Abstract
Protein synthesis, transport and N-glycosylation are coupled at the mammalian endoplasmic reticulum (ER) by complex formation of the ribosome, the Sec61 protein-conducting channel and the oligosaccharyltransferase (OST). Here, we used different cryo-electron microscopy approaches to determine structures of native and solubilized ribosome-Sec61-OST complexes. A molecular model for the catalytic OST subunit revealed how STT3A is integrated into the OST and how STT3 paralog specificity for translocon-associated OST is achieved. The OST subunit DC2 was placed at the interface between Sec61 and STT3A, where it acts as a versatile module for recruitment of STT3A-containing OST to the ribosome-Sec61 complex. This detailed structural view on the molecular architecture of the co-translational machinery for N-glycosylation provides the basis for a mechanistic understanding of glycoprotein biogenesis at the ER.
One Sentence Summary:
Cryo-EM analysis reveals how co-translational protein transport and N-glycosylation are coupled at the mammalian endoplasmic reticulum.
The mammalian translocon is formed by the Sec61 complex, the oligosaccharyltransferase complex (OST) and the translocon-associated protein complex (TRAP) (1). The Sec61 channel enables signal sequence dependent protein translocation for soluble proteins through its central pore as well as integration into the lipid bilayer for transmembrane proteins via a lateral gate (2–5). OST catalyzes asparagine- (N-) linked glycosylation, an essential covalent protein modification (6–8). In higher eukaryotes, the catalytic OST subunit STT3 (Staurosporine and Temperature sensitive 3) is present in two paralogous forms (STT3A and B), assembling with a partially overlapping set of accessory subunits (Fig. 1A): RPN1 (ribophorin I), RPN2 (ribophorin II), OST48 (OST 48 kDa subunit), DAD1 (Defender Against cell Death 1), TMEM258 (transmembrane protein 258) and OST4 (OST 4 kDa subunit) (9). STT3B-specific subunits are the paralogous oxido-reductases TUSC3 (Tumor suppressor candidate 3) and MAGT1 (Magnesium transporter protein 1), whereas DC2 and KCP2 (Keratinocyte-associated protein 2) are found only in STT3A complexes (10). The STT3A complex is thought to act co-translationally and to be stably integrated into the translocon (10). The STT3B complex acts as a proofreader for sites omitted by STT3A (11). Structures of monomeric bacterial and archaeal STT3 homologs provided detailed insights into the catalytic mechanism (12–14). Genetic and biochemical data as well as very recent high-resolution yeast OST structures (15, 16) indicate three sub-complexes of intimately interacting OST subunits, corresponding in the mammalian STT3A complex to RPN1+TMEM258 (subcomplex I), STT3A+OST4+DC2+KCP2 (subcomplex II), and RPN2+DAD1+OST48 (subcomplex III) (7). The overall structure of mammalian OST in a native membrane environment was established by cryo-electron tomography (cryo-ET) at medium resolution (1, 17–19), however neither revealed structural details nor the basis of STT3 paralog specificity.
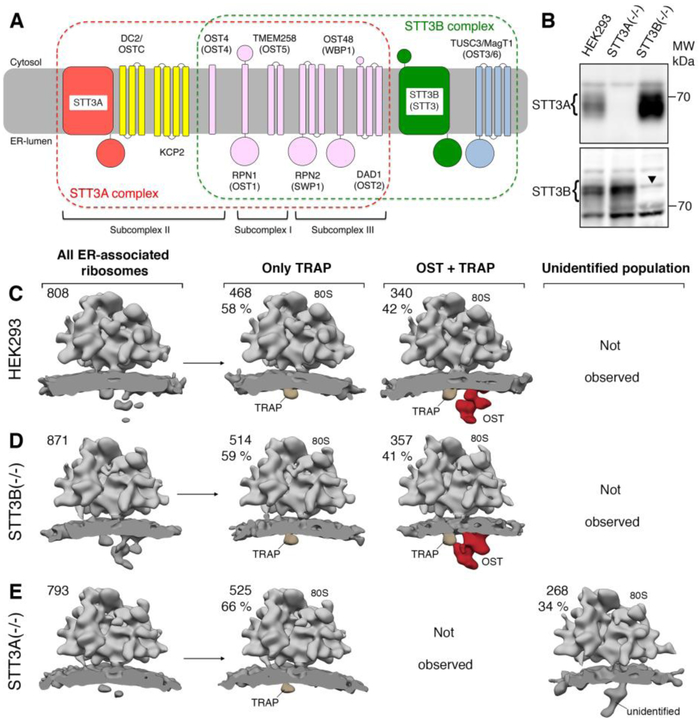
Fig. 1. RTCs harbor exclusively STT3A complexes.
(A) Schematic representation and membrane topology of OST subunits for the STT3A (red frame) and STT3B complexes (green frame, yeast names in parentheses). Shared subunits are depicted in pink. OST subcomplexes are indicated for the STT3A complex. (B) Microsomes from wild type or mutant HEK293 cells were analyzed by immunoblotting using rabbit polyclonal antibodies. The arrowhead in the STT3B blot designates a nonspecific background band. (C)-(E) Ribosome-bound translocon populations observed for microsomes from wild type HEK293 (C), STT3B(−/−) (D) and STT3A(−/−) (E) cell lines after in silico sorting. The absolute number and ratio of subtomograms contributing to each class are given. All densities were filtered to 30 Å resolution.
To confirm STT3 paralog specificity in the ribosome translocon complex (RTC), we analyzed microsomes isolated from established ΔSTT3A and ΔSTT3B HEK cell lines (10) using cryo-ET. Immunoblots confirmed absence of either STT3A or STT3B in the microsomal preparations of knockout cell lines, while both paralogs were present in microsomes prepared from control cells (Fig. 1B). Cryo-ET and in silico analysis of subtomograms showed that control microsomes harbored translocon populations that either included only TRAP (58 %) or TRAP and OST (42 %; Fig. 1C) as expected (17–19). The same populations were found in a similar ratio in microsomes isolated from ASTT3B cells (Fig. 1D), suggesting that translocon-associated OST was not affected by STT3B knockout. In contrast, no translocon-associated OST was observed after STT3A knockout (Fig. 1E), further indicating that RTCs harbor exclusively STT3A complexes (11). Interestingly, instead of the TRAP-OST translocon complexes, a different, possibly partially assembled translocon population was observed after STT3A knockout.
For molecular insights, we employed single particle cryo-EM to visualize solubilized mammalian RTCs translating the well-studied membrane glyocoprotein bovine opsin (20) (Fig. S1, S2). Reconstructions yielded non-programmed and programmed ribosomal complexes showing an overall translocon architecture as observed in the native membrane (18, 19) except for TRAP, which appeared disordered or bound in substoichiometric amounts. Local resolution ranged from 3.5–4.5 Å for Sec61 and adjacent OST transmembrane helices (TMs) to 5–5.5 Å for more peripheral OST-TMs (Fig. S3). In the programmed, peptidyl-tRNA containing complex, the nascent polypeptide density can be traced from the P-site tRNA to the vestibule of the ribosomal tunnel and projects towards the cytoplasmic tip of Sec61a-TM10 (Fig. S4). Sec61 is in a conformation very similar to the previously described ‘primed’ state (Fig. 2A,B; Fig S5) (21) with a closed lateral gate (22, 23) and the plug helix (24) occluding the central pore.
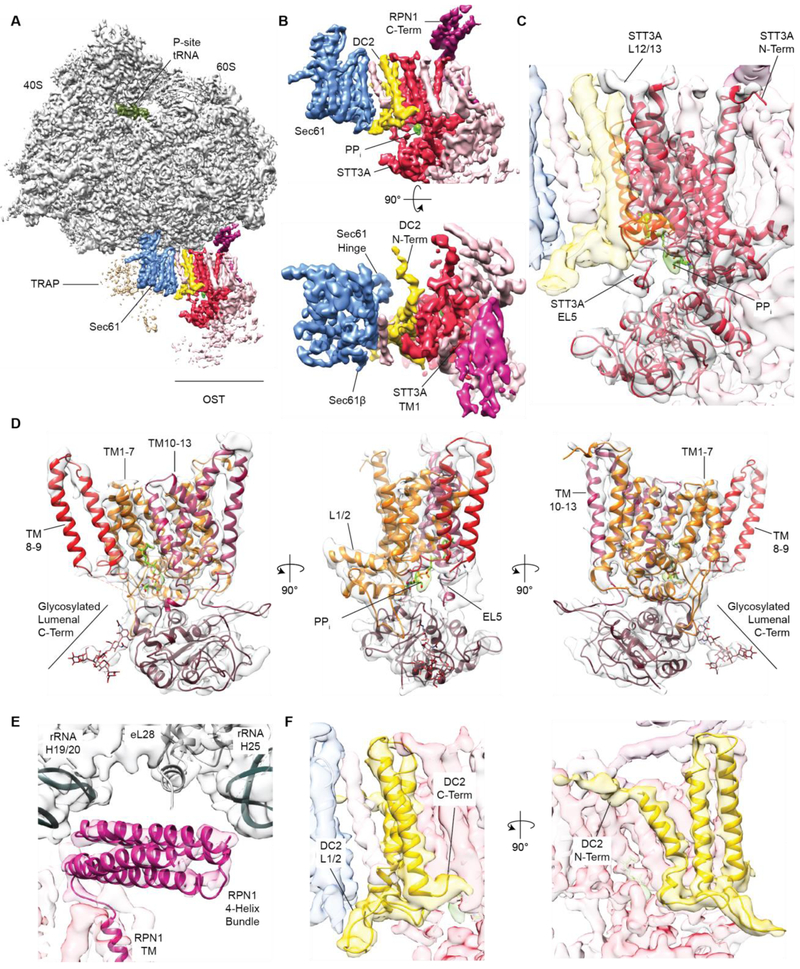
Fig. 2. Localization of STT3A, RPN1 and DC2 in ribosome-bound OST.
(A) Cryo-EM structure of the active solubilized RTC. Ribosome and P-Site tRNA are shown before, the membrane region including Sec61, TRAP and OST after focused refinement (Fig. S2) low-pass filtered to 4 Å. (B) Zoom on the translocon region omitting TRAP as depicted in (A) (upper panel) or rotated by 90° (lower panel). (C, D) Fitted homology model for mammalian STT3A. Density for phosphate groups in the catalytic center is green. (E) Close up view of the cytosolic RPN1 four-helix bundle binding to the ribosome. (F) Zoom on the Sec61-OST interface with a fitted model for the DC2 TMs.
Importantly, 28 additional TMs packed against Sec61 (Fig. 2A, 2B), where OST is positioned in the native translocon (19). We generated a molecular model for mammalian STT3A, revealing high structural similarity to its fungal, archaeal and bacterial homologs (Fig. S6) as well as its orientation in context of the RTC (Fig. 2C, 2D, Fig. S6). Clear density for the pyrophosphate-group of the dolichol carrier was visible in the catalytic site (Fig. 2B-2D, Fig. S3D), suggesting that STT3A was in an active state. Glycosylation of the two consensus motifs in our substrate had already progressed to completion (Fig. S1C) and no peptide substrate density was visible in the catalytic site. The TMs assigned to STT3A were surrounded by 15 additional TMs (Fig. 2B,C, Fig. S7A). Of these TMs, ten were located at the distal side of STT3A, facing away from Sec61. Three of them formed a bundle directly adjacent to STT3A-TMs 1 and 2, whereas another bundle of seven TMs was in proximity to STT3A-TMs 5–8. Based on the three established OST subcomplexes (7) and the number of TMs included in the bundles, we assigned the three-TM bundle to RPN1 and TMEM258 (subcomplex I) and the seven-TM bundle to RPN2, DAD1 and OST48 (subcomplex III). One TM of subcomplex I extended into the metazoan specific cytoplasmic domain of RPN1, which formed a four-helix bundle according to secondary structure predictions (Fig. S7B, C). It was intercalated between the OST TMs and the ribosome and contacted the linker between ribosomal RNA helix H19 and H20, rRNA expansion segment ES7a (H25) and the tail of ribosomal protein eL28 (Fig. 2A, E). Antibodies against the cytosolic RPN1 segment inhibit protein translocation by interfering with ribosome binding to the translocon, confirming direct ribosome-RPNl interaction (25). We further observed four extra TMs tightly associated with STT3A belonging to subcomplex II (STT3A+DC2+OST4+KCP2). One single TM, which we attributed to the single-spanning membrane protein OST4, is tightly intercalated between STT3A-TMs 1, 3, 12 and 13 (Fig. S7). The three remaining TMs located at the interface between STT3A TMs 10–13 and Sec61 were assigned to DC2. We built an atomic model for the three TMs of DC2 de novo based on excellent agreement between features resolved in our map and the predicted length and connectivity of DC2 TMs (Fig. 2F, Fig. S3D). Recent biochemical data (10) show that DC2 assumes a central role in recruiting OST into the translocon complex, and interactions of DC2 with both Sec61 and STT3A have been predicted. Indeed DC2 contacted STT3A via its lumenal C-terminus (to STT3A-TM13) (Fig. 2F), the cytosolic TM2/3-loop (to STT3A-L12/13) and TM2 (close proximity to STT3A-L9/10 also referred to as EL5). The amphipathic DC2 N-terminus projected towards Sec61 on the micelle surface (Fig. 2F) and the lumenal loop of DC2 interacted with the C-termini of Sec61β and Sec61γ (Fig. 2F). We did not observe density for KCP2, likely because it tends to dissociate upon solubilization (10). We observed an additional weaker density for a TM segment intercalated between DC2 and Sec61 in the peptidyl-tRNA containing map, which was absent in the non-programmed map (Fig. S2, Fig. 2B,C) and might correspond to the nascent opsin substrate or a yet unknown translocon component.
We identified two interfaces tying the STT3A complex with the RTC, one between the ribosome and the cytosolic RPN1 domain (Fig. 2A, E) and one between DC2 and Sec61 (Fig. 2B, F), both of which could explain STT3 paralog specificity. First, STT3B possesses a specific 47-amino acid soluble domain extending from STT3-TM1 into the cytosol directly beneath the cytosolic RPN1 helix bundle (Fig. 2B, 2C). The STT3B-specific extension would thus be located in immediate proximity to the ribosome-OST interface where it could interfere with ribosome binding. Second, STT3-TMs 10–13 and the cytosolic STT3-TM12/13 loop, identified as the major contact sites between DC2 and STT3A (Fig. 2C,F), differ considerably between the STT3 paralogs (Table S1, Fig. S8). This suggests that DC2 binds specifically to the STT3A paralog, which would exclude STT3B complexes from the RTC.
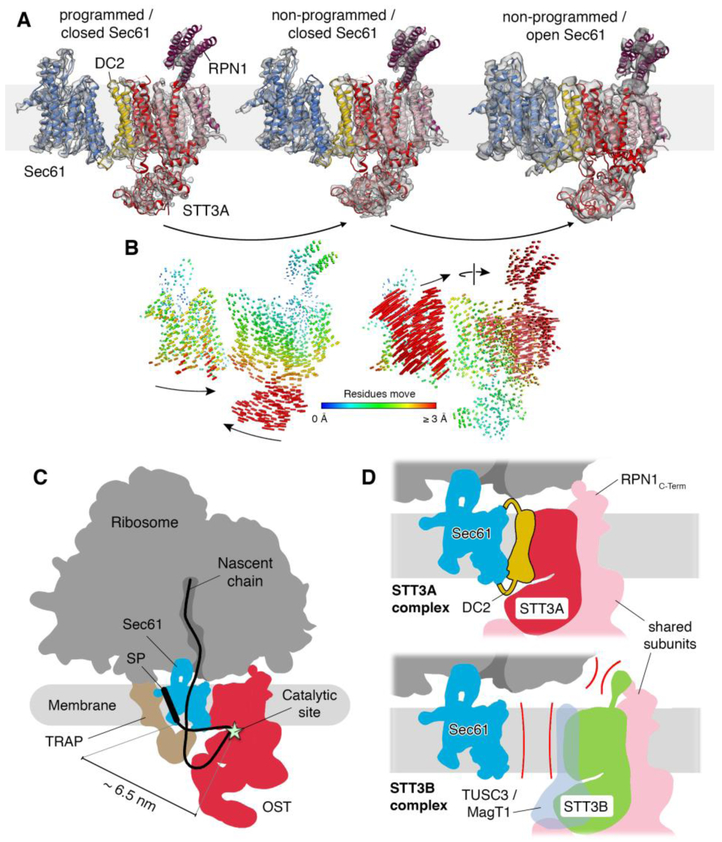
In our second (non-programmed) reconstruction (Fig. S2) the general translocon architecture was very similar to the P-site tRNA containing complex and models for laterally closed Sec61 and OST fitted well as separate rigid bodies (Fig. 3A). Comparison with the model of the programmed RTC revealed a tilting movement between Sec61 and OST with the cytosolic loops of Sec61 and the cytosolic RPN1 domain serving as hinge points on the ribosomal large subunit (Fig. 3B, Movie S1). Furthermore, we improved image processing for an already published cryo-ET dataset (18) of the native RTC with laterally opened Sec61 (Fig. S9) to a resolution allowing rigid body fits of Sec61 and OST (Fig. 3A). Upon opening of the Sec61 lateral gate, the Sec61α N-terminal domain and Sec61β approached DC2. This induced a repositioning of the entire OST complex to accommodate the Sec61 conformational change (Fig. 3B, Movie S1). Although the relative arrangement of DC2 and Sec61 differed significantly between the three observed conformational states, DC2 always stably interacted with Sec61. Thus, DC2 acts as a versatile module that provides robust integration of OST into the translocon complex even under vastly differing conformational states of the translocon complex.
Fig. 3. Translocon dynamics and scheme for cotranslational N-glycosylation.
(A) Models for Sec61 and OST were fitted into the RTC densities with laterally closed (left: programmed, central: non-programmed) and opened Sec61 (right). (B) Trajectories of Cα atoms connecting the observed conformational states with color-coded length. (C) Schematic representation of the RTC with an interpolated example path for a nascent secretory protein. The STT3A catalytic site and a signal peptide (SP) or TM in the Sec61 lateral gate are separated by ~6.5 nm. (D) Molecular basis for STT3 paralog specificity in the RTC. The DC2 and RPN1 subunits tie the STT3A complex into the RTC (upper panel). The lack of DC2 and potential interference of the STT3B-specififc cytosolic domain with ribosome binding exclude STT3B complexes from the RTC.
In conclusion, our cryo-EM reconstructions defined the exact position and orientation of the OST catalytic site in context of the mammalian RTC and enable a detailed dissection of the interface between OST and the ribosome-Sec61 complex. This allowed us to interpolate the path for a nascent glycosylation substrate for co-translational scanning on translocon-associated OST (Fig. 3C) and provided a molecular basis for STT3 paralog specificity in the RTC (Fig. 3D). The minimum distance between a TM segment at the Sec61 lateral gate and the catalytic site of STT3A was about 65 Å explaining why glycosylation sites that are very close to TM segments are often skipped by translocon-associated OST (26).
Supplementary Material
Acknowledgments:
We thank Susanne Rieder and Marko Gogala for technical assistance as well as Christian Schmidt and Jingdong Cheng for critical discussions.
Funding:
This work was supported by funding from the German Research Council (SFB646 to R.B., T.B.; GRK1721 to R.B. and F.F.) and the National Institutes of Health under award number GM35678 (R.G.). R.B. acknowledges support by the Center for Integrated Protein Science Munich (CiPS-M) and the European Research Council (Advanced Grant CRYOTRANSLATION). F.F. received funding from the European Research Council under the European Union’s Horizon2020 Programme (ERC Consolidator Grant Agreement 724425 - BENDER) and the Deutsche Forschungsgemeinschaft (FO 716/4–1). We also acknowledge the support of a Ph.D. fellowship from Boehringer Ingelheim Fonds (to K.B.).
Footnotes
Competing interests:
We declare no competing interests.
Data and material availability:
The cryo-EM density maps and corresponding atomic models reported in this paper have been deposited in the EM Data Bank and Protein Data Bank with the accession codes EMD-4306, EMD-4307, EMD-4308, EMD-4309, EMD-4310, EMD-4311, EMD-4312, EMD-4313, EMD-4314, EMD-4315 and PDB 6FTG for cryo-ET, and EMD-4316, EMD 4317 and PDB 6FTI, 6FTJ for SPA cryo-EM.
References and Notes:
- 1.Pfeffer S, Dudek J, Zimmermann R, Forster F, Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim Biophys Acta 1860, 2122 (October, 2016). [DOI] [PubMed] [Google Scholar]
- 2.Voorhees RM, Hegde RS, Toward a structural understanding of co-translational protein translocation. Curr Opin Cell Biol 41, 91 (August, 2016). [DOI] [PubMed] [Google Scholar]
- 3.Rapoport TA, Li L, Park E, Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol 33, 369 (October 6, 2017). [DOI] [PubMed] [Google Scholar]
- 4.Cymer F, von Heijne G, White SH, Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol 427, 999 (March 13, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann R et al. , Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278, 2123 (December 19, 1997). [DOI] [PubMed] [Google Scholar]
- 6.Mohorko E, Glockshuber R, Aebi M, Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. Journal of inherited metabolic disease 34, 869 (August, 2011). [DOI] [PubMed] [Google Scholar]
- 7.Kelleher DJ, Gilmore R, An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R (April, 2006). [DOI] [PubMed] [Google Scholar]
- 8.Chavan M, Lennarz W, The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem. Sci 31, 17 (January, 2006). [DOI] [PubMed] [Google Scholar]
- 9.Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R, Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell 12, 101 (July, 2003). [DOI] [PubMed] [Google Scholar]
- 10.Shrimal S, Cherepanova NA, Gilmore R, DC2 and KCP2 mediate the interaction between the oligosaccharyltransferase and the ER translocon. J Cell Biol 216, 3625 (November 06, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz-Canada C, Kelleher DJ, Gilmore R, Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272 (January 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lizak C, Gerber S, Numao S, Aebi M, Locher KP, X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350 (June 16, 2011). [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto S et al. , Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl. Acad. Sci. USA 110, 17868 (October 29, 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napiorkowska M et al. , Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat. Struct. Mol. Biol 24, 1100 (December, 2017). [DOI] [PubMed] [Google Scholar]
- 15.Wild R et al. , Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 359, 545 (February 2, 2018). [DOI] [PubMed] [Google Scholar]
- 16.Bai L, Wang T, Zhao G, Kovach A, Li H, The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature, doi: 10.1038/nature25755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahamid J et al. , Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969 (February 26, 2016). [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer S et al. , Structure of the native Sec61 protein-conducting channel. Nat Commun 8403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeffer S et al. , Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun 5, 3072 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Meacock SL, Lecomte FJ, Crawshaw SG, High S, Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell 13, 4114 (December, 2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS, Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 157, 1632 (June 19, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voorhees RM, Hegde RS, Structure of the Sec61 channel opened by a signal sequence. Science 351, 88 (January 1, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gogala M et al. , Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107 (February 6, 2014). [DOI] [PubMed] [Google Scholar]
- 24.Park E, Rapoport TA, Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 473, 239 (May 12, 2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu YH, Sabatini DD, Kreibich G, Antiribophorin antibodies inhibit the targeting to the ER membrane of ribosomes containing nascent secretory polypeptides. J Cell Biol 111,1335 (October, 1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson IM, von Heijne G, Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J Biol Chem 268, 5798 (March 15, 1993). [PubMed] [Google Scholar]
- 27.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36 (October, 2005). [DOI] [PubMed] [Google Scholar]
- 28.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331 (April, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hrabe T et al. , PyTom: a python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J. Struct. Biol 178, 177 (May, 2012). [DOI] [PubMed] [Google Scholar]
- 30.Anger AM et al. , Structures of the human and Drosophila 80S ribosome. Nature 497, 80 (May 2, 2013). [DOI] [PubMed] [Google Scholar]
- 31.Forster F, Pruggnaller S, Seybert A, Frangakis AS, Classification of cryo-electron sub-tomograms using constrained correlation. J. Struct. Biol 161, 276 (March, 2008). [DOI] [PubMed] [Google Scholar]
- 32.Heymann JB, Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol 133, 156 (Feb-Mar, 2001). [DOI] [PubMed] [Google Scholar]
- 33.Zhang K, Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1 (January, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimanius D, Forsberg BO, Scheres SH, Lindahl E, Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, (November 15, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V, Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (August, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126 (December, 2004). [DOI] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 10, 845 (June, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drozdetskiy A, Cole C, Procter J, Barton GJ, JPred4: a protein secondary structure prediction server. Nucleic acids research 43, W389 (July 1, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352 (April, 2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.