Abstract
The glucocorticoid receptor regulates the expression of a large number of genes in mammalian cells. The interaction of this receptor with regulatory elements has been discovered to be highly dynamic, with occupancy states measured in seconds, rather than minutes or hours. This finding has led to a paradigm shift in our understanding of receptor function throughout the genome. The mechanisms involved in these rapid exchange events, as well as the implications for receptor function, are discussed.
Keywords: Glucocorticoid receptor, chromatin, dynamics, gene expression
Introduction
The glucocorticoid receptor (GR), as part of the steroid responsive nuclear receptor family, engages the genomic template to regulate expression of target genes. GR is expressed at moderate to high levels in most tissues in mice at the level of mRNA (Bookout et al. 2006). Its endogenous ligands (the glucocorticoids cortisol in man and corticosterone in rodents) bind to GR, causing translocation to the nucleus from its unliganded cytoplasmic localization. By expression microarray, GR has been shown to regulate several hundred genes in a variety of cell types (Galon et al. 2002; Wang et al. 2003; Planey et al. 2003; Rogatsky et al. 2003; John et al. 2008a). These genes are involved in diverse physiological roles exerted through GR activation, which include tissue development, homeostatic regulation, suppression of inflammation, and stress responses mediated by noise, restraint, hypoglycemic or immune challenge (Borrell et al. 1980; Assenmacher et al. 1995; Karin 1998).
GR activation results in the recruitment of coactivators or corepressors through protein-protein interactions, and the subsequent alteration of gene transcription rates. These interactions include the modulation of transcriptional activity through intrinsic covalent chromatin modifications, including acetylation or methylation (Spencer et al. 1997), or structural modificationsthrough chromatin-remodeling activity (Muchardt and Yaniv 1993; Fryer and Archer 1998).
GR interacts with glucocorticoid responsive elements (GREs) at promoter or enhancer regions in a DNA sequence-specific manner to cause transcriptional induction of target genes. Conversely, repression derives through direct binding to negative GREs (nGREs; Dostert and Heinzel 2004), or transrepression by association with other transcription factors such as activator protein-1 (AP-1), or nuclear factor-kappa B (NF-kB)(De Bosscher K. et al. 2006; Luecke and Yamamoto 2005; Nissen and Yamamoto 2000).
Our understanding of the molecular mechanisms governing GR regulation of gene transcription has been largely based on a few well-characterized model systems. We review our current understanding of GR dynamics in the regulation of transcription in terms of the classic model of static GR binding, and contrast the modern view of rapid dynamics of GR exchange. We discuss transcriptional mechanisms elucidated with studies on the mouse mammary tumor virus promoter, and extend these findings to mechanisms of GR action on a global scale.
Long-term residency of the GR
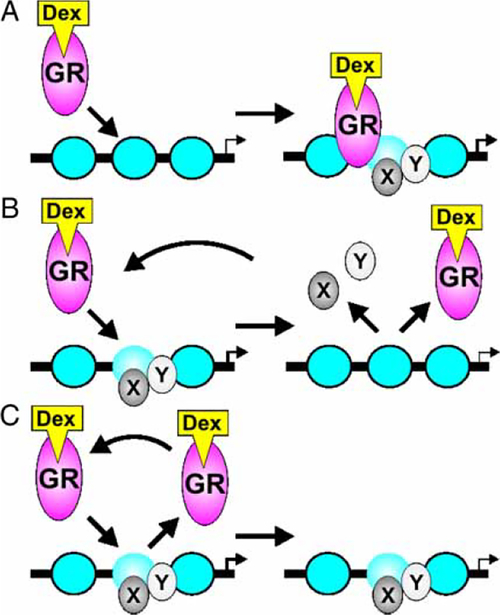
The classic view of transcription factor recognition site binding is that of static or long-term residency on the DNA template for the duration of ligand availability (Becker et al. 1984; Gannon 2005; Shang et al. 2000; Burakov et al. 2002; Schaffner 1988; Wang et al. 2005). Under this model (Figure 1A), the static presence of the receptor at the promoter facilitates recruitment of co-factors, transcription factors and chromatin modifiers to form stable multi-factor complexes for extended periods, with time scales of minutes to hours, consequentially mediating rates of transcriptional initiation.
Figure 1.
Models of glucocorticoid receptor dynamics at hormone response elements. (A) The classic model of receptor binding posits long-term residency of the receptor at the promoter with subsequent recruitment of factors. (B) Direct observation of receptor binding in living cells by Hager and colleagues (McNally et al. 2000) revealed rapid exchange of the receptor (hit-and-run). Most cofactors that have been examined (x, y) also exchange dynamically at sites of receptor binding. (C) Some coregulatory proteins may have longer residence times at selected sites. GR, glucocorticoid receptor; Dex, dexamethasone.
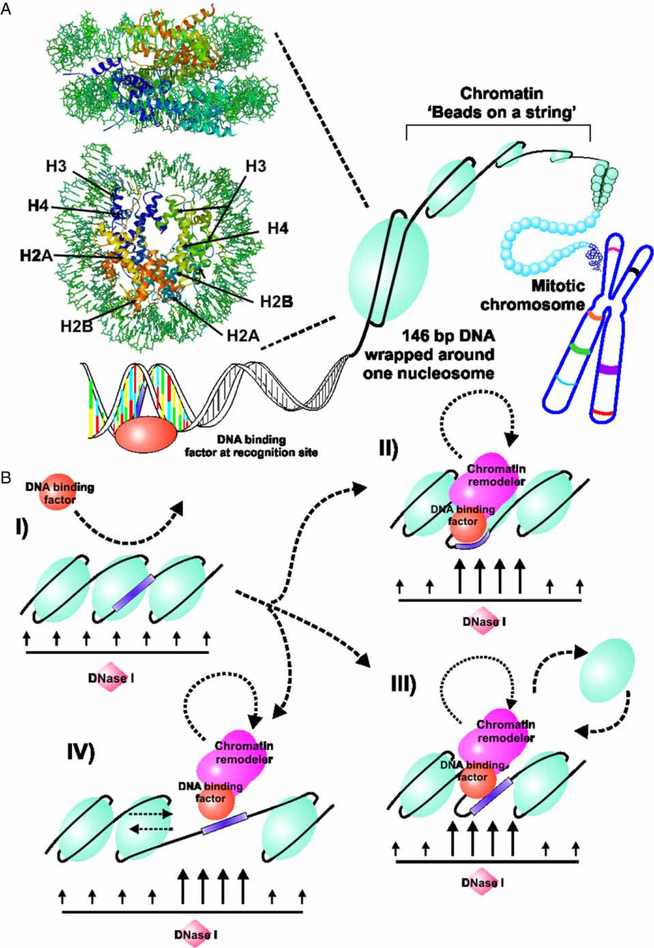
In agreement with the classic model of GR function, GR would act as a pioneer protein, interacting with chromatin through recruitment of multiple protein complexes to induce local chromatin transitions. The modification of chromatin converts a closed conformation to a sustained open conformation that is permissive for binding of the basal transcription apparatus. Chromatin structurally consists of the nucleosome with an approximately 146 base pairs (bp) of DNA wrapped around a histone octamer of core histones: H2A, H2B, H3, and H4 (Wolffe and Hayes 1999; Luger et al. 1997). The histone octamer, with the linker histone H1, mediates DNA compaction for chromosomal formation. However, the compact nature of chromatin thus creates a refractory state to promoter activation (Orphanides and Rein-berg 2000), requiring transcription factor-directed recruitment of chromatin-remodeling proteins to reorganize the local chromatin structure (Figure 2B).
Figure 2.
Interaction of transcription factors with chromatin. (A) DNA in the mammalian cell is organized in a hierarchical series of nucleoprotein structures collectively referred to as chromatin. The basic repeating unit in this structure is the nucleosome, with 146 bp of DNA wrapped on an octameric core of histones (H). The nucleosome is a flattened ellipsoid approximately 110Å on the large axis and 57Å thick. This structure is rendered here in RasMol from the coordinates published for the Xenopus laevis particle by Richmond and colleagues (Ong et al. 2007). Although transcription factors recognize specific sequences in DNA, access to these sites is severely constrained by packaging of the sequences in the nucleosome and higher order structures. (B) Eukaryotic cells have evolved complex systems to reorganize local nucleosome structures during gene regulation. These systems, referred to as remodeling enzymes, are recruited to specific regions by protein–protein interactions with transcription factors, and transiently “open” the nucleosome structure. These molecular reorganizations can be conveniently detected and mapped by the local increase in access to DNA by nucleolytic reagents, including DNaseI (Wu 1980), and chemicals such as methidiumpropyl-EDTA-Fe(II) (Cartwright and Elgin 1989). Multiple mechanisms are considered to be involved in nucleosome remodeling. A pioneer protein, such as GR, can sometimes recognize sites in non-remodeled chromatin (I), and initiate a cascade of binding events. These events can include the transient remodeling of core histone structure (II), the complete or partial eviction of histone components (III), or the “sliding” of nucleosomes to new translational positions on the DNA (IV). The mobility of transcription factors on DNA is now understood to be linked to these chromatin remodeling processes.
GR activation at the MMTV promoter
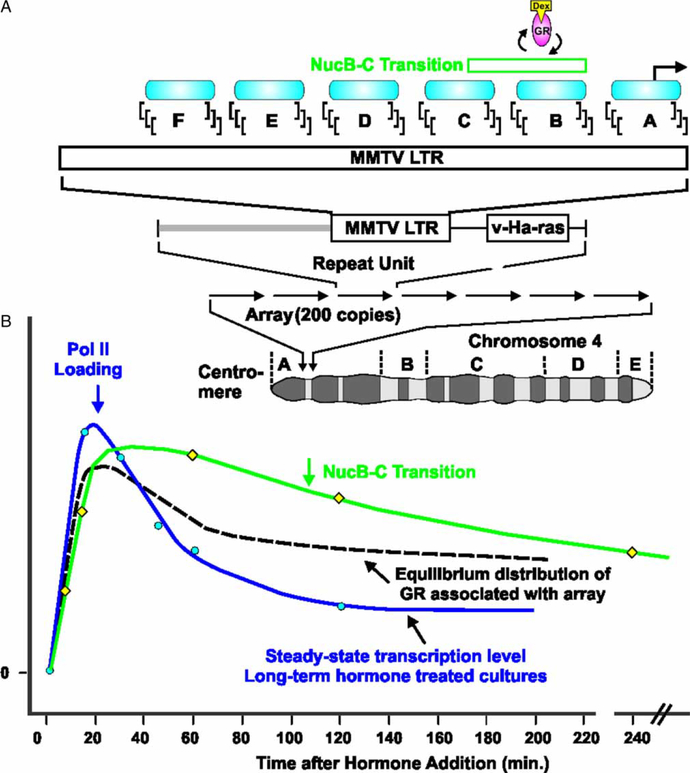
The MMTV promoter, when integrated into cellular-genome, adopts a specific chromatin architecture consisting of six nucleosomes, NucA to NucF (Figure 3A). The second and third nucleosomes, NucB and C, contain six GR binding sites, 270 to 2235 (Payvar et al. 1983; Perlmann and Wrange 1988; Scheidereit et al. 1983; Fletcher et al. 2002). GR action on gene expression was first characterized using the MMTV promoter as a model system (Huang et al. 1981). The transcription rate, measured by quantitative polymerase chain reaction (qPCR), undergoes an initial state of rapid activation, followed by rate declination (at 20 min) to a reduced inductive level maintained over hours (Qiu et al. 2006; John et al. 2008a) (Figure 3B). By chromatin immunoprecipitation (ChIP), GR loading at the MMTV promoter initially tracks with transcription activation, but becomes uncoupled at later times (beyond 20 min) in the activity profile (John et al. 2008a). These changes in transcriptional activity at the MMTV promoter are further corroborated by ChIP RNA polymerase II (Pol II) loading (John et al. 2008a). Thus, in the continuous presence of liganded receptor, the promoter undergoes a complex development of alternate activity states.
Figure 3.
Glucocorticoid receptor induced transcriptional kinetics at a tandem array of GR responsive promoters. (A) The 3134 cell line contains the mouse mammary tumor virus-long terminal repeat (MMTV-LTR)-reporter array integrated at a unique site on chromosome four. Each repeat element (9 kb) includes the MMTV-LTR with 6 positioned nucleosome regions (NucA to NucF). (B) When examined by chromatin immunoprecipitation (ChIP), equilibrium loading of GR and RNA polymerase II (Pol II) at the MMTV promoter is rapidly induced, followed by a strong reduction in RNA polymerase II (Pol II) residency, and a lesser decrease in equilibrium loading of GR which persists for hours. DNaseI hypersensitivity is also rapidly induced, but follows a similar decay over the time period of observation. GR, glucocorticoid receptor; Dex, dexamethasone; v-Ha-ras, Harvey rat sarcoma viral oncogene homolog.
GR activation of the MMTV promoter is associated with a localized chromatin transition, detectable by nuclease sensitivity (Richard-Foy and Hager 1987; Richard-Foy et al. 1987) and restriction enzyme analysis (Archer et al. 1991). A NucB/C DNase hypersensitive site (DHS; between 2109 and 2295) is detectable in the hormone-induced state, concurrent with GR loading at open and accessible chromatin transitions (Fragoso et al. 1998) (Figure 3B). Therefore, in long-term hormone treatment, GR and Pol II loading at the MMTV promoter (as determined by ChIP analysis) manifest an initial increase, followed by a diminution of promoter associated factors. These changes in transcription rate contrast with the continuously sustained chromatin transition at NucB/C, as determined by nuclease hypersensitivity.
ChIP analysis is a widely utilized and powerful technique to observe protein interactions with chromatin and the DNA template at single copy genes in vivo. However, due to the fixation process involving formaldehyde cross-linking, the technique is limited by poor temporal resolution. Indeed, potentially rapid factor associations with the DNA template in vivo would not be detected with this methodology.
GR dynamics in living cells
The view of long-term occupancy of nuclear receptors at promoters and formation of stable multi-factor complexes (Figure 1A) has been challenged by the advent of fluorescent protein tagging for live-cell fluorescence microscopy (Htun et al. 1996). The mouse mammary carcinoma cell line 3134 contains a large tandem array of the MMTV/v-Haras reporter fusion (Figure 3A). The tandem array consists of 1.8 × 106 bp through spontaneous integration of a 9 kb bovine papilloma virus (BPV) multicopy episome (Kramer et al. 1999). The array thus possesses 200 copies of the MMTV long terminal repeat (LTR) conferring 800 to 1200 GR binding sites. The 3617 derivative cell line containing the tandem array has a stably integrated tetracycline-regulated green fluorescent protein-tagged GR (GFPGR), and is thus permissive for fluorescence microscopy in living cells (Walker et al. 1999). This cell line therefore allows high-resolution observation of glucocorticoid receptor loading to the array relative to nuclear background.
In this system, photobleaching techniques, fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) allow the direct measurement of fluorescent protein-tagged factor mobility in living cells. Using these methods it was found, in contrast to the classic view of static binding, that the glucocorticoid receptor undergoes rapid exchange at the MMTV array upon hormone addition at rates (on the order of 10–20 s) neither previously envisaged nor detected by ChIP (McNally et al. 2000). Thus, the receptor undergoes continuous cycling at the MMTV promoter, consistent with a hitand-run model of receptor binding (Figure 1B and C). This concept was first proposed based on in vivo footprinting analysis that suggested GR transiently interacts to promote hepatocyte nuclear factor 5 (HNF5) binding at the rat tyrosine aminotransferase (TAT) gene (Rigaud et al. 1991). This highly dynamic behavior cannot be explained simply by the thermodynamics of DNA binding. Off rates for a number of steroid receptors on DNA have been measured, and are in the range of minutes. As an example, Pandit et al. reported GR t1/2 DNA occupancy times in the range 3.7 –38 min., depending on the ligand (Pandit et al. 2002). Thus, the rapid exchange observed in living cells requires an active mechanism for accelerated association/dissociation.
Cofactors may be recruited during the process of rapid cycling, inducing a chromatin transition as a permissive mechanism for receptor binding and subsequent initiation of transcription. The cofactors may then disengage the template, undergoing independent cycling (Becker et al. 2002)(Figure 1B) or conversely remain bound to the template for longer periods (Figure 1C). This rapid dynamic activity has been recently observed for a number of nuclear transcription factors, including the estrogen receptor (ER) (Reid et al. 2003; Sharp et al. 2006), androgen receptor (AR) (Klokk et al. 2007), nuclear receptor-kB (NF-kB)(Bosisio et al. 2006), Pol II (Dundr et al. 2002) and glucocorticoid receptor-interacting protein 1 (GRIP1)(Becker et al. 2002), albeit at different cycling rates.
While the array represents a non-endogenous system, the receptor-induced transcription kinetics of MMTV in array-containing cell lines and low copy cells are identical (Archer et al. 1994; Smith and Hager 1997), and the GR-induced chromatin transition is also indistinguishable between these two environments (Fragoso et al. 1998). This concurrence implies that the dynamics observed at the array can be extrapolated to endogenous single copy target genes. The phenomenon of dynamic receptor association with genomic sites is likely to confer important regulatory mechanisms in the physiological response of cells to a concomitantly dynamic environment.
Rapid receptor exchange is ATP-dependent
The mechanisms of rapid dynamics of GR and its associated complexes with promoters for transcription initiation are not fully understood. A simple possibility is that ligand dissociation results in removal of GR from the template, possibly by an energy-independent diffusive mechanism. However, ligand dissociation is not required for ejection of the receptor from the template as covalent binding of dexamethasone-21-mesylate to GR does not perturb its recovery by FRAP at the MMTV array (Meijsing et al. 2007). Moreover, factor mobility appears to be highly ATP dependant, suggesting a strong ATPase activity on GR initiation of transcription (Stavreva et al. 2004; Elbi et al. 2004b; Agresti et al. 2005). In living cells, ATP depletion with sodium azide and deoxyglucose significantly reduced the recovery of GFP-GR under FRAP at the MMTV array and in the nucleus (Stavreva et al. 2004). Furthermore, in situ FRAP of transcriptionally competent, permeablized, 3617 cell nuclei revealed a large immobile fraction, attributed to loss of soluble factors required for GR mobility (Elbi et al. 2004a). Reconstitution of the immobile fraction was achieved by incubation with reticulocyte lysate with ATP and an ATP regeneration system, but not with ATP alone, suggesting a requirement for ATPases (Elbi et al. 2004b).
Transcriptional regulation by GR requires chromatin remodeling
Considering that chromatin compaction results in transcriptional silencing, the ability of transcription factors to mediate transcriptional regulation requires a remodeling event. By mechanisms not well understood, transcription factors such as GR bind at recognition sites and recruit remodeling enzymes to facilitate chromatin reorganization. The mechanisms of remodeling can include transient conformational changes in chromatin and the core histone structure, eviction of the histone or components of the octamer and “sliding” of nucleosomes (Figure 2B). Indeed, a subunit [brahma-related gene 1 (Brg1)] of the SWItch/sucrose NonFermentable (Swi/Snf) chromatin-remodeling complex has been shown to coimmunoprecipitate with GR in a hormone-dependent manner (Fryer and Archer 1998). Brg1 mediates chromatin-remodeling through a DNA-dependent ATPase (Khavari et al. 1993), and may thus account for the energy-dependent rapid mobility of GR. Accordingly, activation of the chromatinized MMTV promoter requires GR-mediated chromatin remodeling through active recruitment of Brg1 (Ichinose et al. 1997; Yoshinaga et al. 1992; Fryer and Archer 1998; Fletcher et al. 2002).
Reconstitution of the MMTV promoter as a naked DNA template in the presence of purified GR resulted in binding at GREs. However, GR interaction with the reconstituted chromatinized MMTV promoter in the presence of chromatin remodeler-containing HeLa nuclear extracts and ATP resulted in displacement of both GR and Swi/Snf from the template. This suggests that GR dissociation from DNA is facilitated by transient Swi/Snf activity, which in turn requires ATP (Fletcher et al. 2000). The chromatin remodeling event initiated by GR recruitment of Swi/Snf further mediates the accessibility of secondary transcription factors (Fletcher et al. 2002). In this model, the chromatin transition directed by GR results in receptor destabilization on the template, leading to its dissociation and the development of a chromatin state permissive for binding of secondary factors.
Compared with the low temporal resolution of ChIP, high-intensity UV cross-linking in vitro (Hockensmith et al. 1991; Dimitrov and Moss 2001) with reconstituted chromatinized templates allows detection of rapid interactions between protein factors and the template by applying a brief 5 ns biphotonic pulse (Nagaich and Hager 2004). Using this method, it was found that GR binding is transient with peaks occurring in an oscillatory manner approximately every 5 min. A similar periodic binding was observed for Brg1, although with slight temporal displacement.
The oscillations observed for both GR and Brg1 were ATP-dependent. Elimination of ATP from the reaction results in abolition of the oscillations, indicating that transient GR binding is dependent on Swi/Snf-mediated ATP hydrolysis. The periodicity was also correlated with ATP-dependent oscillations of H2A and H2B cross-linking, associated with nucleosome reorganization events (Nagaich et al. 2004).
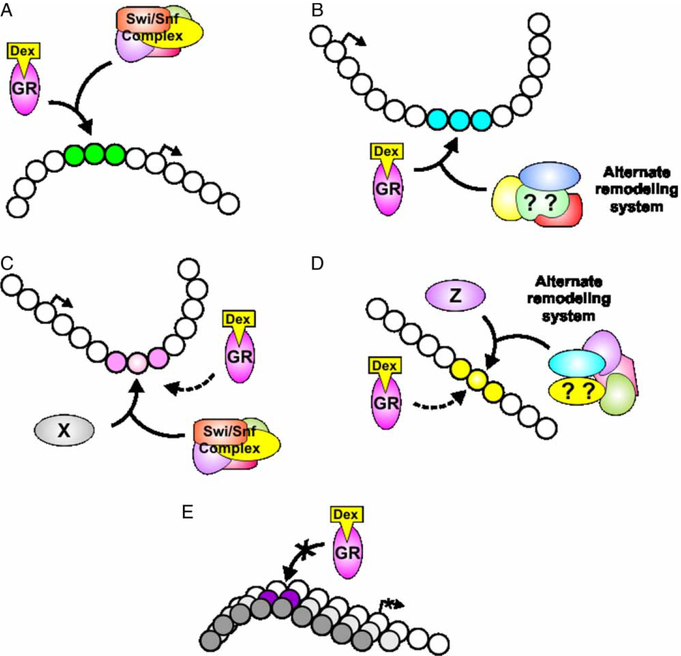
More recently, we have examined genome wide GR binding by ChIP-Chip analysis (ChIP coupled with high-density tiling microarrays) (John et al. 2008,b). We also examined the global distribution of DNase I hypersensitive sites (DHSs) using a novel high-density qPCR method (Sabo et al. 2006; Dorschner et al. 2004). DNase I acts as a preferential cutter of regions of open chromatin and can thus be used to probe local remodeling events as mediated by transcription factor recruitment of chromatin remodeling enzymes (Figure 2B). Sites of GR binding were found to consistently occur at local chromatin transitions, as characterized by the hypersensitivity assay (John et al. 2008,b). The chromatin transitions are not, however, invariably associated with Swi/Snf activity. Indeed, a subset of sites is Brg1-independent. Moreover, GR loading occurs at DHSs that are either hormone inducible or constitutively present in chromatin from cells untreated with hormone. We have shown this phenomenon at a number of endogenous genes [first shown for the TAT gene (Grange et al. 1989)]. These findings are incongruent with the view of GR as a pioneer protein, mediating chromatin remodeling necessary for secondary factor accessibility. Moreover, unremodeled chromatin represents a barrier to transcriptional initiation by GR. Thus, we propose that the chromatin landscape provides a means of mediating cell- and tissue-specificity in response to GR activation. Indeed, we have shown that hormone-inducible and constitutive sites are absent at a number of promoters in a pituitary corticotroph cell line, with correlative lack of GR mediated transcriptional regulation (John et al. 2008,b) (Figure 4).
Figure 4.
Glucocorticoid receptor binding throughout the genome is associated with localized chromatin remodeling. Four classes of remodeling events have been characterized (John et al. 2008,b). GR either induces a de novo chromatin transition (A, B), or binds to a preexisting transition (pre-programmed)(C, D). The de novo transitions are either dependent on the Swi/Snf remodeling complex (A), or independent, requiring alternate remodeling complexes (B). GR also loads at sites of constitutively remodeled chromatin (C, D), which are maintained by other DNA binding proteins (X, Z). These sites are in turn either brahma-related gene 1 (Brg1)-dependent (C) or independent(D). Sites can also be available, or sequestered on a cell specific basis (E). The chromatin landscape thus presents a mechanism of cell and tissue-specific response to GR activation. GR, glucocorticoid receptor; Dex, dexamethasone.
Dynamics of receptor occupancy are modulated by nuclear chaperones and proteasomes
Molecular chaperones are present in both the cytoplasm and the nucleus, forming complexes with unliganded GR (Liu and DeFranco 1999). As the molecular action of the chaperones is ATP-dependent, the dynamic mobility of GR (also ATP-dependent) could in principle be related to chaperone activity. Heat shock protein 90 (hps90) and p23 molecular chaperones, both actively recruited upon GR activation at GREs, have been ascribed to mediate disassembly of transcriptional regulatory complexes (Freeman and Yamamoto 2002). Purified p23 with thyroid hormone (TR)-containing complexes at TR-mediated transcriptional activation in vitro revealed that increasing p23 concentrations led to a reduction in receptor-mediated activation but not basal transcriptional levels (Freeman and Yamamoto 2002). The disassembly capacity of p23 may also be directed at coactivators. Introducing a cognate peptide of the TR interaction surface of the GRIP1 coactivator in vitro at TR-thyroid response element (TRE) complexes resulted in competitive stabilization against p23-mediated dissociation of the complex (Freeman et al. 2000). In vivo, overexpression of full length GRIP1 attenuated a Gal4-p23 and Gal4-Hsp90 construct-mediated inhibition of both TR and GR transcriptional activity (Freeman and Yamamoto 2002). Thus, p23, and to a lesser degree hsp90, appear capable of disassembling receptors from the template to induce dissociation of receptor-coactivator complexes. Thus, molecular chaperones in the nucleus may participate in the rapid dynamic of transcription factors on the DNA template.
Using immunofluorescence, hsp90, hsp70 and p23 were found at the MMTV array, indicating recruitment processes of these chaperones to the array. Inhibition of hsp90 ATPase activity with geldanamycin (Roe et al. 1999; Smith et al. 1995) and radicicol (following dexamethasone and corticosterone pre-treatment) showed increased mobility of GFP-GR, with correlative transcriptional output in the majority of cases examined (Stavreva et al. 2004). Using a ligand-binding domain (LBD)-lacking GR mutant which does not bind p23, exchange rates were also faster (Meijsing et al. 2007). These results contradict the findings of Freeman and Yamamoto that p23 and hsp90 disassemble GR complexes. Also, Elbi et al. showed that the addition of chaperones and cochaper-ones (hsp90, Ydj-1, Hop, hsp70, p23, FKBP51 and CHIP E3 ligase) in situ resulted in a significant recovery of the immobile, ATP-dependent GR (Elbi et al. 2004b; Liu and DeFranco 1999).
The discrepancy between the function of molecular chaperones as stabilizers or dissassemblers of GR complexes at the template remains unresolved and may depend on the cellular milieu and the ligand state of the GR complex. Chaperones by dogma form complexes with unliganded GR as a prerequisite for ligand binding. In contrast to this accepted role of chaperones however, overexpressed hsp90 was found to bind liganded GR and result in nuclear localization and inhibition of GR-mediated transcriptional activity (Kang et al. 1999). These findings in aggregate suggest that the role of molecular chaperones in the nucleus is not just to form complexes with unliganded GR, but to concomitantly orchestrate factor movement within the nucleus by assembly or disassembly of liganded and unliganded GR complexes under different cellular states (Liu and DeFranco 1999).
Proteasomes have also been implicated in the regulation of transcriptional activation through proteolysis of steroid receptors (Molinari et al. 1999; Reid et al. 2003). These complexes have also been proposed to facilitate mobility of transcriptional complexes for rapid association with the template (Collins and Tansey 2006; Baker and Grant 2005; Lipford and Deshaies 2003). The 26S proteasome consists of the proteolytic 20S subunit and two 19S subunits which contain ATPase activity (Baumeister et al. 1998). Concurrently, inhibition of proteasomes with MG132 resulted in a reduction in GFP-GR mobility by FRAP (Stavreva et al. 2004; Elbi et al. 2004b) and is correlated with increased transcriptional output of the MMTV reporter (Deroo et al. 2002). However, proteasomes may also modify the chromatin landscape and increase the pool of phosphorylated Pol II to mediate increased transcription (Kinyamu and Archer 2007). The role of chaperones and proteasomes in regulating transcriptional factors is clearly very complex, and remains poorly understood. However, these activities are likely involved in the rapid dynamics of GR and its interaction with the chromatin landscape. The balance and orchestration of chaperones and proteasomes in the modification of receptor dynamics and transcriptional activity however remains to be elucidated.
Nuclear receptors recruit protein complexes for promoter progression
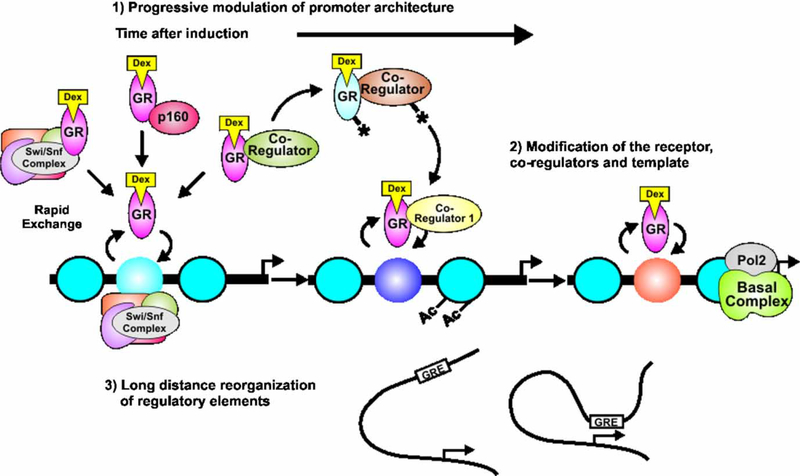
The rapid cycling of nuclear receptors posits that the dynamic process results in a high frequency of transient interaction events, with a fraction of these events leading to successful transcription initiation. An opposing argument would propose that a majority of dynamic cycling events are nonproductive, and transcription initiation only occurs in a minority of cases when the receptor returns to the template for long-term stable interactions with the template. The results obtained from fluorescent microscopy provide a modern model of receptor binding to the promoter, and contradict the concept of stable long-term template interactions derived from ChIP data. To reconcile the opposing models, an integrative model of promoter progression (Hager et al. 2006; Hager and Nagaich 2006) hypothesizes a step-wise process of recruitment to prime a promoter for transcriptional initiation and elongation of the basal apparatus over time (Figure 5).
Figure 5.
The promoter progression model proposes an integrative view of fast and slow dynamics of receptors and cofactors at the promoter. Receptors and cofactors exchange rapidly with the template at response elements, allowing the regulatory sites to constantly sample receptor/cofactor complexes present in the nucleus. Promoter activity can be modulated in a time-dependent manner by multiple mechanisms, including (1) progressive modification of promoter chromatin structure, and (2) post-translational modification of factors involved in transcription initiation. Alteration in promoter modification and structure may include long distance architectural transfiguration to facilitate transcriptional initiation (3). GR, glucocorticoid receptor; Dex, dexamethasone; Pol II, RNA polymerase II; GRE, glucocorticoid responsive element; Ac, acetylation; p160, Figure 4, oactivator family.
The phenomenon of promoter progression should be considered in two temporal planes. The first involves rapid dynamics of receptors and cofactors on the order of seconds, occurring upon initial ligand binding to the receptor for chromatin binding. The second involves the accumulated effects over time, from minutes to hours, of many hundreds of receptor template interactions. As the receptor returns to the template with a large variety of cofactors, the receptor, the template and receptor associated factors are all subject to gradual modification, leading to time-dependent modification of activity states at the promoter. This results in complex kinetics of induction, as exemplified by the transcription profile for MMTV (Figure 3B) (Qiu et al. 2006).
Temporal recruitment and modifications of factors and template
For ER, it was found by ChIP analysis that the receptor mediates a cyclical assembly of transcriptional complexes at promoters (Shang et al. 2000). The cyclical recruitment involved sequential recruitment of cofactors and post-translational modifications (methylation, acetylation and phosphorylation) of histones and the transcriptional apparatus (Burakov et al. 2002; Metivier et al. 2003). While the temporal resolution of ChIP failed to detect rapid dynamics, the findings strongly suggest that the promoter undergoes a step-wise assembly of cofactors and concomitant post-translational modifications initiated by receptor binding to confer regulated transcriptional output. Thus, the promoter progression model suggests the rapid dynamics of receptors on the promoter coincide with recruitment of cofactors that consummate in equilibria states of template-bound and template-unbound receptors and cofactor complexes over time in favor of an activity state, as observed through ChIP.
At the MMTV promoter, the binding of liganded GR facilitates Brg1-dependent chromatin transitions. These events are coupled to transient reorganization and modification of histones H2A and H2B, and enhance the binding of factors such as Oct1 and TFIID (Fletcher et al. 2002; Nagaich et al. 2004; Keeton et al. 2002). Interestingly, chromatin remodeling was thought to be required for nuclear factor 1 (NF1) to bind (Fletcher et al. 2002). Recent evidence however suggests NF1 is required for GR binding to organized chromatin, but also appears to mediate stronger GR-dependent chromatin remodeling synergistically on subsequent rounds of template interaction at the MMTV and 11bHSD2 promoter, but not the Sgk promoter (Hebbar and Archer 2007). Octamer-binding factor 1 (Oct1) was shown to bind weakly at the MMTV (Georgel et al. 2003) in cooperation with NF-1 in the basal transcriptional state and potentiates GR binding and gene transcription following hormone exposure (Belikov et al. 2004a) (Belikov et al. 2004b). Therefore, the temporal order of binding events may require multiple factors to modify the chromatin template, which may vary under physiological conditions.
GR regulated genes proceed through a continuum of activity states
Steroid receptors act to orchestrate cofactor recruitment and confer time-dependent activity states of the promoter. A global and temporal analysis of GR regulated genes by nascent RNA levels has revealed distinct classes of regulation, highlighting the kinetic complexity of responsiveness to GR activation (John et al. 2008a). With constant hormone treatment, the transcription rate of MMTV undergoes initial rapid induction and declination to lower inductive levels: A refractory phase. The transcriptionratethusundergoes alternate phases of activity with corresponding Pol II loading (Figure 3B). This complex kinetic profile of gene activity, with varying induced and repressed states, is likely to be physiologically important in the variety of GR actions throughout the mammalian organism.
During these phases of activity, the promoter may undergo dynamic changes in architecture to reflect the transcriptional state. Histone deacetylases (HDACs) have been argued to mediate deacetylation of histones, leading to a closed chromatin structure, and the inhibition of factor binding to promoters. Recently, HDAC1 has been shown to associate with liganded GR and act as a coactivator for transcription, rather than a corepressor (Qiu et al. 2006). Furthermore, HDAC1 becomes acetylated during the refractory phase of the MMTV promoter. HDAC1 thus participates in induction of MMTV during the early phase, but becomes acetylated by histone acetylases (HATs) over time, resulting in reduced enzymatic activity and contributing to the refractory phase (Qiu et al. 2006). This mechanism illustrates the central concept of promoter progression. We have found that different tissue-derived cell lines have divergent kinetic profiles, with dissimilar refractory phases in the presence of constant hormone (S.C. Biddie, S. John and G.L. Hager, unpublished observation). Thus, programmed promoter progression, dependent on the cellular environment, may give a kinetic profile of a gene that is physiologically relevant for tissue-specificity.
Rapid dynamics of the glucocorticoid receptor is permissive for factor binding
The AF-1 or τ1 domain of GR at the amino-terminal region (Godowski et al. 1987; Giguere et al. 1986; Hollenberg and Evans 1988) has been demonstrated to bind the Tata binding protein (TBP) (Ford et al. 1997), TFIID (McEwan et al. 1993), and CREB binding protein (CBP) (Almlof et al. 1997). τ1 was also reported to interact with Swi/Snf (Wallberg et al. 2000). More recent findings however challenge the requirement of t1 for chromatin remodeling at the MMTV. Using the C656G GR mutant, which manifests higher ligand affinity (Chakraborti et al. 1991), mutation of τ1 did not affect chromatin remodeling, and may rather participate in later stages of promoter progression through interaction with NF1 and the basal transcriptional apparatus (Keeton et al. 2002).
The GR (LBD) binds the molecular chaperone complex, including immunophilins, dimerized hsp90, p23 (Meijsing et al. 2007; Freeman et al. 2000), and GRIP1 (Freeman and Yamamoto 2002). Given the diversity of protein–protein interactions with GR, the rapid dynamic model would provide a means to sample or scan the nuclear milieu and the genomic template (Meijsing et al. 2007). This rapid sampling confers a complexity through a combinatorial and stochastic plethora of protein–protein interactions, which would result in productive binding events with the template (Voss et al. 2006).
The binding of a given GR complex must however coincide, with the temporal activity state of a promoter which would result in progression towards transcriptional initiation and elongation. A rapid and stochastic approach would therefore accommodate the diverse requirements for promoter progression through specific temporally organized states. The rapid dynamics may also be permissive for regulation of genes which require specific programmed events for transcriptional initiation.
Discerning physiological roles for rapid dynamics
The rapid dynamics of transcription factors requires a paradigm shift to understand the physiological relevance of such activity. Rapid mobility may provide a mechanism of sensing the local environment, conferring adaptive modifications of either the template or its interacting proteins to changes in the cellular milieu. The high mobility of GR suggests the capability to continuously sense equally dynamic fluctuations in ligand concentrations within a cell (Stavreva et al. 2004; Meijsing et al. 2007; Freeman and Yamamoto 2002; Reid et al. 2003; Shang et al. 2000; McNally et al. 2000).
Although the mechanisms of rapid factor mobility are not fully understood, it is clear that the balance of chaperones and cofactors plays an important role. Moreover, the continuous post-translational modifications of the template in various cellular states may modify the promoter progression for that gene, thus affecting the transcriptional output. Such alterations to the environment have been reported to modify NF-kB-dependent gene activation following bacterial lipopolysaccharide stimulation (Saccani and Natoli 2002; De Santa et al. 2007) and AR-target promoters in prostate cancer (Zhu et al. 2007). It is reasonable to suggest that disruption of template modifications may also inherently affect the mobility of transcription factors and promoter progression at target genes.
Glucocorticoid receptor response to physiological and pathological states
Secretion of endogenous glucocorticoids occurs with circadian (Weitzman et al. 1971; Gallagher et al. 1973; Veldhuis et al. 1989) and ultradian rhythmicity (Windle et al. 1997). Thus, the ability of GR to modify transcription rates relative to the cellular concentration of ligand may provide an important regulatory mechanism to maintain homeostasis. Indeed, GR nuclear translocation and GRE binding correlate with hormone levels during the circadian cycle in the rat brain (Kitchener et al. 2004). However, our current understanding of how physiological rhythms confer homeostasis via GR through rapid dynamics and promoter progression is not well understood. The role of rapid GR dynamics and promoter activity states during the ultradian rhythm (Atkinson et al. 2006; Windle et al. 1997; Cabrera et al. 2000) and psychological, physiological and immunological stress-induced elevations in hormone concentration also remains to be resolved.
Perspectives
The classic model of long-term and static binding of transcription factors to the template, forming stable complexes to initiate and elongate transcription is a concept that persisted for more than two decades. With the direct observation of receptor/template binding in living cells (McNally et al. 2000), this view has been drastically revised to a new understanding of rapid factor cycling at regulatory elements. These developments profoundly alter our understanding of transcription regulation. This modern model conflicts with findings largely derived from ChIP which inherently possesses lower temporal resolution. We therefore propose an integrative view of promoter progression, whereby the promoter undergoes a temporally specific sequence of programmed events, resulting in a shift in the equilibrium state of factors associating with the promoter. During this process, factors continuously cycle concurrently at the promoter resulting in a scanning process across the genome. How these phenomena traverse from single cells to the organism is not understood and difficult to resolve. Nevertheless, these events likely represent physiologically important activities which provide diverse and complex hormone responsive regulatory mechanisms for transcriptional activity.
Acknowledgements
S.C.B was supported, in part, by The Needham Cooper Postgraduate Medicine Scholarship held by Stafford L. Lightman and the Faculty of Medicine, University of Bristol. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME. 2005. GR and HMGB1 interact only within chromatin and influence each other’s residence time. Mol Cell 18:109–121. [DOI] [PubMed] [Google Scholar]
- Almlof T, Gustafsson JA, Wright AP. 1997. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol Cell Biol 17:934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer TK, Cordingley MG, Wolford RG, Hager GL. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol 11:688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer TK, Lee H-L, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL. 1994. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol 8(5):568–576. [DOI] [PubMed] [Google Scholar]
- Assenmacher I, Barbanel G, Gaillet S, Givalois L, Ixart G, Malaval F, Mekaouche M, Siaud P, Szafarczyk A. 1995. Central regulation of ACTH release in stress. Ann NY Acad Sci 771: 41–54. [DOI] [PubMed] [Google Scholar]
- Atkinson HC, Wood SA, Kershaw YM, Bate E, Lightman SL. 2006. Diurnal variation in the responsiveness of the hypothalamic-pituitary-adrenal axis of the male rat to noise stress. J Neuroendocrinol 18:526–533. [DOI] [PubMed] [Google Scholar]
- Baker SP, Grant PA. 2005. The proteasome: Not just degrading anymore. Cell 123:361–363. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. 1998. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 92: 367–380. [DOI] [PubMed] [Google Scholar]
- Becker PB, Renkawitz R, Schutz G. 1984. Tissue-specific DNaseI hypersensitive sites in the 5′-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J 3:2015–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Baumann CT, John S, Walker D, Vigneron M, McNally JG, Hager GL. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep 3:1188–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Astrand C, Holmqvist PH, Wrange O. 2004a. Chromatin-mediated restriction of nuclear factor 1/CTF binding in a repressed and hormone-activated promoter in vivo. Mol Cell Biol 24:3036–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Holmqvist PH, Astrand C, Wrange O. 2004b. Nuclear factor 1 and octamer transcription factor 1 binding preset the chromatin structure of the mouse mammary tumor virus promoter for hormone induction. J Biol Chem 279: 49857–49867. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell J, Torrellas A, Guaza C, Borrell S. 1980. Sound stimulation and its effects on the pituitary-adrenocortical function and brain catecholamines in rats. Neuroendocrinology 31:53–59. [DOI] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. 2006. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J 25:798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov D, Crofts LA, Chang CP, Freedman LP. 2002. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem 277:14359–14362. [DOI] [PubMed] [Google Scholar]
- Cabrera R, Korte SM, Lentjes EG, Romijn F, Schonbaum E, de Nicola A, de Kloet ER. 2000. The amount of free corticosterone is increased during lipopolysaccharide-induced fever. Life Sci 66:553–562. [DOI] [PubMed] [Google Scholar]
- Cartwright IL, Elgin SC. 1989. Nonenzymatic cleavage of chromatin. Methods Enzymol 170:359–369. [DOI] [PubMed] [Google Scholar]
- Chakraborti PK, Garabedian MJ, Yamamoto KR, Simons SS Jr. 1991. Creation of “super” glucocorticoid receptors by point mutations in the steroid binding domain. J Biol Chem 266: 22075–22078. [PubMed] [Google Scholar]
- Collins GA, Tansey WP. 2006. The proteasome: A utility tool for transcription? Curr Opin Genet Dev 16:197–202. [DOI] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Haegeman G. 2006. Cross-talk between nuclear receptors and nuclear factor kappaB. Oncogene 25:6868–6886. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083–1094. [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Rentsch C, Sampath S, Young J, DeFranco DB, Archer TK. 2002. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol Cell Biol 22:4113–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Moss T. 2001. UV laser-induced protein-DNA cross-linking. Methods Mol Biol 148:395–402. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, Kohli A, Li Q, McArthur M, Stamatoyannopoulos JA. 2004. High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods 1:219–225. [DOI] [PubMed] [Google Scholar]
- Dostert A, Heinzel T. 2004. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des 10:2807–2816. [DOI] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623–1626. [DOI] [PubMed] [Google Scholar]
- Elbi C, Walker DA, Lewis M, Romero G, Sullivan WP, Toft DO, Hager GL, DeFranco DB. 2004a. A novel in situ assay for the identification and characterization of soluble nuclear mobility factors. Science STKE 2004:PL10. [DOI] [PubMed] [Google Scholar]
- Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL, DeFranco DB. 2004b. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc Natl Acad Sci USA 101: 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Ryu B-W, Baumann CT, Warren BS, Fragoso G, John S, Hager GL. 2000. Structure and dynamic properties of the glucocorticoid receptor-induced chromatin transition at the MMTV promoter. Mol Cell Biol 20:6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS, Hager GL. 2002. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol 22:3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, McEwan IJ, Wright AP, Gustafsson JA. 1997. Involvement of the transcription factor IID protein complex in gene activation by the N-terminal transactivation domain of the glucocorticoid receptor in vitro. Mol Endocrinol 11:1467–1475. [DOI] [PubMed] [Google Scholar]
- Fragoso G, Pennie WD, John S, Hager GL. 1998. The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol 18: 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Felts SJ, Toft DO, Yamamoto KR. 2000. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev 14: 422–434. [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Yamamoto KR. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296: 2232–2235. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. 1998. Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88–91. [DOI] [PubMed] [Google Scholar]
- Gallagher TF, Yoshida K, Roffwarg HD, Fukushima DK, Weitzman ED, Hellman L. 1973. ACTH and cortisol secretory patterns in man. J Clin. Endocrinol Metab 36:1058–1068. [DOI] [PubMed] [Google Scholar]
- Galon J, Franchimont D, Hiroi N, Frey G, Boettner A, Ehrhart-Bornstein M, O’Shea JJ, Chrousos GP, Bornstein SR. 2002. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J 16:61–71 [DOI] [PubMed] [Google Scholar]
- Gannon F. 2005. Cycles EMBO Rep 6:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel PT, Fletcher TM, Hager GL, Hansen JC. 2003. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev 17(13):1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V, Hollenberg SM, Rosenfeld MG, Evans RM. 1986. Functional domains of the human glucocorticoid receptor. Cell 46:645–652. [DOI] [PubMed] [Google Scholar]
- Godowski PJ, Rusconi S, Miesfeld R, Yamamoto KR. 1987. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement [published erratum appears in Nature 1987 Mar 5–11;326(6108):105]. Nature 325:365–368. [DOI] [PubMed] [Google Scholar]
- Grange T, Roux J, Rigaud G, Pictet R. 1989. Two remote glucocorticoid responsive units interact cooperatively to promote glucocorticoid induction of rat tyrosine aminotransferase gene expression. Nucleic Acids Res 17:8695–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, Elbi C, Johnson TA, Voss TC, Nagaich AK, Schiltz RL, Qiu Y, John S. 2006. Chromatin dynamics and the evolution of alternate promoter states. Chromosome Res 14:107–116. [DOI] [PubMed] [Google Scholar]
- Hager GL, Nagaich AK. 2006. Transcription factor dynamics Gene expression and regulation. In: Ma J, editor. Higher education press. New York, Beijing: Springer; p 493–502. [Google Scholar]
- Hebbar PB, Archer TK. 2007. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem 282:8284–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockensmith JW, Kubasek WL, Vorachek WR, Evertsz EM, von Hippel PH. 1991. Laser cross-linking of protein-nucleic acid complexes. Methods Enzymol 208:211–236. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Evans RM. 1988. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell 55:899–906. [DOI] [PubMed] [Google Scholar]
- Htun H, Barsony J, Renyi I, Gould DJ, Hager GL. 1996. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA 93:4845–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AL, Ostrowski MC, Berard D, Hager GL. 1981. Glucocorticoid regulation of the Ha-MuSV p21 gene conferred by sequences from mouse mammary tumor virus. Cell 27: 245–255. [DOI] [PubMed] [Google Scholar]
- Ichinose H, Garnier JM, Chambon P, Losson R. 1997. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene 188:95–100. [DOI] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Biddie SC, Trump S, Koch-Paiz CA, Davis SR, Walker R, Meltzer P, Hager GL. 2008a. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. 2008b.. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 29:611–624. [DOI] [PubMed] [Google Scholar]
- Kang KI, Meng X, vin-Leclerc J, Bouhouche I, Chadli A, Cadepond F, Baulieu EE, Catelli MG. 1999. The molecular chaperone Hsp90 can negatively regulate the activity of a glucocorticosteroid-dependent promoter. Proc Natl Acad Sci USA 96: 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M 1998. New twists in gene regulation by glucocorticoid receptor: Is DNA binding dispensable? Cell 93:487–490. [DOI] [PubMed] [Google Scholar]
- Keeton E, Fletcher TM, Baumann CT, Hager GL, Smith CL. 2002. Glucocorticoid receptor domain requirements for chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter in different nucleoprotein contexts. J Biol Chem 277:28247–28255. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174. [DOI] [PubMed] [Google Scholar]
- Kinyamu HK, Archer TK. 2007. Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol Cell Biol 27:4891–4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchener P, Di BF, Borrelli E, Piazza PV. 2004. Differences between brain structures in nuclear translocation and DNA binding of the glucocorticoid receptor during stress and the circadian cycle. Eur J Neurosci 19:1837–1846. [DOI] [PubMed] [Google Scholar]
- Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang CY, Hager GL, Saatcioglu F. 2007. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol Cell Biol 27:1823–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer P, Fragoso G, Pennie WD, Htun H, Hager GL, Sinden RR. 1999. Transcriptional state of the mouse mammary tumor virus promoter can effect topological domain size in vivo. J Biol Chem 274:28590–28597. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Deshaies RJ. 2003. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol 5:845–850. [DOI] [PubMed] [Google Scholar]
- Liu J, DeFranco DB. 1999. Chromatin recycling of glucocorticoid receptors: Implications for multiple roles of heat shock protein90. Mol Endocrinol 13:355–365. [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. 2005. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev 19:1116–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution [see comments]. Nature 389:251–260. [DOI] [PubMed] [Google Scholar]
- McEwan IJ, Wright APH, Dahlman-Wright K, Carlstedt-Duke J, Gustafsson J-Å . 1993. Direct interaction of the t1 transactivation domain of the human glucocorticoid receptor with the basal transcriptional machinery. Mol Cell Biol 13:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. 2000. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Elbi C, Luecke HF, Hager GL, Yamamoto KR. 2007. The ligand binding domain controls glucocorticoid receptor dynamics independent of ligand release. Mol Cell Biol 27: 2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763. [DOI] [PubMed] [Google Scholar]
- Molinari E, Gilman M, Natesan S. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J 18:6439–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12:4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Hager GL. 2004. UV laser cross-linking: A real-time assay to study dynamic protein/DNA interactions during chromatin remodeling. Sci STKE 256:PL13. [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford RG, Hager GL. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14:163–174. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 14:2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong MS, Richmond TJ, Davey CA. 2007. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol 368: 1067–1074. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. 2000. RNA polymerase II elongation through chromatin. Nature 407:471–475. [DOI] [PubMed] [Google Scholar]
- Pandit S, Geissler W, Harris G, Sitlani A. 2002. Allosteric effects of dexamethasone and RU486 on glucocorticoid receptor-DNA interactions. J Biol Chem 277:1538–1543. [DOI] [PubMed] [Google Scholar]
- Payvar F, DeFranco DB, Firestone GL, Edgar B, Wrange O, Okret S, Gustafsson JA, Yamamoto KR. 1983. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell 35(2 Pt 1): 381–392. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Wrange O. 1988. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J 7: 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planey SL, Abrams MT, Robertson NM, Litwack G. 2003. Role of apical caspases and glucocorticoid-regulated genes in glucocorticoid-induced apoptosis of pre-B leukemic cells. Cancer Res 63:172–178. [PubMed] [Google Scholar]
- Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, Adkins NL, Stavreva DA, Wiench M, Georgel PT, Schiltz RL, Hager GL. 2006. HDAC1 acetylation is linked to progressive modulation of steroid receptor induced gene transcription. Mol Cell 22: 669–679. [DOI] [PubMed] [Google Scholar]
- Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11:695–707. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H, Hager GL. 1987. Sequence specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6:2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H, Sistare FD, Riegel AT, Simons SS Jr, Hager GL. 1987. Mechanism of dexamethasone 21-mesylate antiglucocorticoid action: II. Receptor-antiglucocorticoid complexes are unable to interact productively with MMTV LTR chromatin in vivo. Mol Endocrinol 1:659–665. [DOI] [PubMed] [Google Scholar]
- Rigaud G, Roux J, Pictet R, Grange T. 1991. In vivo footprinting of rat TAT gene: Dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell 67:977–986. [DOI] [PubMed] [Google Scholar]
- Roe SM, Prodromou C, O’Brien R, Ladbury JE, Piper PW, Pearl LH. 1999. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J Med Chem 42:260–266. [DOI] [PubMed] [Google Scholar]
- Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR. 2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA 100:13845–13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, Stamatoyannopoulos JA. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3:511–518. [DOI] [PubMed] [Google Scholar]
- Saccani S, Natoli G. 2002. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflamma-tory genes. Genes Dev 16:2219–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W 1988. Gene regulation. A hit-and-run mechanism for transcriptional activation? [news]. Nature 336:427–428. [DOI] [PubMed] [Google Scholar]
- Scheidereit C, Geisse S, Westphal HM, Beato M. 1983. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature 304(5928):749–752. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852. [DOI] [PubMed] [Google Scholar]
- Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE, Mancini MA. 2006. Estrogen-receptor-alpha exchange and chromatin dynamics are ligandand domain-dependent. J Cell Sci 119:4101–4116. [DOI] [PubMed] [Google Scholar]
- Smith CL, Hager GL. 1997. Transcriptional regulation of mammalian genes in vivo: A tale of two templates. J Biol Chem 272:27493–27496. [DOI] [PubMed] [Google Scholar]
- Smith DF, Whitesell L, Nair SC, Chen S, Prapapanich V, Rimerman RA. 1995. Progesterone receptor structure and function altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol 15:6804–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194–198. [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. 2004. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol 24:2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis JD, Iranmanesh A, Lizarralde G, Johnson ML. 1989. Amplitude modulation of a burstlike mode of cortisol secretion subserves the circadian glucocorticoid rhythm. Am J Physiol 257:E6–14. [DOI] [PubMed] [Google Scholar]
- Voss TC, John S, Hager GL. 2006. Single cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligand-specific transcription. Mol Endocrinol 20:2641–2655. [DOI] [PubMed] [Google Scholar]
- Walker D, Htun H, Hager GL. 1999. Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods (Companion to Methods in Enzymology) 19:386–393. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Neely KE, Hassan AH, Gustafsson J-Å , Workman JL, Wright APH. 2000. Recruitment of the SWI-SNF chromatin remodeling complex as a mechanism of gene activation by the glucocorticoid receptor tau1 activation domain. Mol Cell Biol 20:2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642. [DOI] [PubMed] [Google Scholar]
- Wang Z, Malone MH, Thomenius MJ, Zhong F, Xu F, Distelhorst CW. 2003. Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J Biol Chem 278:27053–27058. [DOI] [PubMed] [Google Scholar]
- Weitzman ED, Fukushima D, Nogeire C, Roffwarg H, Gallagher TF, Hellman L. 1971. Twenty-four hour pattern of the episodic secretion of cortisol in normal subjects. J Clin Endocrinol Metab 33:14–22. [DOI] [PubMed] [Google Scholar]
- Windle RJ, Shanks N, Lightman SL, Ingram CD. 1997. Central oxytocin administration reduces stress-induced corticosterone release and anxiety behavior in rats. Endocrinology 138: 2829–2834. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. 1999. Chromatin disruption and modification.Nucleic Acids Res 27:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C 1980. The 5’ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286(5776): 854–860. [DOI] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. 1992. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258:1598–1604. [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. 2007. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 27:609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]