composition and Immune Response in Experimental Steatohepatitis Mouse Models. Dig. Dis. Sci 62, 396–406 (2017). [DOI] [PubMed] [Google Scholar]
- 240.Lieber CS & DeCarli LM The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcohol. Clin. Exp. Res 6, 523–31 (1982). [DOI] [PubMed] [Google Scholar]
- 241.Tsukamoto H, Reidelberger RD, French SW & Largman C Long-term cannulation model for blood sampling and intragastric infusion in the rat. Am. J. Physiol 247, R595–9 (1984). [DOI] [PubMed] [Google Scholar]
- 242.Ronis MJJ et al. Increased 4-hydroxynonenal protein adducts in male GSTA4–4/PPAR-α double knockout mice enhance injury during early stages of alcoholic liver disease. Am. J. Physiol. Gastrointest. Liver Physiol 308, G403–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ericsson AC et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10, e0116704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Stappenbeck TS & Virgin HW Accounting for reciprocal host-microbiome interactions in experimental science. Nature 534, 191–9 (2016). [DOI] [PubMed] [Google Scholar]
- 245.Nakagawa H et al. Loss of liver E-cadherin induces sclerosing cholangitis and promotes carcinogenesis. Proc. Natl. Acad. Sci 111, 1090–1095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Etienne-Mesmin L, Vijay-Kumar M, Gewirtz AT & Chassaing B Hepatocyte Toll-Like Receptor 5 Promotes Bacterial Clearance and Protects Mice Against High-Fat Diet-Induced Liver Disease. Cell. Mol. Gastroenterol. Hepatol 2, 584–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wu T et al. Multimodal imaging of a humanized orthotopic model of hepatocellular carcinoma in immunodeficient mice. Sci. Rep 6, 35230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Round JL & Mazmanian SK The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol 9, 313–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]